Abstract
A chemical derivatization approach has been developed for the enrichment of O-GlcNAc modified proteins. The procedure is based on the isolation technique used for N-glycoproteins with appropriate modifications because of the differences in the two types of glycosylation: a prolonged periodate oxidation is followed by hydrazide resin capture, on-resin proteolytic digestion and release of the modified peptides by hydroxylamine. This enrichment strategy offers a fringe benefit in mass spectrometry analysis. Upon collisional activation the presence of the open carbohydrate ring leads to characteristic fragmentation facilitating both glycopeptide identification and site assignment. The enrichment protocol was applied to the Drosophila proteasome complex previously described as O-GlcNAc modified. The O-GlcNAc modification was located on proteasome interacting proteins, deubiquitinating enzyme Faf (CG1945) and a ubiquitin-like domain containing protein (CG7546). Three other proteins were also found GlcNAc modified, a HSP70 homologue (CG2918), scribbled (CG5462) and the 205 kDa microtubule-associated protein (CG1483). Interestingly, in the HSP70 homologue the GlcNAc modification is attached to an asparagine residue of a N-glycosylation motif.
Keywords: post-translational modification, O-glycosylation, O-GlcNAc, glycopeptides, enrichment, mass spectrometry, CID, ETD
Introduction
O-linked β-N-acetylglucosamine (O-GlcNAc) is a monomeric carbohydrate modification attached to serine and threonine residues of cytosolic and nuclear proteins1,2. It is a dynamic modification incorporated by O-GlcNAc transferase and removed by O-GlcNAcase3. This post-translational modification (PTM) is involved in a number of cellular processes, e.g. signal transduction3–5, transcription1,6–9, translation3,7 or nuclear transport10. Its importance in the brain was shown by extensive modification of pre- and postsynaptic proteins involved in vesicle cycling, dendritic spine formation, cytoskeletal organization or microtubule assembly11,12. Perturbations in O-GlcNAc levels have been related to diseased states. Increased levels of O-GlcNAc modification have been associated with diabetes, whereas decreased level of O-GlcNAc glycosylation of tau protein is assumed to contribute to Alzheimer’s disease3,13.
The O-GlcNAc modification has also been linked to the proteasomal degradation pathway14,15. The ubiquitin-proteasome system (UPS) is responsible for the removal of damaged or unfolded proteins. In addition, by the degradation of key regulatory proteins it modulates cell cycle, cell differentiation or apoptosis16–18. UPS comprises the ubiquitination machinery tagging the proteins for degradation and the 26S proteasome as the executive unit. In the ubiquitination pathway, the ubiquitin activating enzyme E119 and a deubiquitinating enzyme UCH-L120 has been shown as O-GlcNAc modified. In the 26S proteasome, different subunits of the 20S catalytic core particle as well as of the 19S regulatory complex were found to be O-GlcNAc modified14,15. An inverse relationship between the modification and proteasome function has been demonstrated, increased levels of O-GlcNAc modification decreased proteasomal activity15. Additionally, a number of interacting proteins co-purify with the proteasome21–23, of which HSP7024 has been shown to carry this PTM. However, the exact site of the modification could not be located on any of these proteins.
Being a substoichiometric modification, the analysis of O-GlcNAc glycosylation necessitates enrichment. The isolation techniques so far utilize chemical25 or enzymatic derivatization12,26,27 and affinity chromatography11,28. In the chemical derivatization approach the sugar moiety is eliminated under alkaline conditions, the resulting double bond is then used to introduce a thiol function or a biotin tag for the enrichment on a thiol reactive resin or by avidin chromatography, respectively. In chemoenzymatic labeling a ketone or azido moiety is added enzymatically to the O-GlcNAc modification. Then a biotin tag is attached through this ketone or azido functionality that enables affinity purification of the modification. Lectin weak affinity chromatography (LWAC) is based on the weak affinity of O-GlcNAc to the lectin wheat germ agglutinin (WGA). An adequately long WGA column ensures separation of nonglycosylated peptides from O-GlcNAc modified ones.
Here, we propose an enrichment method that is analogous to the isolation of N-glycosylated proteins by the hydrazide resin capture approach29–31. N-linked complex glycoconjugates are readily oxidized by periodate mainly due to their sialic acid and mannose content, the resulting aldehydes are then selectively bound to a solid support with hydrazide functional groups and the modified peptides released enzymatically by PNGase F. The method has recently been extended for the enrichment of sialylated O-linked glycoproteins by replacing the PNGase F cleavage with acid hydrolysis of the sialic acid glycosidic bond32. Though the periodate oxidation is mostly effective for exocyclic vicinal diols and cyclic cis-diol compounds, cellulose33 – a linear polymer of β-D-glucose – and cyclodextrins34,35 – cyclic oligomers of α-D-glucose –, both containing trans-diol moieties, were also found to be oxidized by periodate. This observation encouraged us to experiment with the periodate oxidation - hydrazide resin capture approach for the enrichment of O-GlcNAc modified proteins.
The enrichment method was developed using an O-GlcNAc modified peptide standard and α-crystallin. Then the strategy was applied to the 26S proteasome purified from Drosophila melanogaster to locate the modification on proteins associated with the UPS.
Materials and methods
Chemicals
Sequencing grade side-chain protected porcine trypsin (modified by reductive methylation) was ordered from Promega (Madison, WI, USA). ZipTip C18 tips were from Millipore (Billerica, MA, USA), OMIX C18 tips were from Varian (Lake Forest, CA, USA). Click-iT O-GlcNAc peptide standard was obtained from Invitrogen (Eugene, OR, USA). Affi-Gel Hz was from BioRad (Hercules, CA, USA). Amino-functionalized controlled pore glass (LCA-CPG, pore size 513 Å) was ordered from CPG Inc. (Lincoln Park, NJ, USA). High purity solvents (HPLC grade) were purchased from Sigma (Steinheim, Germany) and Merck (Darmstadt, Germany). All other chemicals were obtained from Sigma (Steinheim, Germany).
