Abstract
The metabolic syndrome is highly complex disease and has become one of the major public-health challenges worldwide. We sought to identify genetic loci with potential influence on multiple metabolic factors in a white population in Beaver Dam, Wisconsin, and to explore the possibility of genetic heterogeneity by family history of diabetes (FHD). Three metabolic factors were generated using principal component factor analysis, and they represented: 1) glycemia, 2) blood pressure, and 3) combined (body mass index, high-density lipoprotein cholesterol and serum uric acid) factors. Multipoint model-free linkage analysis of these factors with 385 microsatellite marker was performed on 1,055 sib-pairs, using Haseman-Elston regression. Genome-wide suggestive evidence of linkage was found at 30cM on chromosome 22q (empirical P [Pe] = 0.0002) for the glycemia factor, at 188–191 cM on chromosome 1q (Pe = 0.0007) for the blood pressure factor, and at 82 cM on chromosome 17q (Pe = 0.0007) for the combined factor. Subset analyses of the families by FHD showed evidence of genetic heterogeneity, with divergent linkage signals in the subsets on at least 4 chromosomes. We found evidence of genetic heterogeneity by FHD for the three metabolic factors. The results also confirmed findings of previous studies that mapped components of the metabolic syndrome to a chromosome 1q region.
Keywords: linkage mapping, factor analysis, genetic heterogeneity, metabolic syndrome, diabetes mellitus
INTRODUCTION
The metabolic syndrome is characterized by clustering of obesity, dyslipidemia, hyperglycemia, and hypertension within individuals. It is highly prevalent and has become one of the major public-health challenges worldwide. According to the third National Health and Nutrition Examination Survey, the prevalence of metabolic syndrome was 24% in US adults (1). This is cause for concern, as adults with metabolic syndrome are atgreater risk for type 2 diabetes (2, 3), cardiovascular disease and stroke (4, 5). Similarly, we previously found that components of the metabolic syndrome predicted incident cardiovascular diseases and diabetes in an adult population in Beaver Dam, Wisconsin, and persons with a cluster of the components were associated with an increased risk of diabetes, such that those with four or more components were 35 times as likely to develop the disease as those with none of the components (6).
The development of metabolic syndrome clearly involves both environmental factors and largely unknown genetic factors. Data from the National Heart, Lung, and Blood Institute (NHLBI) Family Heart Study indicated that individual metabolic traits were highly heritable (e.g. heritability of 46% for body mass index [BMI] and 63% for high-density lipoprotein [HDL] cholesterol), and genetic correlations between individual traits were significant (7). These provide evidence that genetic factors contribute to the manifestations of metabolic syndrome, and also suggest that its genetic mechanisms are complex in nature. It is likely that each individual trait is influenced by several genes, and the genes affecting one trait may also affect some of the other traits (7).
To understand why these metabolic syndrome-related traits cluster in individuals and families, a central question would be whether there is one, or more likely, several genes that influence metabolic traits. To answer this question, investigators have used factor analysis to identify the underlying genetic basis of metabolic syndrome (8–12). Factor analysis can identify the presence of unrelated underlying factors from many inter-correlated traits related to metabolic syndrome. The identified factors may capture the dimensions of the entire syndrome, and each individual factor may be reflective of the “underlying biology”.
In the present study, we attempted 1) to identify genetic loci with potential influence on multiple metabolic factors, obtained from principal-component factor analysis, and 2) to investigate genetic heterogeneity of the factors in an adult white population in Beaver Dam, Wisconsin.
METHODS AND PROCEDURES
Study population
The study families were recruited from a population-based study, which was initially designed to determine prevalence, incidence and risk factors for age-related eye disease. Methods used to identify the study participants have been described previously (13). Briefly, a private census of the Beaver Dam population was performed and 5,924 eligible persons aged 43–86 years were identified. From 1988 to 1990, a baseline examination was completed for 4,926 (83.2%) persons, of whom 98.8% were white. Informed consent was obtained from each participant and the study protocol was approved by the Wisconsin-Madison Institutional Review Board.
