Summary
Deletion 13q14 on fluorescence in situ hybridization (FISH) analysis is the most common cytogenetic abnormality in chronic lymphocytic leukemia (CLL), and is a favorable prognostic biomarker when detected as a sole abnormality. We intensively interrogated clinical outcome in 323 consecutive, untreated CLL patients with isolated 13q- identified within two years of diagnosis. We also analyzed outcome in 217 additional patients with deletion 11q22.3 or 17p13.1, or trisomy 12 based on whether these occurred in isolation or in conjunction with 13q-. Patients with a heterozygous 13q- and those with a homozygous deletion had similar time to first treatment (TFT) and overall survival (OS). In contrast, a higher percentage of 13q- nuclei was associated with significantly shorter TFT (p<0.001). The 5-year untreated rate was 79% for patients with isolated 13q- in ≤65.5% of nuclei compared to 38% among those with 13q- in >65.5% of nuclei (p<0.001). The percentage of nuclei exhibiting 13q- remained an independent predictor of TFT after controlling for ZAP-70, IgVH, or CD38 (all p<0.001). Among patients with 13q- plus one other FISH abnormality, concomitant 13q- appeared to attenuate the shorter survival associated with 17p- (p=0.019). The clinical implications of 13q- in CLL appear more complex than originally appreciated.
Keywords: Chronic lymphocytic leukemia, 13q deletion, 17p deletion
Introduction
A deletion of chromosome band 13q14 (13q-) detected by fluorescence in situ hybridization (FISH) analysis is the single most common cytogenetic abnormality in patients with chronic lymphocytic leukemia (CLL). In their seminal study of 325 treated and untreated patients, Dohner and colleagues (2000) observed a 13q deletion in 55% of patients, and 13q- was the only FISH abnormality in 36% of patients. When detected as a sole abnormality, the presence of 13q– conferred a favorable outcome, with 60% of patients alive after 5 years as compared with 27% for patients with a normal FISH analysis. In a Mayo Clinic study of 159 previously untreated, early stage CLL patients (Shanafelt et al., 2006), 13q- was the sole FISH abnormality in 45% of patients. The median survival for these patients was 17 years, compared with 13.2 years for patients with a normal FISH pattern.
Despite several studies validating the prognostic utility of the hierarchical Dohner classification, important questions remain about the 13q- category. First, it is unknown if there is a difference in clinical outcome based on whether patients have a heterozygous (13q-x1) or homozygous (13q-x2) deletion of 13q. Although we previously postulated that 13q-x2 may be a more aggressive anomaly than 13q-x1 (Dewald et al., 2003), a preliminary study of 122 patients with 13q- as the sole abnormality found similar treatment free survival among CLL patients with one versus two 13q- chromosomes (Fink et al., 2004). Second, while the percentage of interphase nuclei that exhibit the cytogenetic defect has been shown to be of prognostic importance in deletion 17p13.1 (17p-) (Catovsky et al., 2007; Tam et al., 2009), it is unknown whether this characteristic influences the risk of progression among patients with other cytogenetic findings such as isolated 13q-. Third, although 13q- is frequently observed in combination with other cytogenetic defects, it is unknown whether the presence of 13q- in any way modulates the risk associated with deletion 11q or 17p, or trisomy 12, where the present hierarchical classification assumes it has no impact. To address these questions, we have conducted detailed analysis of the clinical outcome of 323 CLL patients with 13q- as their sole cytogenetic abnormality on FISH analysis conducted within two years of diagnosis and prior to any chemotherapeutic treatment. In addition, we analyzed the clinical outcome of 217 CLL patients with deletion 11q22.3 (11q-), 17p-, or trisomy 12 based on whether these defects occurred in isolation or in conjunction with 13q-.
Methods
The standard CLL FISH panel at Mayo Clinic tests for 6q, 11q, 13q, and 17p deletion or monosomy, trisomy 12, and IGH gene rearrangement (200 interphase nuclei each; for chromosome 13, our target probe is D13S319 our internal control probe is LAMP1) (Dewald et al., 2003). Among 2243 patients diagnosed with CLL at Mayo Clinic between 10/26/1992 – 5/10/2008, we identified 323 who had a 13q- as the sole FISH-detected abnormality on analysis of uncultured cells from a whole blood or bone marrow specimen obtained within two years of diagnosis and prior to any chemotherapeutic treatment. No cell separation method was employed. For patients diagnosed prior to 2002, archived fixed cell pellets were studied to establish the FISH pattern within two years of diagnosis and prior to therapy. To evaluate whether the presence of 13q- modulates the clinical outcome of patients with a 2nd cytogenetic abnormality, we also identified patients with defects of 11q-, 17p-, and trisomy 12 either in isolation or in combination with 13q-. Patients with more complex defects (e.g., 11q- with trisomy 12) were excluded to isolate the influence of 13q- on the 2nd abnormality. All of the research for this study was approved by the Mayo Clinic institutional review board and all bloods were obtained according to the Helsinki convention.