Controlled pore glass hydrazide (H-CPG) was synthesized from LCA-CPG. First, the amino functions of LCA-CPG were derivatized to yield free carboxyls36, then the resulting carboxyls were reacted with hydrazine37.
26S proteasome subcomplexes were purified from Drosophila embryos as previously described38.
O-GlcNAc enrichment by hydrazide resin capture
The proteins were solubilized with 0.5% SDS in 50 mM sodium acetate. The pH was adjusted to pH 5–6 and sodium periodate was added at a final concentration of 20 mM. The oxidation was performed at 37 °C for 6 h in the dark, then it was terminated by 5 equivalents of sodium sulfite solution (freshly prepared) for 20 min. If needed, the pH of the solution was readjusted to pH 5–6. The hydrazide resin (stored in isopropanol) was washed three times with water and added to the reaction mixture. The settled slurry was ca. 1/8 to 1/10 of the total volume. The coupling proceeded overnight with gentle agitation by vertical rotation. The supernatant was discarded and the resin was washed five times with 0.5 M triethylamine phosphate, pH 8.5 in 30% acetonitrile, then with 100 mM triethylamine phosphate, pH 8. The disulfide bridges were reduced with dithiothreitol (20 mM in 100 mM triethylamine phosphate, pH 8) at 56 °C for 1 h and the free sulfhydryls derivatized with iodoacetamide at a final concentration of 50 mM at pH 8 for 1 h in the dark. The resin was washed three times with 50 mM triethylamine phosphate, pH 7.5. Then trypsin was added, and the digestion proceeded at 37 °C overnight. Nonspecifically bound tryptic peptides were removed by washing the resin five times with each 0.5 M triethylamine phosphate, pH 8.5 in 30% acetonitrile and 0.1% trifluoroacetic acid in 50% acetonitrile; then three times with isopropanol and finally with 50 mM sodium acetate. The modified peptides were cleaved by the addition of 200 mM hydroxylamine hydrochloride in 50 mM sodium acetate, pH 5 overnight with vertical rotation. The eluate was collected, then the resin washed with 50 mM sodium acetate and added to the eluate. The sample was desalted on a C18 ZipTip prior to the MS analysis.
BEMAD of O-GlcNAc modified peptides
The sample was dried and redissolved in a solution of 250 mM sodium hydroxide and 50 mM cysteamine hydrochloride. The reaction proceeded at 37 °C for 3 h. Then the sample was desalted on a C18 ZipTip.
MALDI-TOF MS
Data were acquired on a Bruker Reflex III mass spectrometer in reflectron mode. 2,5-dihydroxy-benzoic acid served as the matrix.
LC-MS/MS
CID analysis was performed on a Premier Q-TOF mass spectrometer on-line coupled to a nanoAQUITY UPLC system (Waters Micromass). HPLC conditions: Samples were injected onto a Symmetry trap column (C18, 5 μm, 180 μm × 20 mm) at a flow rate of 15 μL/min for 3 min in 3% of solvent B and separated on an AQUITY UPLC BEH C18 column (C18, 1.7 μm, 75 μm × 200 mm) at a flow rate of 300 nL/min with a gradient of 10–40% B in 90 min. Solvent A was 0.1% formic acid in water, solvent B 0.1% formic acid in acetonitrile. MS conditions: The spray voltage was set at 3–3.5 kV and the cone voltage at 26–28 V. Desolvation temperature was 180 °C. Data were acquired in a data-dependent fashion: 1 sec MS surveys were followed by 5 sec CID experiments on computer-selected multiply charged ions. Collision energy was adjusted to the m/z and charge of the precursor ion. A 30 sec dynamic exclusion was used. Data were processed by Mascot Distiller (v2.2.1.0). In ETD analysis peptide fractionation was performed similarly as above, only a 60 min gradient was used. The data acquisition was carried out in a linear ion trap – Orbitrap hybrid mass spectrometer. MS survey measurements were performed in the Orbitrap at a resolution of 60000, CID and ETD experiments were carried out in the linear trap. Ion populations within the trap were controlled by integrated automatic gain control (AGC). For CID, AGC target was set to 10000, with dissociation at 35% of normalized collision energy, activation time: 30 ms. For ETD, the AGC target values were set to 10000 and 100000 for the isolated precursor cations and fluoranthene anions, respectively, and allowing 200 ms of ion/ion reaction time. Supplemental activation for the ETD experiments was enabled. A 60 sec dynamic exclusion was used. Data were processed by an in-house peak-picking software: PAVA.
Database search
Q-TOF CID data were searched against Drosophila melanogaster proteins in the NCBI 20080718 database (52783 sequences) using in-house Mascot (v2.2.05) and Protein Prospector (v5.2.2, prospector.ucsf.edu) search engines. Monoisotopic masses with precursor mass tolerance of ± 50 ppm and fragment mass tolerance of ± 0.1 Da were used and semitryptic cleavages were allowed. ETD data were searched against the UniProtKB 20090707 database (32646 Drosophila melanogaster entries) using Protein Prospector. Monoisotopic masses with precursor mass tolerance of ± 15 ppm and fragment mass tolerance of ± 0.6 Da were used. Only tryptic peptides were considered, with one missed cleavage permitted.