Based on information collected from the baseline examination, family relationships were identified and extended pedigree information was confirmed during follow-up examinations (14, 15). To be a member of a pedigree, an individual needed to be a participant in the baseline examination and be related to another eligible individual through a sibling, parent, child, first cousin or avuncular relationship. There were no inclusions/exclusions based on prior or current evidence of prevalent metabolic or cardiovascular diseases at the time of family ascertainment. After excluding pedigreesnot informative for linkage (ie, parent-offspring pairs), there were 2,068 people who were members of 486 pedigrees (of 602 original pedigrees), consisting of 1,055 sib-pairs who had complete data on all relevant metabolic traits and have been genotyped for genome-wide microsatellite markers. The mean family size was 10.5 ± 12.5 members.
Metabolic phenotypic data collection
Participants responded to an interview during which they were asked whether they had ever been told by their physicians that they had any of the following: diabetes, angina, a heart attack, a transient ischemic attack, a stroke, and/or hypertension. If participants self-reported diabetes, they were asked if they were currently using insulin, oral hypoglycemia agents, and/or a special diet. Weight and height were measured on a standard scale, with subjects not wearing shoes, and then BMI was calculated. Blood pressure was measured using a random-zero sphygmomanometer following the Hypertension Detection and Follow-up Program protocol (16). Following the examination, casual blood specimens were obtained and serum total cholesterol (17), HDL cholesterol (18), serum glucose, glycosylated hemoglobin (HbA1c) (19), and serum uric acid (20) were assayed according to standard protocols.
Diabetes was defined as either 1) a previous history of diabetes treated with insulin, oral hypoglycemic agents and/or diet, or without treatment but presence of hyperglycemia at the baseline examination, or 2) the presence of elevated HbA1c >2 SDs above the mean of the relevant age-sex group at the baseline examination in persons with no previous medical history of diabetes (i.e., newly diagnosed diabetes) (21). Hyperglycemia was defined as HbA1c >2 SDs above the mean of the relevant age-sex group or a casual serum glucose level >200 mg/dl. Primary care providers were consulted whenever there was doubt about the diagnosis.
Principal-component factor analysis
Multiple metabolic factors were derived using factor analysis on seven metabolic traits in the Beaver Dam study population, after adjusting for age, age-squared and gender (age and age squared were included in the regression models because these provided a better fit than age alone). The seven traits were BMI, systolic and diastolic blood pressure, serum glucose, HbA1c, serum HDL cholesterol, and serum uric acid. The principal-components factor method (22) was used to analyze the correlation matrix between these traits. The factors with eigenvalues ≥1 were retained. Data were then rotated using a varimax rotation to generate interpretable components. Factor loadings were examined and loadings ≥0.40 were used to characterize the factor. The scoring coefficients, generated during the factor analysis, were then used to compute factor scores for each individual subject. All principal-component factor analysis was performed using Stata software (version 9.2; Stata Corp., College Station, TX).
Genotyping and genetic analyses
Correlations of the factors obtained from factor analysis between all possible family pairs in the pedigrees were calculated, using the FCOR program of the Statistical Analysis for Genetic Epidemiology (S.A.G.E.) software package (version 5.0). Heritability was estimated for each factor using parent-offspring correlations (r) by the equation h2 = 2r.
A genome-wide scan with 385 microsatellite markers on 22 chromosomes (modified Weber panel 8 marker set) was performed by the Center for Inherited Disease Research (Johns Hopkins University, Baltimore), using automated fluorescent microsatellite analysis. The average spacing of markers was 9 cM throughout the genome and there were no gaps in the map >18 cM. Map distances were based on the Marshfield map (23). Mendelian inconsistencies were checked using MARKERINFO (S.A.G.E) and misspecified family relationships were identified using RELTEST (S.A.G.E.) and RELCHECK (24). Allele frequencies of all the markers were estimated with the FREQ program (S.A.G.E.).