For initial analysis, patients with a sole 13q- were segregated into one of three groups: heterozygous 13q-(13q-x1), homozygous 13q- (13q-x2), or mosaic deletions with some cells having 13q-x1 and some cells 13q-x2. These groups were then analyzed for differences in clinical characteristics using either chi-squared or Fisher’s exact statistics for qualitative variables and analysis of variance for the quantitative variables. Analysis of overall survival (OS) and time to first treatment (TFT) were assessed using Kaplan-Meier curves and p-values were calculated using a log-rank test. Additionally, we investigated whether any differences existed between 13q-x1 alone versus 13q-x2 when mosaic patients were included with the 13q-x2 group. We next investigated whether the percentage of cells (as a continuous variable) with a 13q deletion was associated with OS or TFT, followed by analysis to determine the threshold of percent abnormal cells that best predicted both OS and TFT (Contal & O’Quigley, 1999) Using the threshold value, we ran Cox proportional hazards analyses that included the dichotomized threshold value along with prognostic factors to see if the percentage of abnormal nuclei retained its predictive ability independent of each of the prognostic factors. Finally, patients with 11q-, 17p-, or trisomy 12 were investigated to compare OS and TFT for those with and without a concomitant 13q deletion. For these three groups, OS and TFT results were displayed using Kaplan-Meier curves and p-values were calculated using a log-rank test. All statistical tests were two-sided and considered significant at the alpha=0.05 level; all analyses were performed in SAS version 9.1 (SAS Institute, http://www.sas.com).
Results
Comparison of 13q-x1, 13q-x2 and mosaic 13q deletion groups
We identified 323 patients with an isolated 13q- on FISH testing. After a median follow-up of 2.1 years, 52 patients (16%) have been treated and 30 (9%) have died with a median TFT and OS of 6.9 and 9.3 years, respectively. When patients with 13q- were classified as having heterozygous, homozygous, or mosaic deletion of 13q, 194, 61, and 68 patients were in the 13q-x1, 13q-x2, and mosaic 13q- groups, respectively. The mean and range percentage of abnormal nuclei in each group was 43.7% (8–98.5%), 47.2% (3.5–93.5%), and 54.0% (15.5–98%), respectively (p=0.02). All of the patients had some cells with an apparently normal chromosome 13 pair.
There was no significant difference among the three 13q- groups with respect to sex, Rai stage or other important clinical variables (Table I). Age was significantly different with the 13q-x2 group being four years older than the 13q-x1 group (p=0.04). When patients with mosaic and nonmosaic 13q-x2 were combined and compared to the 13q-x1 group, results were the same as comparing the three groups except that age was no longer significantly different (Table II). The median TFT and median OS were similar in both the three group 13q- comparison and when comparing the combined mosaic and 13q-x2 groups to the 13q-x1 group. No differences in TFT or OS were observed for 13q- patients relative to 179 contemporary patients with a normal FISH pattern fulfilling the eligibility criteria (Figure 1).
Table I.
Mayo Clinic CLL cases with a 13q- FISH abnormality, comparing 13q-x1 versus 13q-x2, versus mosaic FISH patterns.
| 13q-x1 (n=194) | 13q-x1/13q-x2 (n=68) | 13q-x2 (n=61) | p-value | ||||
|---|---|---|---|---|---|---|---|
| N | % | N | % | N | % | ||
| Age | 0.04 | ||||||
| Mean (std dev) | 61.3 | 11.1 | 61.6 | 10.5 | 65.3 | 10.3 | |
| Median, range | 61 | 37–87 | 60.5 | 43–90 | 66 | 46–87 | |
| Gender | 0.08 | ||||||
| Female | 74 | 38 | 32 | 47 | 17 | 28 | |
| Male | 120 | 62 | 36 | 53 | 44 | 72 | |
| Rai Stage | 0.30 | ||||||
| Low (0) | 98 | 56 | 44 | 70 | 37 | 66 | |
| Intermediate (1 or 2) | 65 | 37 | 18 | 29 | 17 | 30 | |
| High (3 or 4) | 11 | 6 | 1 | 2 | 2 | 4 | |
| Unknown | 20 | 5 | 5 | ||||
| CD38 | 0.17 | ||||||
| Positive | 24 | 13 | 5 | 8 | 3 | 5 | |
| Negative | 161 | 87 | 61 | 92 | 55 | 95 | |
| Missing | 9 | 2 | 3 | ||||
| ZAP-70 | 0.79 | ||||||
| Positive | 38 | 25 | 12 | 21 | 11 | 22 | |
| Negative | 112 | 75 | 45 | 79 | 38 | 78 | |
| Missing | 44 | 11 | 12 | ||||
| IgVH | 0.28 | ||||||
| Unmutated | 33 | 26 | 7 | 15 | 9 | 21 | |
| Mutated | 95 | 74 | 41 | 85 | 33 | 79 | |
| Missing | 66 | 20 | 19 | ||||
Table II.