Cysteine carbamidomethylation was considered as fixed modification, and acetylation of protein N-termini, methionine oxidation and pyroglutamic acid formation from N-terminal glutamine residues as variable modifications. Additionally, a new modification with a mass of +231 Da on serine and threonine residues was defined considering oxidation of the O-GlcNAc ring and conversion of the aldehyde groups to oximes, enabling neutral loss of 126 or 231 Da (see detailed structural description in Figure 2 in Results and Discussion). This modification was included into the modifications list and used as a variable modification. Conversion of serine and threonine residues to S-aminoethyl-cysteine and β-methyl-S-aminoethyl-cysteine, respectively, was considered as a variable modification in BEMAD experiments. N-terminal serine/threonine oxidation and conversion to oxime (−16 and −30 Da, respectively) was also taken into account as variable modification. Results of the database searches were manually validated.
Figure 2.
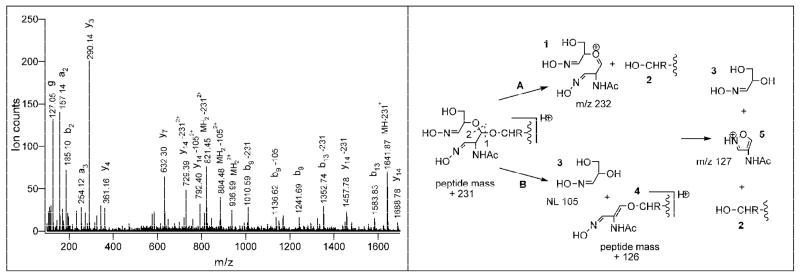
Fragmentation of the oxime derivatives of oxidized O-GlcNAc modified peptides. Left panel: CID spectrum of the oxime derivative of the O-GlcNAc modified α-crystallin peptide AIPVSgREEKPSSAPSS; precursor ion at m/z 936.98 (2+); acquired in a Q-TOF mass spectrometer; g denotes a sugar related product ion. Right panel: Fragmentation scheme. Pathway A: The sugar is eliminated in the same manner, as from O-glycosylated peptides in general. An oxonium ion (1) is observed at m/z 232 and the “unmodified” peptide is detected of the same charge as or one charge less (2) than the precursor ion. Pathway B: Upon cleavage of the adjacent acetal bond only half of the carbohydrate derivative is released as a neutral (3; 105 Da) and a mass signature of 126 Da is left behind on the modified amino acid (4). Further fragmentation leads to a product ion of m/z 127, we hypothesize an isoxasolium structure (5) for this product.
Results and Discussion
Adaptation of the periodate oxidation - hydrazide resin capture approach to the O-GlcNAc modification
Based on the original isolation technique developed for N-linked glycoproteins the method consists of the following steps: i) periodate oxidation of the carbohydrate moiety, ii) capture to hydrazide functionalized solid phase, iii) removal of nonglycosylated proteins, iv) enzymatic digestion, v) removal of nonglycopeptides and vi) release of the modified peptides. However, appropriate changes had to be introduced because of the differences between N-linked complex glycoconjugates and O-GlcNAc modification.
The first main difference is the reduced reactivity of the O-GlcNAc moiety in the periodate oxidation due to the trans configuration of the vicinal hydroxyls at positions C3 and C4. However, at elevated temperatures and longer incubation times the GlcNAc ring can also be oxidized to the dialdehyde derivative. Unfortunately, N-terminal serine and threonine residues – being vicinal amino alcohols – are also oxidized by the periodate to the respective glyoxylyl derivatives39 and will be captured by the hydrazide resin. Even if the oxidation is performed at the protein level the N-terminal peptides of proteins starting with serine or threonine will contribute to the nonglycosylated background.
Another important difference to the original protocol is in the release of the modified peptides. In the enrichment of N-glycoproteins, the release of the modified peptides is performed by PNGaseF treatment cleaving the peptide from the hydrazide captured sugar structure29–31. The site of the modification is “labeled” by the conversion of the respective asparagine to aspartic acid. The oxidized and hydrazide captured O-GlcNAc cannot be removed by an enzyme. At the same time the liberation of the Ser/Thr-modified peptides by β-elimination is analogous to the PNGaseF treatment of the N-linked glycopeptides. This approach provides two advantages: i) the modified amino acid is marked and ii) peptides attached N-terminally to the hydrazide resin are unaffected. However, the relatively harsh conditions that have to be used for the reaction may lead to partial degradation of the sample. That is especially likely when a threonine is modified, since this residue is much less prone to β-elimination than a glycosylated serine. Moreover, β-elimination of unmodified serine, threonine or alkylated cystein residues of nonspecifically bound peptides can lead to false positive results. Therefore, nondestructive alternatives were considered: peptides are released by the cleavage of the hydrazone bond with the sugar modification still attached, thus providing a direct evidence of glycosylation. However, peptides captured N-terminally to the resin are also released by this method impairing selectivity.