We used the factor scores as continuous traits to localize chromosomal regions responsible for each factor. Prior to linkage analysis, data normality was checked. George-Elston transformation (25) (λ1 = −1.287, λ2 = 0.432) was performed on factor 1 because of its highly kurtotic distribution. The coefficients of skewness and kurtosis were 0.12 and −0.07 for factor 1 (after transformation), 0.42 and 0.63 for factor 2, 0.18 and 0.23 for factor 3, respectively. These properties of the factor score distributions minimally violated the normality assumption.
Multipoint model-free linkage analyses were performed using Haseman-Elston regression in SIBPAL (S.A.G.E.), which models quantitative sib-pair traits as a function of marker allele identity-by-descent sharing (26). We used a modified Haseman-Elston method, in which the dependent variable is weighted average of the squared trait difference and the squared mean-corrected trait sum, further adjusted for the non-independence of the sibling pairs (option W4 in SIBPAL) (27). This approach has been shown to have more power than the original Haseman-Elston method and its other extensions (27). The nominal P (Pn) values were obtained assuming the asymptotic distribution of the likelihood-ratio test statistics. To verify all multipoint Pn values (<0.01), we performed up to 1,000,000 permutations to obtain empirical P (Pe) values.
In consideration of the potential effects of diabetes and taking hypertension medication on metabolic traits (28–30), Haseman-Elston linkage of each factor was repeated, including diabetes affection status and taking hypertension medication (yes vs. no) as a covariate in the analyses. Furthermore, recognizing that genetic heterogeneity of common complex traits is a thorny issue for linkage analysis, we performed linkage analysis of the factors in two subsets of the families defined by the presence or absence of family history of diabetes (FHD). For a given family, the presence of FHD was defined as at least one member enrolled in the linkage study having been diagnosed with diabetes or having newly diagnosed diabetes at the baseline examination. For a given family, if none of the enrolled family members had diabetes, that family was defined as not having FHD.
RESULTS
Phenotypic characteristics of the Beaver Dam study population and the family members enrolled in the linkage scan are presented in Table 1. The age and sex distribution and the levels of metabolic traits of the family members were similar to those of the entire population. The absolute values of the pairwise correlation coefficients among the seven traits range from 0.756 between HbA1c and serum glucose levels to 0.006 between HbA1c and diastolic blood pressure (Supplementary Table S1). Three factors with eigenvalues ≥1 were identified (Table 2). The first factor correlated highly with serum glucose and HbA1c, and thus was interpreted as a “glycemia” factor. The second factor had high loadings of systolic and diastolic blood pressures, and was labeled as a “blood pressure” factor. The third factor correlated at least moderately with BMI, serum HDL cholesterol and serum uric acid; therefore, it was labeled as a “combined” factor. The three factors together explained 70.9% of the total variance.
Table 1.
Characteristics of the Beaver Dam study population and the family members enrolled in the linkage study
| Characteristics | Entire study population (n = 4,926) | Members of families (n = 2,068) |
|---|---|---|
| Age (years) | 62.0 ± 11.2 | 63.1 ± 10.9 |
| Gender (% female) | 56.1 | 54.7 |
| Diabetes (%) | 9.1 | 9.2 |
| Serum glucose (mg/dl) | 106.7 ± 39.4 | 107.1 ± 39.7 |
| HbA1c (%) | 6.1 ± 1.6 | 6.1 ± 1.6 |
| Serum HDL cholesterol (mg/dl) | 52.0 ± 17.6 | 51.4 ± 17.3 |
| Serum uric acid (mg/dl) | 6.0 ± 1.9 | 6.0 ± 1.8 |
| BMI (kg/m2) | 28.8 ± 5.4 | 29.1 ± 5.6 |
| Systolic blood pressure (mmHg) | 132.1 ± 20.5 | 133.8 ± 20.8 |
| Diastolic blood pressure (mmHg) | 77.3 ± 11.0 | 77.6 ± 11.1 |
HbA1c, glycosylated hemoglobin; HDL, high-density lipoprotein; BMI, body mass index.
Data are reported as means ± SD, except for gender and diabetes, which are expressed as percentage of subjects.
Table 2.