Mayo Clinic CLL cases with a 13q-x1 versus pooled cases with either a 13q-x2 or a mosaic pattern.
| 13q-x1 (n=194) | 13q-x2 and 13q-x1/13q-x2 (n=129) | p-value | |||
|---|---|---|---|---|---|
| N | % | N | % | ||
| Age | 0.10 | ||||
| Mean (std dev) | 61.3 | 11.1 | 63.3 | 10.5 | |
| Median, range | 61 | 37–87 | 63 | 43–90 | |
| Gender | 0.98 | ||||
| Female | 74 | 38 | 49 | 38 | |
| Male | 120 | 62 | 80 | 62 | |
| Rai Stage | 0.08 | ||||
| Low (0) | 98 | 56 | 81 | 68 | |
| Intermediate (1 or 2) | 65 | 37 | 35 | 29 | |
| High (3 or 4) | 11 | 6 | 3 | 3 | |
| Unknown | 20 | 10 | |||
| CD38 | 0.07 | ||||
| Positive | 24 | 13 | 8 | 6 | |
| Negative | 161 | 87 | 116 | 94 | |
| Missing | 9 | 5 | |||
| ZAP-70 | 0.50 | ||||
| Positive | 38 | 25 | 23 | 22 | |
| Negative | 112 | 75 | 83 | 78 | |
| Missing | 44 | 23 | |||
| IgVH | 0.16 | ||||
| Unmutated | 33 | 26 | 16 | 18 | |
| Mutated | 95 | 74 | 74 | 82 | |
| Missing | 66 | 39 | |||
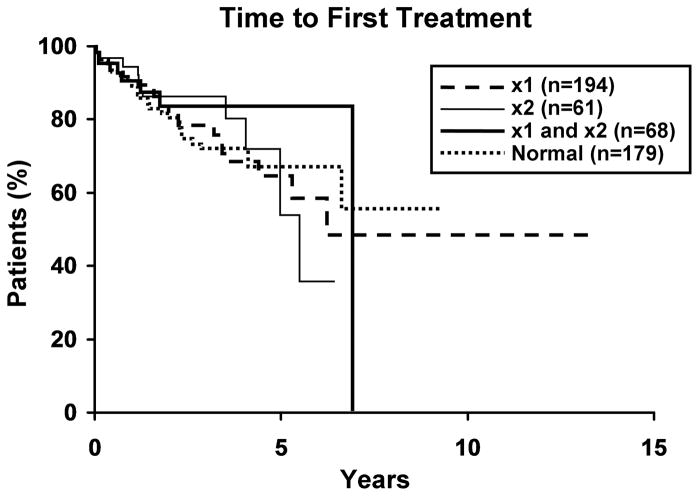
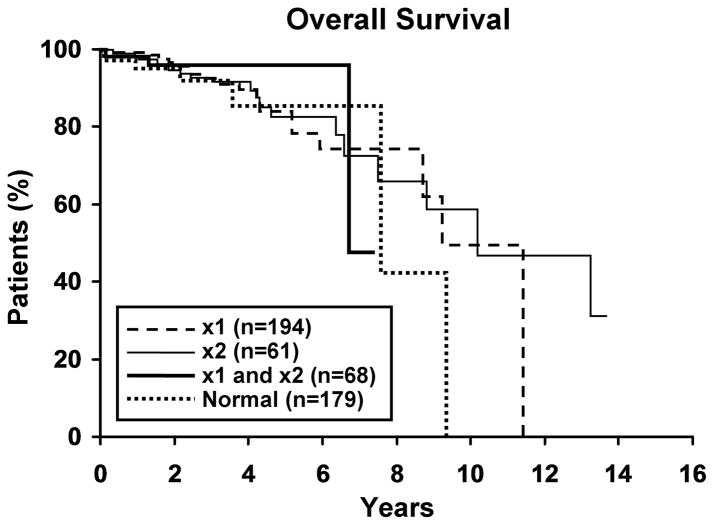
Figure 1. Clinical outcome for patients with 13q- and normals.
1a. Time to first treatment (TFT) for three 13q- groups and normals. There was no significant difference (p=0.84) in TFT among the three 13q-groups, nor between the three groups and normals (p=0.91). Median TFT for 13q-x1, 13q-x2 and both 13q-x1/13q-x2 are 6.1, 5.5 and 6.9 years, respectively. More than 50% of the normal group remain untreated, thus a median value is not reported. 1b. Overall survival (OS) for three 13q- groups and normals. There was no difference (p=0.38) in OS between the three groups, nor between the three groups and normals (p=0.49). Median OS for 13q-x1, 13q-x2, both 13q-x1/13q-x2, and normals are 10.1, 6.7, 7.5, and 9.2 years, respectively.