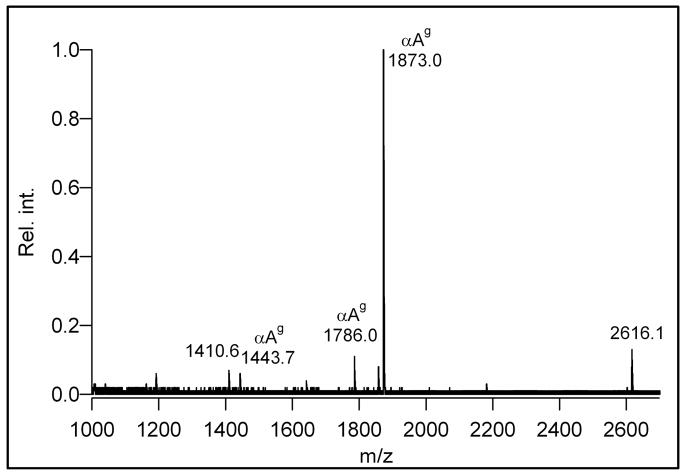
The enrichment protocol was developed using a standard peptide and then optimized on α-crystallin, known to be O-GlcNAc modified. In the periodate oxidation performed with the O-GlcNAc modified peptide standard several conditions with respect to periodate concentration, time, temperature and pH were tested. The initial oxidation product was α-N-glyoxylyl-APTSgTIAPG, however, a minor peak appeared at −2 Da within a few hours at room temperature indicating partial oxidation of the carbohydrate ring. The ring opening was complete in an overnight reaction. At elevated temperatures the reaction proceeded faster, at 37 °C the sugar ring was completely oxidized to the dialdehyde derivative in 4–6 h (Supplemental Figure 1). The reaction took place only under acidic conditions (pH 3–6.5), under alkaline conditions (pH~9) no oxidation of the sugar structure was observed even in a 24 h reaction. Periodate oxidation at the protein level was performed in the presence of denaturing agents, GuHCl or SDS. Both were found to be compatible with the enrichment, however, SDS proved to be superior over GuHCl probably due to electrostatic interactions with the resin. Oxidative damage of the peptides was not investigated, however, partial oxidation of cysteine and methionine residues was observed. The oxidation reaction was terminated by sulfite31, then oxidized O-GlcNAc was reacted with hydrazide resin overnight. Two types of solid support were tested, agarose-bound hydrazide (Affi-Gel Hz) and hydrazide coupled to silica (H-CPG). The Affi-Gel is widely used in the enrichment of N-linked glycopeptides29,31. The H-CPG resin has been applied to the selective isolation of oxidative stress related 4-hydroxynonenal modified proteins36. In the O-GlcNAc enrichment no significant difference in the performance of the two resins was observed. After removal of the nonglycosylated proteins, the resin-bound proteins were digested with trypsin and the nonglycosylated peptides were also removed. The modified peptides were released either by β-elimination or hydrazone bond cleavage. For the latter approach, three reactions were tested: periodate40, acidic36,39 or hydroxylamine cleavage. The β-elimination strategy (coupled to Michael-addition) gave moderate yields. The hydrazone bond was found to be cleaved by periodate in solution, however, this approach was inefficient when applied to solid-phase bound crystallin. The acidic cleavage was also omitted because a partial sugar loss was observed resulting in decreased sensitivity. For the cleavage of the hydrazone bond the overnight hydroxylamine treatment under mildly acidic conditions (pH 5) was found to be the most efficient converting the O-GlcNAc modified peptides to oxime derivatives. The MALDI-TOF mass spectrum of the enriched α-crystallin peptides is shown in Figure 1. The oxime derivative of the α-crystallin A glycopeptide AIPVSgREEKPSSAPSS is observed at m/z 1873.0 as the most abundant peak. Signals at m/z 1443.7 and 1786.0 represent the truncated forms AIPVSgREEKPS and AIPVSgREEKPSSAPS, respectively. The AIPVSgREEKPSSAPS peptide represents the shorter isoform of α-crystallin A, the AIPVSgREEKPS peptide is either a degradation product or belongs to an even shorter isoform. The oxime derivative of the α-crystallin B glycopeptide EEKPAVTgAAPK was observed only in the LC-MS/MS analysis (Supplemental Figure 2).
Figure 1.
MALDI-TOF mass spectrum of the isolated α-crystallin glycopeptides. The peptides at m/z 1443.7, 1786.0 and 1873.0 denoted as αAg point to the same glycosylation site at Ser-162 in the α-crystallin A chain and correspond to peptides AIPVSgREEKPS, AIPVSgREEKPSSAPS and AIPVSgREEKPSSAPSS, respectively.
CID fragmentation of the oxime derivatives of O-GlcNAc modified peptides
In general, O-glycosylated peptides show characteristic fragmentation upon collisional activation. Since the glycosidic bond is much more susceptible to fragmentation than the amide bond, extensive sugar loss from the precursor ion is observed in the CID spectrum along with abundant oxonium ion at the corresponding m/z. Moreover, due to a gas-phase rearrangement the sugar is eliminated without “marking” the originally modified residue in the process. The oxime derivatives of the oxidized O-GlcNAc modified peptides show slightly different fragmentation as demonstrated by the CID spectrum of the α-crystallin A peptide AIPVSgREEKPSSAPSS, m/z 936.98 acquired on a Q-TOF mass spectrometer (Figure 2, left panel). Cleavage of the glycosidic bond results in product ions observed at m/z 821.78 and 1641.87 corresponding to neutral loss and charged loss of the sugar derivative (−231 Da), respectively. As a new feature, an additional characteristic loss from the precursor ion is observed at m/z 884.48 corresponding to the neutral loss of 105 Da. No oxonium ion is detected at m/z 232.09, instead a sugar related product ion is observed at m/z 127.05. Note that this product ion and the neutral loss of 105 Da add up to the mass of the oxonium ion. Since the crystallin peptide contains multiple prolines and the amide bond N-terminally to proline is also relatively facile, most of the sequence ions detected correspond to these cleavages, either retaining the modification or having lost the above mentioned 105 or 231 Da. This distinct fragmentation of the oxime derivatives of the oxidized O-GlcNAc modified peptides is summarized in the right panel of Figure 2. Since the carbohydrate ring is split open in the periodate oxidation, cleavage of the C-O bond on either side of the carbonyl C results in two characteristic losses. In pathway A, the sugar is eliminated in the same manner as intact carbohydrates from O-glycosylated peptides in general, i.e. without any indication where the modification was originally located. Depending on where the charge is retained, an oxonium ion is observed at m/z 232 and the “unmodified” peptide is detected of the same charge as (constant neutral loss) or one charge less than (constant charged loss) the precursor. If the adjacent acetal bond is cleaved (pathway B), only half of the carbohydrate derivative is released. The result is a neutral loss of 105 Da and a mass signature of 126 Da on the modified amino acid. Further fragmentation leads to a product ion of m/z 127, we hypothesize an isoxasolium structure for this product. The fragmentation pattern of the oxime derivatives of the O-GlcNAc modified peptides largely depends on the type of the activation. In ion traps, due to the single activation of the precursor ion mainly the neutral loss of 231 and 105 Da from the precursor ion is observed along with the oxonium ion at m/z 232 indicating a peptide being O-GlcNAc modified. As a consequence, ion trap CID spectra may not contain sufficient information for peptide identification and/or site assignment. In Q-TOF instruments multiple collisions in the collision cell lead to additional peptide backbone cleavages and also to the splitting of the oxonium ion.