Factor loadings of principal component analysis after varimax rotation
| Traitsa | Factor 1 (glycemia) | Factor 2 (blood pressure) | Factor 3 (combined) |
|---|---|---|---|
| Serum glucose | 0.93 | 0.04 | −0.04 |
| HbA1c | 0.93 | 0.01 | −0.05 |
| Serum HDL cholesterol | −0.18 | 0.13 | 0.70 |
| Serum uric acid | −0.10 | 0.09 | −0.74 |
| BMI | 0.17 | 0.27 | −0.68 |
| Systolic blood pressure | 0.09 | 0.88 | −0.06 |
| Diastolic blood pressure | −0.03 | 0.89 | −0.04 |
| Eigenvalue | 2.09 | 1.61 | 1.26 |
| Total variance (%) | 29.8 | 23.0 | 18.1 |
| Accumulate variance (%) | 29.8 | 52.8 | 70.9 |
HbA1c, glycosylated hemoglobin; HDL, high-density lipoprotein; BMI, body mass index.
Data in bold denote absolute values of loadings ≥0.4.
Adjusted for age, age-squared and gender.
Familial correlation among possible relative pairs for each of the three factors is shown in Table 3. Estimates of the heritability were 48% for the glycemia factor, 40% for the combined factor and 17% for the blood pressure factor.
Table 3.
Familial correlations of the three metabolic factors
| Glycemia factor |
Blood pressure factor |
Combined factor |
||
|---|---|---|---|---|
| Relationship | n | Correlation ± S.E. | Correlation ± S.E. | Correlation ± S.E. |
| Full Sibling | 1,061 | 0.074 ± 0.034 | 0.120 ± 0.035 | 0.207 ± 0.036 |
| Parent-offspring | 368 | 0.240 ± 0.053 | 0.085 ± 0.056 | 0.199 ± 0.056 |
| Half-sibling | 61 | −0.002 ± 0.133 | 0.193 ± 0.130 | 0.194 ± 0.134 |
| Avuncular | 686 | 0.174 ± 0.041 | 0.020 ± 0.045 | 0.036 ± 0.047 |
| Half-avuncular | 9 | 0.405 ± 0.302 | 0.763 ± 0.163 | −0.276 ± 0.352 |
| Cousin | 1,559 | −0.010 ± 0.027 | 0.062 ± 0.031 | −0.021 ± 0.030 |
| Half-cousin | 10 | −0.000 ± 0.212 | −0.004 ± 0.253 | −0.022 ± 0.323 |
See Table 2 for the constructs of the three factors.
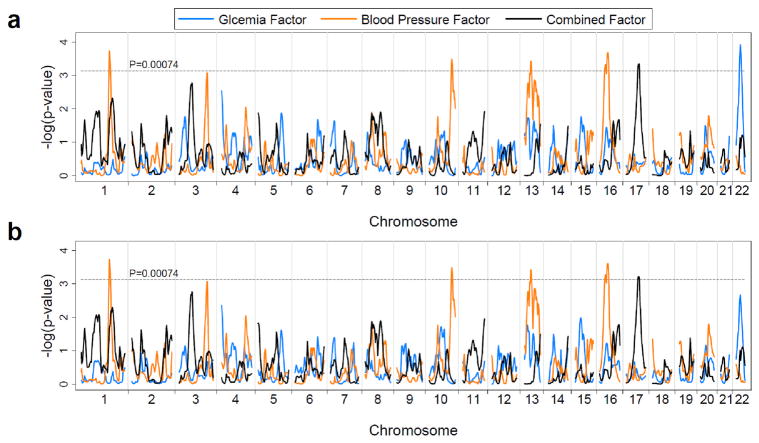
Multipoint linkage analysis of the three factors was performed on the 1,055 sib-pairs. In the Haseman-Elston regression model without covariates (Figure 1a and Table 4), we observed several regions with a multipoint Pn <0.00074, the Lander and Kruglyak’s criterion for suggestive linkage (31), on chromosomes 1, 10, 13, 16, 17, and 22. The glycemia factor had its strongest linkage at 30 cM on chromosome 22 (near maker D22S689). The blood pressure had its strongest linkage signal at 188–191 cM on chromosome 1 (between marker D1S1619 and D1S1589). Other linkage signals for the blood pressure factor with a Pn <0.00074 were observed in three regions on chromosomes 10q, 13q and 16p. The strongest signal for the combined factor was detected at 82 cM on chromosome 17 (marker D17S1290). Overall, the linkage signals of the three individual factors did not cluster together, except that regions linked with the blood pressure factor and the combined factor modestly overlapped on chromosome 1q (Figure 1a). The linkage results that included diabetes (Figure 1b) or taking hypertension medication (data not shown) as a covariate were essentially similar to these models.