Clinical prognosis and percentage of abnormal nuclei
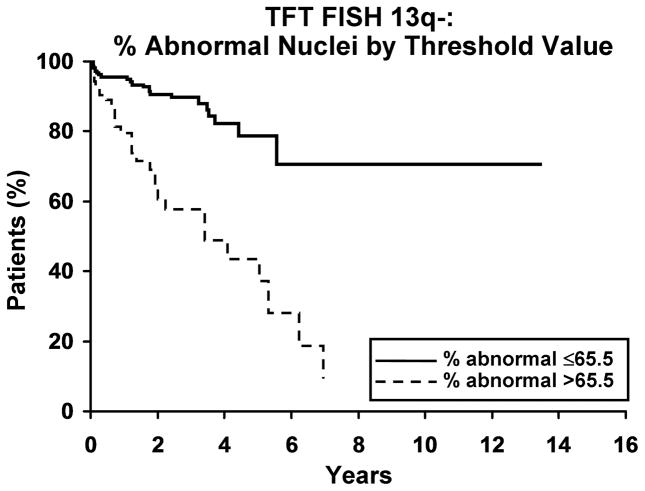
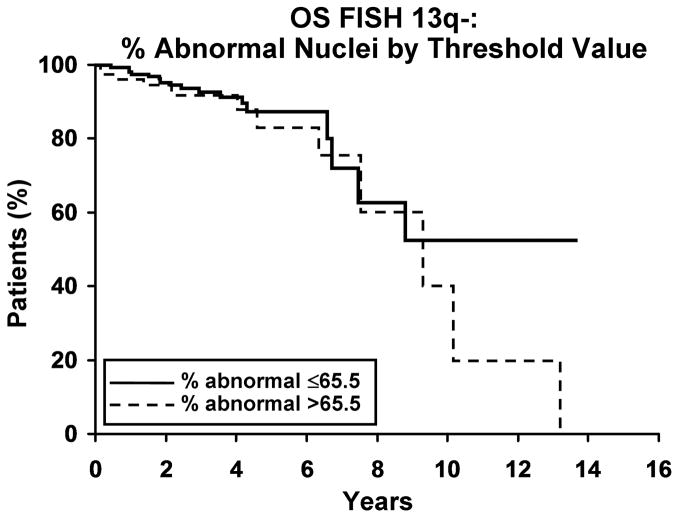
We next evaluated whether the percentage of abnormal nuclei with a defect of 13q- was related to clinical outcome. As a continuous variable, the percentage of nuclei with 13q- was related to TFT but not OS. For each 1% increase in the percentage of abnormal nuclei with 13q- the hazard ratio for treatment was 1.03 (95% confidence interval 1.02 to 1.04; p<0.001); for OS, the hazard ratio was 1.01 (95% C.I. 0.99–1.02; p=0.65). We determined that a value of 65.5 abnormal nuclei was the threshold that best predicts OS and TFT. Patients with >65.5% abnormal nuclei exhibited a shorter TFT than those with ≤65.5% abnormal nuclei (median 3.4 years vs not reached; HR=4.4 (95% C.I. 2.5 to 7.7); p<0.001, Figure 2a). The 5-year untreated rate for those with >65.5% abnormal nuclei is 38% compared to 79% among those with ≤65.5% of abnormal cells. The association of the 65.5% threshold value with OS was not significant (median 9.3 years vs not reached; HR=1.5 (95% C.I. 0.7, 3.2); p=0.28, Figure 2b). In multivariable analyses, having >65.5% abnormal nuclei with 13q- predicted shorter TFT independent of CD-38, ZAP 70 and IgVH mutation status (Table III).
Figure 2. Clinical outcome for patients with 13q- by threshold value.
2a. Time to first treatment (TFT) and percentage of abnormal nuclei by threshold. A higher percentage of abnormal nuclei, with either 13q-x1 or 13q-x2, is associated with a significantly shorter TFT (p<0.001, HR=4.4 (2.5–7.7)). Median time to treatment for % abnormal ≤ 65.5% and >65.5% was not reached and 3.4 years, respectively. The 5-year untreated rate for % abnormal ≤65.5% and >65.5% was 79% and 38%, respectively. 2b. Overall survival (OS) and percentage of abnormal nuclei by threshold. OS appears not to be affected by the percentage of abnormal nuclei (p=0.28, HR=1.5 (0.7–3.2)). Median OS for % abnormal ≤65.5 and % abnormal >65.5 are not reached and 9.3 years, respectively. The 5-year survival rate for % abnormal ≤65.5% and >65.5% was 88% and 83%, respectively.
Table III.