Proteasome interacting proteins are O-GlcNAc modified
The O-GlcNAc enrichment protocol was applied to the subcomplexes of the proteasome purified from Drosophila melanogaster in two fractions. One of the fractions contained mainly the subunits of the catalytic core particle while the other one the subunits of the regulatory complex. Fractions were concentrated by acetone treatment, then protein precipitate was solubilized in SDS containing sodium acetate buffer at pH 7 and the pH adjusted to pH 5–6 for the periodate oxidation and the subsequent coupling to the Affi-Gel. For the release of the O-GlcNAc modified peptides the hydroxylamine cleavage was applied. Since oximes are acid labile, the desalted and dried samples were dissolved in water prior to LC-MS/MS analysis on a Q-TOF mass spectrometer and a linear ion trap - Orbitrap tandem mass spectrometer equipped with ETD. Additionally, a portion of the samples was subjected to BEMAD experiments to assist site assignment. The database searches were performed supplementing appropriate variable modifications on serine/threonine residues, i.e. opening of the sugar ring and conversion to oxime in the hydroxylamine cleaved samples or conversion of the modified serine/threonine to the corresponding cysteine derivative in BEMAD experiments. Additionally, several subunits of the proteasome start with serine or threonine due to processing of the N-terminal methionine or a pre-sequence. N-terminal oxidation and hydrazide capture of these proteins was also taken into account by conversion of peptide N-terminal serine/threonine to glyoxylyl oxime used as variable modification.
Altogether, a total of twelve GlcNAc modified peptides were observed in the enriched fractions. The modified peptides generally eluted in the low organic part of the chromatography preceding the nonglycosylated peptides. This chromatographic separation is due to the increased hydrophilic character of the O-GlcNAc modified peptides, a consequence of the sugar modification as well as the amino acids surrounding the modification site28. The nonspecific background mostly corresponded to N-terminal oxidation and hydrazide capture of some proteasome subunits (Supplemental Figure 3). The O-GlcNAc modified peptides represented approximately 4% of the total MS/MS spectra, which was less than the 8.5% obtained by lectin weak affinity chromatography in a post-synaptic density preparation28.
Seven GlcNAc modified peptides detected could be identified unambiguously, representing six different glycosylation sites as summarized in Table 1. The modification sites were determined from CID, ETD or BEMAD experiments. No O-GlcNAc modification was located on proteasomal subunits. Instead, interacting partners of the proteasome were found GlcNAc modified.
Table 1.
O-GlcNAc modified peptides identified from Drosophila melanogaster proteins.
| FlyBase Symbol | Name | Function | m/za | Sequence | O-GlcNAc site | Method for site assignment |
|---|---|---|---|---|---|---|
| CG19475 | Faf/fat facets | ubiquitin-specific protease | 881.43 (2+) | 2614TPTTg SSPSTAAWPAR2628 | Thr-2617 | CID |
| CG7546 | CG7546 | apoptosis | 668.10 (4+) | 776STSTAPAGGATVVPPTSg AAVTRPVTTGR803 | Ser-792 | BEMAD |
| b753.3821 (2+) | 713ANTLPTTg ATQTR724 | Thr-719 | ETD | |||
| CG2918 | CG2918 | HSP70 chaperone | 723.67 (3+) | 613SEESTKQDTEAKNg ETIK629 | Asn-625 | CID |
| 745.8537 (2+) | 619qDTEAKNg ETIK629 | Asn-625 | CID | |||
| CG5462 | scribbled | polarization of embryonic epithelia | 511.7723 (2+) | 1209VTETITK1215 + GlcNAc | Thr-1210 or Thr-1212 or Thr-1214 | None |
| CG1483 | MAP205 | microtubule binding | 908.9448 (2+) | 1026NTSSTTg TSTATATITK1041 | Thr-1031 | ETD |
denotes the glycosylated residue.
‘q’ stands for pyroglutamic acid.
The m/z value is given for the precursor of the respective (either the oxime or the BEMAD) derivative used for the modification site assignment.
Precursor masses detected in the Orbitrap mass spectrometer were within 5 ppm of the calculated value.
In the deubiquitinating enzyme Faf (CG1945) O-GlcNAc glycosylation at Thr-2617 was determined from the CID spectrum of the precursor at m/z 881.43 (2+) (Supplemental Figure 4). Faf belongs to the ubiquitin processing protease (UBP/USP) class of deubiquitinating enzymes41,42. It fulfills an editing role by hydrolyzing the ubiquitin tag from target proteins thus preserving them from proteasomal degradation41,43. Faf is essential for proper development of the eye: by deubiquitination of its substrate liquid facets it assists delta signaling and cell fate determination44.