Figure 1.
Genome-wide multipoint linkage analyses for the three metabolic factors: nominal P values of Haseman-Elston regression without (a) and with (b) diabetes as a covariate.
Table 4.
Genome-wide linkage results for the three metabolic factors with a multipoint nominal P value <0.00074a
| All families (n of sib-pairs = 1,055) |
Families with history of diabetes (n of sib-pairs = 519) |
Families without history of diabetes (n of sib-pairs = 531) |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Cytogenic location | Position (cM)b |
Closest maker | Nominal P value |
Empirical P value |
Nominal P value |
Empirical P value |
Nominal P value |
Empirical P value |
P for heterogeneityc |
| Glycemia factor | |||||||||
| 22q12.3 | 30 | D22S689 | 0.0001a | 0.0002a | 0.0009 | 0.001 | 0.166 | – | 0.029 |
| Blood pressure factor | |||||||||
| 1q24.3 | 188–191 | D1S1619, D1S1589 | 0.0002a | 0.0007a | 0.007 | 0.009 | 0.008 | 0.022 | 0.907 |
| 7q32.3 | 137 | D7S1804 | 0.089 | – | 0.0004a | 0.0006a | 0.776 | – | 0.004 |
| 10q26.13-10q26.3 | 149–166 | D10S1656, D10S1248 | 0.0003a | 0.001 | 0.006 | 0.008 | 0.003 | 0.012 | 0.636 |
| 13q21.32 | 50 | D13S788 | 0.0004a | 0.001 | 0.0004a | 0.0007a | 0.012 | – | 0.753 |
| 13q32.2 | 78 | D13S793 | 0.001 | 0.003 | 0.129 | – | 0.00002a | 0.0002a | 0.003 |
| 16p12.3-16p12.1 | 32–56 | D16S3103, D16S540 | 0.0002a | 0.0006a | 0.009 | 0.010 | 0.001 | 0.004 | 0.555 |
| Combined factor | |||||||||
| 17q23.2 | 82 | D17S1290 | 0.0005a | 0.0007a | 0.147 | – | 0.0001a | 0.0001a | 0.048 |
See Table 2 for the constructs of the three factors.
“−” indicates the empirical P values were not obtained because of their nominal P values ≥0.01.
P <0.00074, the Lander and Kruglyak’s criteria for genome-wide suggestive linkage.
Map distances were based on the Marshfield map.
By Z test for difference between the two Haseman-Elston regression coefficients from the two subsets of families.
A total of 189 persons in these analyses had diabetes at the time of the baseline examination, and 25 (13.2%) of them were newly diagnosed. We stratified families by whether they had FHD. There were 139 families (n of sib-pairs = 519) with FHD, and 338 families (n of sib-pairs = 531) without FHD; nine families with a member whose diabetes status was missing were excluded from the subset analyses. The mean age (63.5 ± 10.8 years) of members in the families with FHD was not significantly different from that (62.9 ± 10.9 years) in the families without FHD (P = 0.277).