Stratified time to first treatment analysis based on percentage of 13q- nuclei and other prognostic factors
| Stratification factor | N | Variable | Hazard Ratio (HR) | 95% confidence interval for HR | p-value |
|---|---|---|---|---|---|
| CD38 | 308 | % 13q->65.5 | 4.1 | 2.3, 7.4 | <0.001 |
| CD38 (positive) | 2.5 | 1.2, 4.8 | 0.01 | ||
| ZAP-70 | 256 | % 13q->65.5 | 5.2 | 2.5, 10.5 | <0.001 |
| ZAP-70 (positive) | 2.9 | 1.5, 5.7 | 0.002 | ||
| IgVH | 218 | % 13q->65.5 | 3.6 | 1.9, 6.9 | <0.001 |
| IgVH (unmutated) | 3.5 | 1.8, 6.6 | <0.001 |
Effect of 13q- on clinical course for CLL patients with a second FISH abnormality
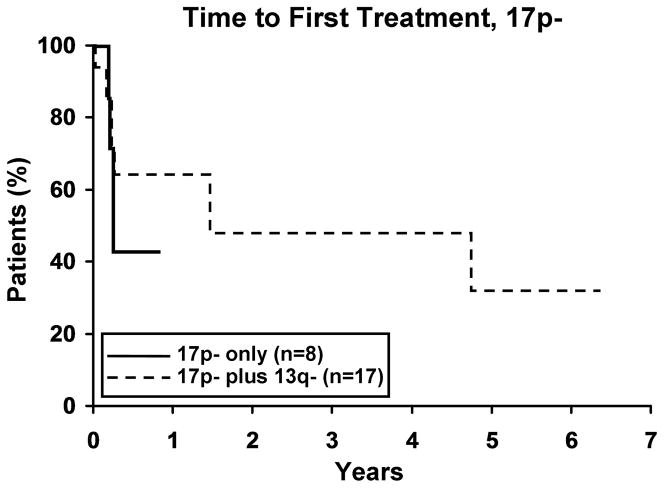
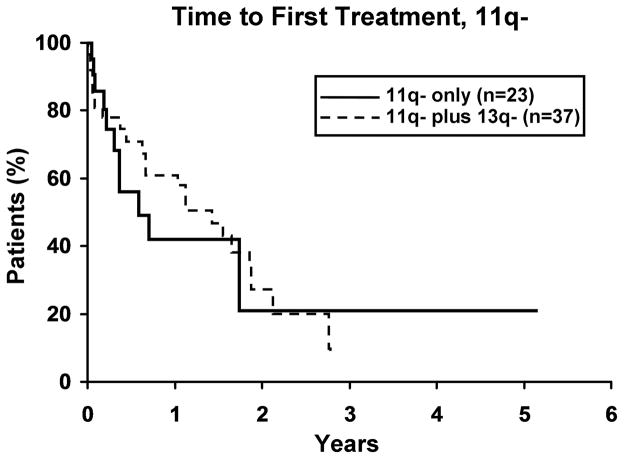
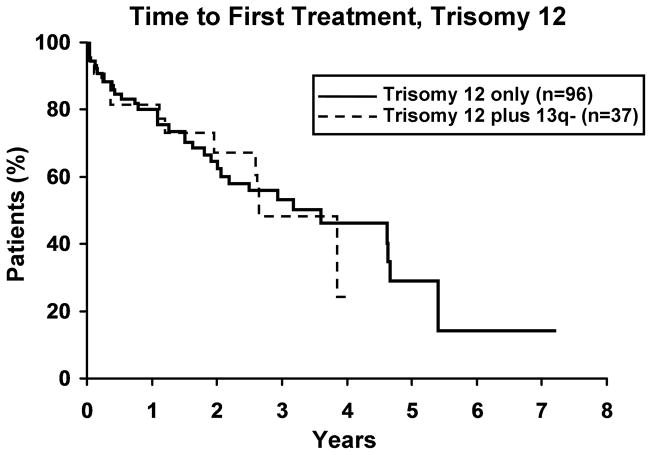
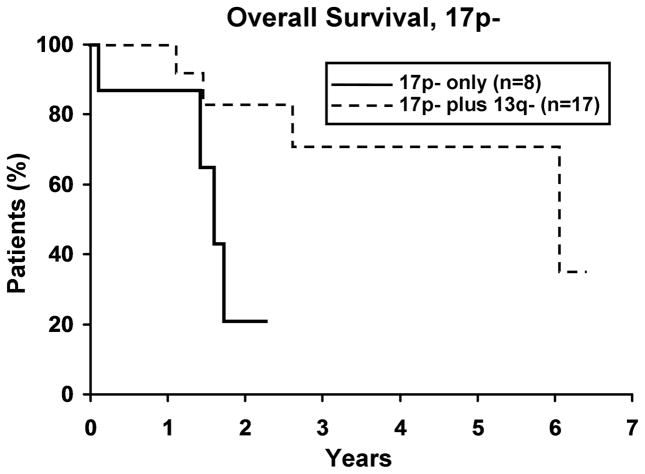
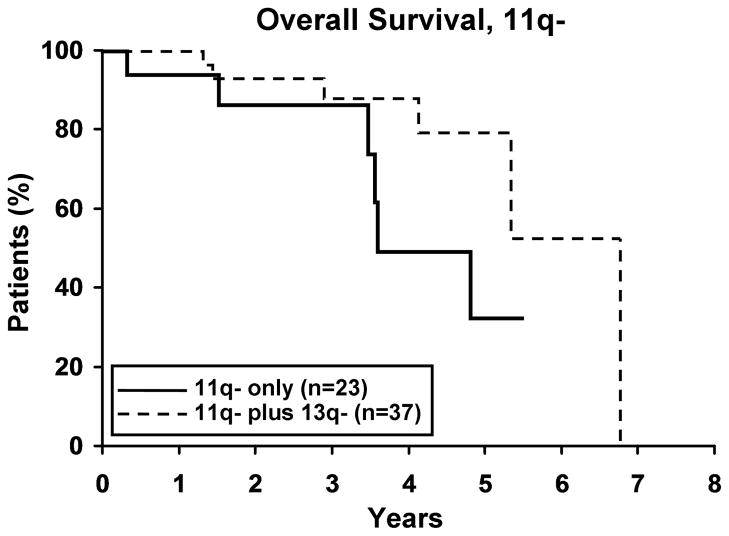
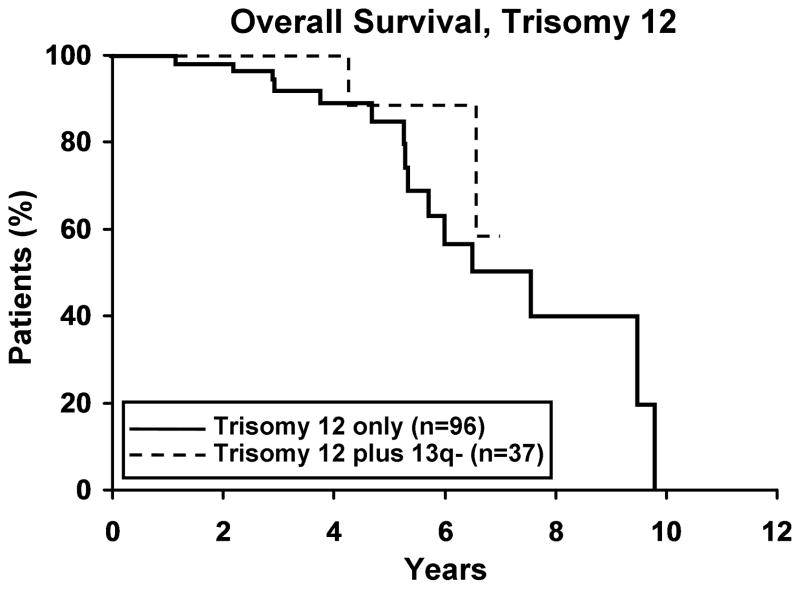
To evaluate whether the presence of 13q- modulates the clinical outcome of patients with a 2nd cytogenetic abnormality, we next identified patients with 11q -, 17p -, or trisomy 12 in either isolation or combination with 13q-. Among those with 11q- (n=59), 17p- (n=25), and trisomy 12 (n=133), 37, 17 and 37 respectively, also had a 13q-. No difference in TFT was observed for any defect based on whether or not it occurred in combination with 13q- (Figure 3). Despite the small sample size for 17p- cases (Figure 4a), OS was significantly longer (p=0.019) for 17p- patients with a concomitant 13q-. No significant difference was observed in OS for patients with 11q- or trisomy 12 (Figure 4b, c), based on whether or not there was a concomitant 13q-.