Two O-GlcNAc sites were identified in CG7546, Thr-719 and Ser-792. Modification site assignment in the 713ANTLPTTATQTR724 peptide was accomplished from the ETD spectrum of the precursor at m/z 753.3821 (2+) (Figure 3), the mass difference between z5* and z6 unambiguously confirmed modification at Thr-719. For Ser-792 only the BEMAD experiment provided sufficient information for site assignment (Supplemental Figure 5). CG7546 has a conserved ubiquitin-like domain (UBL) on its N-terminus. A homologous UBL domain is found in Xenopus laevis or human Scythe/Bat3 proteins. Moreover, in Xenopus Scythe tandem UBL domains (UBL1 and UBL2) were found that redundantly interact with the embryonic Xrpn10c subunit of the proteasome45. Scythe was also demonstrated to interact with the ATPase domain of HSC70/HSP70 via its C-terminal BAG domain inducing inhibition of the chaperone function of HSP7046. HSP70/HSC70 proteins are also known interactors of the proteasome22,23.
Figure 3.
ETD spectrum of the “O-GlcNAc-oxime” modified 713ANTLPTTgATQTR724 peptide; precursor ion at m/z 753.3821 (2+); Thr-719 was identified as the site of modification. Asterisks indicate ions produced by hydrogen migration, i.e. z+1 fragments.
Interestingly, the protein CG2918 found glycosylated in this study shows HSP70 homology. Manual validation of the CID spectrum of the precursor at m/z 745.86 (2+) revealed that in this protein the GlcNAc modification is attached to Asn-625, as confirmed by fragments y5 (m/z 730.37) and b8 (m/z 1025.43) retaining the 126 Da mass signature (Figure 4). The N-glycosidic bond is fairly stable upon collisional activation while the C-O bond on the other side of the carbonyl C is easily fragmented, hence the abundant 105 losses and the retention of the 126 Da sugar fraction on the peptide fragments. This modification site fits to the Asn-X-Ser/Thr consensus sequence of N-glycosylation. Moreover, the protein has on the C-terminus a retention signal for the endoplasmic reticulum where N-glycosylation takes place. A single N-GlcNAc modification on transmembrane proteins has already been demonstrated28, however, there is no information about the role of this modification and the enzymes involved. Additionally, another N-glycosylation site has previously been identified in CG2918 at Asn-49747 which is conserved in its human ortholog (HYOU1) and is also glycosylated29.
Figure 4.
CID spectrum of the “GlcNAc-oxime” modified 619qDTEAKNgETIK629 peptide; precursor ion at m/z 745.86 (2+). The carbohydrate is attached to Asn-625. ‘q’ denotes pyroglutamic acid; 0 stands for water loss.
Two other proteins not connected to the proteasomal degradation pathway were also present in the mixture and were found O-GlcNAc modified. Scribbled (CG5462) is a scaffolding, multi-PDZ domain protein required for the polarization of embryonic epithelia48 and involved in synapse formation49. In this protein the modification was located on the 1209VTETITK1215 peptide, however, the exact site of the modification could not be determined from either CID or ETD of the precursor at m/z 511.77 (2+). In the CID spectrum no mass signature (either 126 or 231 Da) was retained on the sequence ions (Supplemental Figure 6), and the ETD spectrum was low quality for site assignment. In the 205 kDa microtubule-associated protein (MAP205, CG1483) the glycosylated peptide 1026NTSSTTTSTATATITK1041 contains a multitude of potential modification sites. Here, ETD spectrum of the precursor at m/z 908.9448 (2+) revealed modification at Thr-1031 (Supplemental Figure 7). High molecular weight MAPs isolated from rat brain have already been described as O-GlcNAc modified12,50.
Conclusions
An enrichment protocol utilizing periodate oxidation, hydrazide capture and release of the modified peptides by hydroxylamine cleavage was developed for the analysis of the O-GlcNAc modification. Using this approach and mass spectrometry analysis, four Drosophila proteins co-purified with the 26S proteasome were identified as O-GlcNAc modified. Similarly to other glycosylation studies11,32,51 the combination of different fragmentation techniques, i.e. CID and ETD, was applied for modification site assignment, but the use of further chemical derivatization (BEMAD) also proved beneficial. Interestingly, N-linked glycosylation of the HSP70 chaperone like CG2918 with a single GlcNAc in the proper consensus position was also detected, just like in a recently published study28.
At the present stage N-terminal oxidation of serine/threonine residues compromises the use of this enrichment method in large scale analysis. However, N-terminal derivatization is expected to overcome this shortcoming, which may also enable enrichment at the peptide level. Since the method also enriches N-glycopeptides, prior PNGase F treatment has to be performed when applied to complex mixtures containing extracellular or membrane proteins.
Our O-GlcNAc enrichment procedure provides an alternative to the existing methods. As demonstrated earlier in a phosphoproteomic study52 the different enrichment techniques yield complementary information. Similarly, the different O-GlcNAc enrichment methods may identify different modified peptide populations.
Supplementary Material
Acknowledgments
This work was supported by Hungarian Science Foundation grant OTKA T60283, National Office for Research and Technology grant OMFB-00467/2009 (to KFM) and by NIH grant NCRR P41RR001614 to the UCSF MS Facility (director A.L. Burlingame). We thank Lajos Kovács for useful discussion.
Footnotes
Supporting Information Available. MALDI-TOF mass spectra of the periodate oxidized O-GlcNAc peptide standard; base peak chromatogram of an enriched fraction of the proteasome; extracted ion chromatogram and full-scan mass spectrum of a selected glycopeptide (m/z 745.85); CID and ETD spectra of the oxime or BEMAD derivatives of O-GlcNAc modified peptides. This material is available free of charge via the Internet at http://pubs.acs.org.
Contributor Information
Eva Klement, Email: klement@brc.hu.
Zoltán Lipinszki, Email: lzoltan@brc.hu.
Zoltán Kupihár, Email: kupi@ovrisc.mdche.u-szeged.hu.