Figure 3. Time to first treatment (TFT) for 13q- with a second FISH abnormality.
3a. Isolated 17p-(n=8) vs 17p- plus 13q- (n=17). There was no difference in TFT between the two 17p- groups (p=0.41). Median TFT for 17p- and 17p- plus 13q- was 0.2 and 1.5 years, respectively. 3b. Isolated 11q- (n=23) vs 11q- plus 13q- (n=37). There was no difference in TFT between the two 11q- groups (p=0.71). Median TFT for 11q- and 11q- plus 13q- was 0.6 and 1.4 years, respectively. 3c. Isolated trisomy 12 (n=96) vs trisomy 12 plus 13q- (n=37). There was no difference in TFT between the two trisomy 12 groups (p=0.89). Median TFT for trisomy 12 and trisomy 12 plus 13q- was 3.5 and 2.6 years, respectively.
Figure 4. Overall Survival (OS) for 13q- with a second FISH abnormality.
4a. Isolated 17p- (n=8) vs 17p- plus 13q- (n=17). There was a significant difference in OS between the two 17p- groups with the 17p-plus 13q- patients surviving longer (p=0.019). Median OS for 17p- and 17p- plus 13q- was 1.6 and 6.1 years, respectively. 4b. Isolated 11q- (n=23) vs 11q- plus 13q- (n=37). Though not significant, there appears to be a trend in OS between the two 11q- groups with the 11q- plus 13q- surviving longer (p=0.07). Median OS for 11q- and 11q- plus 13q- was 3.6 and 6.7 years, respectively. 4c. Isolated trisomy 12 (n=96) vs trisomy 12 plus 13q- (n=37). There was no difference in OS between the two trisomy 12 groups (p=0.39). Median OS for trisomy 12 and trisomy 12 plus 13q- was 7.5 years and not reached, respectively.
Discussion
FISH is one of the most powerful and widely used prognostic tools in patients with CLL even though it evaluates a small number of genetic defects. Further dissection of the clinical usefulness of this methodology in CLL is an important and ongoing process. In our study therefore we focused on the association of a variety of 13q defect parameters and the clinical course of CLL patients. The major conclusions here are that patients with heterozygous (13q-x1) and homozygous (13q-x2) deletion of 13q had similar OS and TFT, but that patients with a higher percentage of nuclei exhibiting a 13q deletion had significantly shorter TFT. The relationship between TFT and percent nuclei affected by 13q- persisted as both a continuous and categorical variable and remained an independent predictor of TFT after controlling for ZAP-70, IgVH, or CD38 status (all p<0.001). The presence of a concomitant 13q- also appears to attenuate the shorter survival associated with 17p- (p=0.019).
These observations have important clinical implications. First, and consistent with Dohner et al (2000), the results support grouping of CLL patients with isolated 13q- into a single category regardless of whether or not they have a heterozygous or homozygous deletion. Second, the results suggest that patients with isolated 13q- in >65.5% of nuclei experience a more aggressive clinical course than those patients with ≤65.5% of nuclei affected (38% untreated at 5 years vs. 79%, respectively; p<0.001). The magnitude of this effect is such that the 5-year untreated rate for patients with >65.5% of nuclei affected by 13q- is less favorable than patients with a normal FISH analysis (38% untreated at 5 years vs 67%; p=0.002) who are classified as “intermediate” risk under the Dohner classification. The mechanism for this more unfavorable progression with larger percentages of 13q- nuclei is unclear but may relate to the known deletion of miR-15a and miR-16-1 within the 13q deletion region. These microRNAs are known to regulate at least the anti-apoptotic protein Bcl-2 levels in CLL B cells and thus the CLL B cell clones with more prevalent 13q- cells will have more resistance to cell death (Cimmino et al., 2005).
How do these findings compare with the results of other studies? Dohner et al (2000) described a median TFT and OS for patients with 13q- of 92 and 133 months as compared to 49 and 111 months for patients with normal FISH. Similarly, in our original prospective study of untreated, predominantly early stage CLL patients (Shanafelt et al., 2006) we reported a median OS of 204 months and 158 months for 13q- and normal FISH groups, respectively. Neither of these studies compared the outcome of patients with heterozygous 13q- to those with homozygous 13q-. In a small series of 38 patients with isolated 13q-, Chena et al (2008) reported higher Rai stage and shorter progression-free survival in the 6 patients with 13q-x2 (with or without some 13q-x1 cells) as compared to 32 patients with 13q-x1. We are unable to confirm this finding in our series of 323 patients. In another recent study, Hernandez et al (2009) evaluated clinical outcome of 109 patients with 13q- as the sole cytogenetic abnormality based on the percentage of abnormal nuclei. Patients with 80% or more nuclei with 13q- had shorter TFT and OS than those with fewer than 80% abnormal nuclei. The relationship between the higher percentage of 13q deletion nuclei and TFT is confirmed in the present cohort, although no difference in OS was observed. The observation that the percent of nuclei harboring 13q- influences its prognostic significance is also conceptually similar to the relationship observed for other FISH categories such as 17p- (Catovsky et al., 2007; Tam et al., 2009). We did observe that the co-existence of 13q- with 17p- may attenuate some of the disease progression risk known to be associated with the latter cytogenetic defect. The exact reason for this is not clear but if there are two separate clones this could be related to dominance of the 13q- clone in terms of its growth potential. If the 13q- and 17p- defects are present in the same clone it is possible that differences in gene expression profiles associated with the double defect may ultimately result in a less aggressive clone. We also realize that larger studies are needed to evaluate these original findings and in particular to address the relationship between 13q deletion and OS.
In summary almost all studies of FISH and prognosis in CLL have examined each FISH abnormality as classified under the hierarchical Dohner classification, with little attention paid to the effect of the potential interaction of multiple FISH detectable abnormalities. This is due in part to the requirement for extremely large sample sizes to evaluate all possible permutations and the belief that the highest risk abnormalities, such as 17p-, overshadow the clinical implications of other abnormalities. The results of the present analysis challenge this dogma. The clinical implications of an isolated 13q- defect in patients with CLL appear more complex than originally appreciated. Although patients with heterozygous and homozygous 13q- appear to have similar clinical outcome, individuals with a high percentage of leukemic cells carrying the 13q- defect appear to experience a more aggressive clinical course equivalent to those in the “intermediate” risk FISH categories. We believe that the continued investigation of the relationship between specific FISH defects and clinical outcome will lead to improved risk stratification. The molecular basis for such clinical differences in outcome may provide important insights into disease biology and is the subject of ongoing investigation.
Acknowledgments
This work was supported in part by NIH/National Cancer Institute, CA95241, to NEK. We also thank Mr. Edson Spencer and the Donner Family Foundation for their philanthropic support.