Andor Udvardy, Email: udvardy@brc.hu.
Katalin F. Medzihradszky, Email: folkl@cgl.ucsf.edu.
References
- 1.Hart GW. Annu Rev Biochem. 1997;66:315–335. doi: 10.1146/annurev.biochem.66.1.315. [DOI] [PubMed] [Google Scholar]
- 2.Roquemore EP, Chou TY, Hart GW. Guide to Techniques in Glycobiology. 1994;230:443–460. doi: 10.1016/0076-6879(94)30028-3. [DOI] [PubMed] [Google Scholar]
- 3.Comer FI, Hart GW. J Biol Chem. 2000;275:29179–29182. doi: 10.1074/jbc.R000010200. [DOI] [PubMed] [Google Scholar]
- 4.Wells L, Vosseller K, Hart GW. Science. 2001;291:2376–2378. doi: 10.1126/science.1058714. [DOI] [PubMed] [Google Scholar]
- 5.Vosseller K, Wells L, Lane MD, Hart GW. Proc Natl Acad Sci U S A. 2002;99:5313–5318. doi: 10.1073/pnas.072072399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jackson SP, Tjian R. Cell. 1988;55:125–133. doi: 10.1016/0092-8674(88)90015-3. [DOI] [PubMed] [Google Scholar]
- 7.Comer FI, Hart GW. Biochim Biophys Acta, Gen Subj. 1999;1473:161–171. doi: 10.1016/s0304-4165(99)00176-2. [DOI] [PubMed] [Google Scholar]
- 8.Lefebvre T, Planque N, Leleu D, Bailly M, Caillet-Boudin ML, Saule S, Michalski JC. J Cell Biochem. 2002;85:208–218. [PubMed] [Google Scholar]
- 9.Vosseller K, Sakabe K, Wells L, Hart GW. Curr Opin Chem Biol. 2002;6:851–857. doi: 10.1016/s1367-5931(02)00384-8. [DOI] [PubMed] [Google Scholar]
- 10.Guinez C, Morelle W, Michalski JC, Lefebvre T. Int J Biochem Cell Biol. 2005;37:765–774. doi: 10.1016/j.biocel.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 11.Vosseller K, Trinidad JC, Chalkley RJ, Specht CG, Thalhammer A, Lynn AJ, Snedecor JO, Guan SH, Medzihradszky KF, Maltby DA, Schoepfer R, Burlingame AL. Mol Cell Proteomics. 2006;5:923–934. doi: 10.1074/mcp.T500040-MCP200. [DOI] [PubMed] [Google Scholar]
- 12.Khidekel N, Ficarro SB, Peters EC, Hsieh-Wilson LC. Proc Natl Acad Sci U S A. 2004;101:13132–13137. doi: 10.1073/pnas.0403471101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dias WB, Hart GW. Mol BioSyst. 2007;3:766–772. doi: 10.1039/b704905f. [DOI] [PubMed] [Google Scholar]
- 14.Sumegi M, Hunyadi-Gulyas E, Medzihradszky KF, Udvardy A. Biochem Biophys Res Commun. 2003;312:1284–1289. doi: 10.1016/j.bbrc.2003.11.074. [DOI] [PubMed] [Google Scholar]
- 15.Zhang FX, Su KH, Yang XY, Bowe DB, Paterson AJ, Kudlow JE. Cell. 2003;115:715–725. doi: 10.1016/s0092-8674(03)00974-7. [DOI] [PubMed] [Google Scholar]
- 16.Hershko A, Ciechanover A. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 17.Pickart CM. Mol Cell. 2001;8:499–504. doi: 10.1016/s1097-2765(01)00347-1. [DOI] [PubMed] [Google Scholar]
- 18.Konstantinova IM, Tsimokha AS, Mittenberg AG. Int Rev Cell Mol Biol. 2008;267:59–124. doi: 10.1016/S1937-6448(08)00602-3. [DOI] [PubMed] [Google Scholar]
- 19.Guinez C, Mir AM, Dehennaut V, Cacan R, Harduin-Lepers A, Michalski JC, Lefebvre T. FASEB J. 2008;22:2901–2911. doi: 10.1096/fj.07-102509. [DOI] [PubMed] [Google Scholar]
- 20.Cole RN, Hart GW. J Neurochem. 2001;79:1080–1089. doi: 10.1046/j.1471-4159.2001.00655.x. [DOI] [PubMed] [Google Scholar]
- 21.Guerrero C, Tagwerker C, Kaiser P, Huang L. Mol Cell Proteomics. 2006;5:366–378. doi: 10.1074/mcp.M500303-MCP200. [DOI] [PubMed] [Google Scholar]
- 22.Wang XR, Huang L. Mol Cell Proteomics. 2008;7:46–57. doi: 10.1074/mcp.M700261-MCP200. [DOI] [PubMed] [Google Scholar]
- 23.Bousquet-Dubouch MP, Baudelet E, Guerin F, Matondo M, Uttenweiler-Joseph S, Burlet-Schiltz O, Monsarrat B. Mol Cell Proteomics. 2009;8:1150–1164. doi: 10.1074/mcp.M800193-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guinez C, Losfeld ME, Cacan R, Michalski JC, Lefebvre T. Glycobiology. 2006;16:22–28. doi: 10.1093/glycob/cwj041. [DOI] [PubMed] [Google Scholar]
- 25.Wells L, Vosseller K, Cole RN, Cronshaw JM, Matunis MJ, Hart GW. Mol Cell Proteomics. 2002;1:791–804. doi: 10.1074/mcp.m200048-mcp200. [DOI] [PubMed] [Google Scholar]
- 26.Khidekel N, Arndt S, Lamarre-Vincent N, Lippert A, Poulin-Kerstien KG, Ramakrishnan B, Qasba PK, Hsieh-Wilson LC. J Am Chem Soc. 2003;125:16162–16163. doi: 10.1021/ja038545r. [DOI] [PubMed] [Google Scholar]
- 27.Wang Z, Udeshi ND, O’Malley M, Shabanowitz J, Hunt DF, Hart GW. Mol Cell Proteomics. doi: 10.1074/mcp.M900268-MCP200. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chalkley RJ, Thalhammer A, Schoepfer R, Burlingame AL. Proc Natl Acad Sci U S A. 2009;106:8894–8899. doi: 10.1073/pnas.0900288106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang H, Li XJ, Martin DB, Aebersold R. Nat Biotechnol. 2003;21:660–666. doi: 10.1038/nbt827. [DOI] [PubMed] [Google Scholar]
- 30.Pan S, Wang Y, Quinn JF, Peskind ER, Waichunas D, Wimberger JT, Jin JH, Li JG, Zhu D, Pan C, Zhang J. J Proteome Res. 2006;5:2769–2779. doi: 10.1021/pr060251s. [DOI] [PubMed] [Google Scholar]
- 31.Sun BY, Ranish JA, Utleg AG, White JT, Yan XW, Lin BY, Hood L. Mol Cell Proteomics. 2007;6:141–149. doi: 10.1074/mcp.T600046-MCP200. [DOI] [PubMed] [Google Scholar]
- 32.Nilsson J, Ruetschi U, Halim A, Hesse C, Carlsohn E, Brinkmalm G, Larson G. Nat Methods. 2009;6:809–811. doi: 10.1038/nmeth.1392. [DOI] [PubMed] [Google Scholar]
- 33.Hou QX, Liu W, Liu ZH, Bai LL. Ind Eng Chem Res. 2007;46:7830–7837. [Google Scholar]
- 34.Kobayashi M, Urayama T, Suzawa I, Takagi S, Matsuda K, Ichishima E. Agric Biol Chem. 1988;52:2695–2702. [Google Scholar]
- 35.Pumera M, Jelinek I, Jindrich J, Coufal P, Horsky J. J Chromatogr A. 2000;891:201–206. doi: 10.1016/s0021-9673(00)00628-2. [DOI] [PubMed] [Google Scholar]
- 36.Roe MR, Xie HW, Bandhakavi S, Griffin TJ. Anal Chem. 2007;79:3747–3756. doi: 10.1021/ac0617971. [DOI] [PubMed] [Google Scholar]
- 37.Zhang X, Breslav M, Grimm J, Guan K, Huang A, Liu F, Maryanoff CA, Palmer D, Patel M, Qian Y, Shaw C, Sorgi K, Stefanick S, Xu D. J Org Chem. 2002;67:9471–9474. doi: 10.1021/jo026288n. [DOI] [PubMed] [Google Scholar]
- 38.Udvardy A. J Biol Chem. 1993;268:9055–9062. [PubMed] [Google Scholar]
- 39.Geoghegan KF, Stroh JG. Bioconjugate Chem. 1992;3:138–146. doi: 10.1021/bc00014a008. [DOI] [PubMed] [Google Scholar]
- 40.Enders D, Wortmann L, Peters R. Acc Chem Res. 2000;33:157–169. doi: 10.1021/ar990062y. [DOI] [PubMed] [Google Scholar]
- 41.Huang Y, Baker RT, Fischer-Vize JA. Science. 1995;270:1828–1831. doi: 10.1126/science.270.5243.1828. [DOI] [PubMed] [Google Scholar]
- 42.Wing SS. Int J Biochem Cell Biol. 2003;35:590–605. doi: 10.1016/s1357-2725(02)00392-8. [DOI] [PubMed] [Google Scholar]
- 43.Wu Z, Li Q, Fortini ME, Fischer JA. Dev Genet. 1999;25:312–320. doi: 10.1002/(SICI)1520-6408(1999)25:4<312::AID-DVG5>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 44.Overstreet E, Fitch E, Fischer JA. Development. 2004;131:5355–5366. doi: 10.1242/dev.01434. [DOI] [PubMed] [Google Scholar]
- 45.Kikukawa Y, Minami R, Shimada M, Kobayashi M, Tanaka K, Yokosawa H, Kawahara H. FEBS J. 2005;272:6373–6386. doi: 10.1111/j.1742-4658.2005.05032.x. [DOI] [PubMed] [Google Scholar]
- 46.Thress K, Song J, Morimoto RI, Kornbluth S. EMBO J. 2001;20:1033–1041. doi: 10.1093/emboj/20.5.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koles K, Lim JM, Aoki K, Porterfield M, Tiemeyer M, Wells L, Panin V. Glycobiology. 2007;17:1388–1403. doi: 10.1093/glycob/cwm097. [DOI] [PubMed] [Google Scholar]
- 48.Bilder D, Li M, Perrimon N. Science. 2000;289:113–116. doi: 10.1126/science.289.5476.113. [DOI] [PubMed] [Google Scholar]
- 49.Roche JP, Packard MC, Moeckel-Cole S, Budnik V. J Neurosci. 2002;22:6471–6479. doi: 10.1523/JNEUROSCI.22-15-06471.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ding M, Vandre DD. J Biol Chem. 1996;271:12555–12561. doi: 10.1074/jbc.271.21.12555. [DOI] [PubMed] [Google Scholar]
- 51.Darula Z, Medzihradszky KF. Mol Cell Proteomics. 2009;8:2515–2526. doi: 10.1074/mcp.M900211-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bodenmiller B, Mueller LN, Mueller M, Domon B, Aebersold R. Nat Methods. 2007;4:231–237. doi: 10.1038/nmeth1005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.