Footnotes
Authorship
Contribution: DLVD, TDS, TGC, CSZ, SAS, and NEK performed the research. DLVD, TDS, CSZ, SAS, and NEK designed the research and KGR, SMS and SLS analyzed the data. DLVD, TDS, CSZ, SAS, KGR, SLS, and NEK wrote the manuscript.
Conflict-of-Interest disclosure: The authors declare no conflict of interests.
References
- Catovsky D, Richards S, Matutes E, Oscier D, Dyer MJS, Bezares RF, Pettitt AR, Hamblin T, Milligan DW, Child JA, Hamilton MS, Dearden DE, Smith AG, Bosanquet AG, Davis Z, Brito-Babapulle V, Else M, Wade R, Hillmen P on behalf of the UK National Cancer Research Institute (NCRI) Haematological Oncology Clinial Studies Group and NCRI Chronic Lymphocytic Laukaemia Working Group. Assessment of fludarabine plus cyclophosphamide for patients with chronic lymphocytic leukaemia (the LRF CLL4 Trial): a randomised controlled trial. The Lancet. 2007;370:230–239. doi: 10.1016/S0140-6736(07)61125-8. [DOI] [PubMed] [Google Scholar]
- Chena C, Avalos JS, Bezares RF, Arrossagaray K, Turdo K, Bistmans A, Slavutsky I. Biallelic deletion 13q14.3 in patients with chronic lymphocytic leukemia: cytogenetic, FISH and clinical studies. European Journal of Haematology. 2008;81:94–99. doi: 10.1111/j.1600-0609.2008.01086.x. [DOI] [PubMed] [Google Scholar]
- Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojic SE, Aqeilan RI, Zupo S, Dono M, Rassenti L, Alder H, Volinia S, Liu C, Kipps TJ, Croce CM. \miR-15 and miR-16 induce apoptosis by targeting BCL2. Proceedings of the National Academy of Science. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contal C, O’Quigley J. An application of changepoint methods in studying the effect of age on survival in breast cancer. Computational Statistics and Data Analysis. 1999;30:253–270. [Google Scholar]
- Dewald GW, Brockman SR, Paternoster SF, Bone ND, O’Fallon JR, Allmer C, James CD, Jelinek DF, Tschumper RC, Hanson CA, Pruthi RK, Witzig TE, Call TG, Kay NE. Chromosome anomalies detected by interphase fluorescence in situ hybridization: correlation with significant biological features of B-cell chronic lymphocytic leukaemia. British Journal of Haematology. 2003;121:287–295. doi: 10.1046/j.1365-2141.2003.04265.x. [DOI] [PubMed] [Google Scholar]
- Dohner H, Stilgenbauer S, Benner A, Leupolt E, Krober A, Bullinger L, Dohner K, Bentz M, Lichter P. Genomic aberrations and survival in chronic lymphocytic leukemia. New England Journal of Medicine. 2000;343:1910–1916. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- Fink S, Geyer S, Shanafelt T, Smoley S, Paternoster S, Giordano K, Tschumper R, Jelinek D, Bone N, Kabat B, Call T, Phyliky R, Kay N, Dewald G. Clinical Significance of Homozygous D13S319 Deletion in B-Cell Chronic Lymphocytic Leukemia (B-CLL) Blood. 2004;104 A:2799. [Google Scholar]
- Hernandez JA, Rodriguez AE, Gonzalez M, Benito R, Fontanillo C, Sandoval V, Romero M, Martin-Nunez G, Garcis de Coca A, Fisac R, Galende J, Recio I, Ortuno F, Garcia JD, de las Rivas J, Gutierrez NC, San Miguel JF, Hernandez JM. A high number of losses in 13q14 chromosome band is associated with a worse outcome and biological differences in patients with B-cell chronic lymphoid leukemia. Haematologica. 2009;94:364–371. doi: 10.3324/haematol.13862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanafelt TD, Witzig TE, Fink SR, Jenkins RB, Paternoster SF, Smoley SA, Stockero KJ, Nast DM, Flynn HC, Tschumper RC, Geyer S, Zent CS, Call TG, Jelinek DF, Kay NE, Dewald GW. Prospective evaluation of clonal evolution during long-term follow-up of patients with untreated early-stage chronic lymphocytic leukemia. Journal of Clinical Oncology. 2006;24:4634–4641. doi: 10.1200/JCO.2006.06.9492. [DOI] [PubMed] [Google Scholar]
- Tam CS, Shanafelt TD, Wierda WG, Abruzzo LV, Van Dyke DL, O’Brien S, Ferajoli A, Lerner SA, Lynn A, Kay NE, Keating MJ. De novo deletion 17p13.1 chronic lymphocytic leukemia shows significant clinical heterogeneity: the MD Anderson and Mayo Clinic experience. Blood. 2009;114:957–964. doi: 10.1182/blood-2009-03-210591. [DOI] [PMC free article] [PubMed] [Google Scholar]