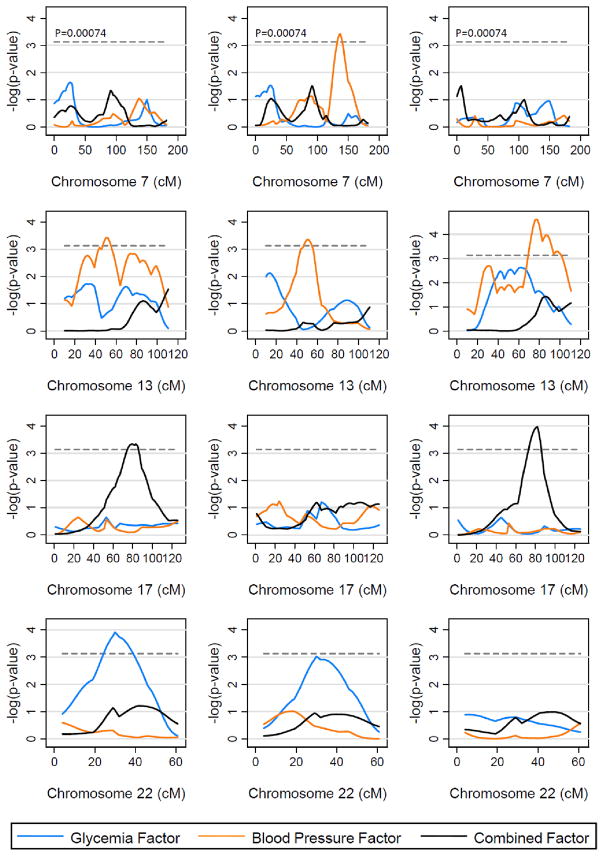
As shown in Table 4 and Figure 2, the families with and without FHD gave different linkage results for the three factors on at least 4 chromosomes. For several regions (ex., chromosome 22q for the glycemia factor, 13q for the blood pressure factor, and 17q for the combined factor), the stratification of families by FHD indicated which subset of families were contributing to the original linkage signal. Moreover, additional linkage peaks were identified after stratification by FHD. For the blood pressure factor, in the families with FHD, a region with genome-wide suggestive evidence of linkage was mapped to 137 cM on chromosome 7 (marker D7S1804, Pn = 0.0004; Pe = 0.0006), which was not detected when all families were analyzed as a whole. Interestingly, chromosome 13 showed a relatively complex pattern of linkage peaks in all families over an extensive distance from 50 cM to 78 cM, which may be actually composed of two peaks derived respectively from the two subsets of families. We observed genome-wide suggestive evidence of linkage for the blood pressure factor to one region at 50 cM on chromosome 13 in the families with FHD (near maker D13S788, Pn = 0.0004; Pe = 0.0007), whereas another region with a stronger evidence of linkage at 78 cM was identified in the families without FHD (near maker D13S793, Pn = 0.00002; Pe = 0.00002). Four of these regions showed significant genetic heterogeneity (P <0.05) between the families with FHD and without FHD (Table 4). Linkage peaks that occurred on or very close to telomeres are reported in Supplementary Table S2. It has been suggested that intervals without flanking markers (as at telomeres) cause an inflation of Type 1 error and thus should be interpreted with caution (32).
Figure 2.
Nominal P values of linkage analyses for the three metabolic factors on four chromosomes in all families (n of sib-pairs = 1,055; the left column), the subset of families with history of diabetes (n of sib-pairs = 519; the middle column), and the subset of families without history of diabetes (n of sib-pairs = 531, the right column).
To determine whether the evidence of linkage to chromosome 22q for the glycemia factor might reflect linkage to diabetes, we performed additional linkage analysis on diabetes as a discrete phenotype using the SIBPAL program (26). In the 486 families, there were 1,072 sib-pairs with information on both diabetes status and genotypes. The results of the multipoint linkage scan revealed one region with a Pn value <0.00074 (Supplementary Figure S1); it was on chromosome 22q12 (near maker D22S689, Pn = 0.0005; Pe = 0.004). It is noteworthy that this peak was coincident with the peak for glycemia factor in the families with FHD.
DISCUSSION
We used a factor analysis approach with a genome-wide scan to map loci of multiple metabolic factors. Because we only had restricted set of metabolic syndrome traits, we were not aimed to identify genetic loci for metabolic syndrome itself. However, our findings can still be compared to those from studies using an established definition of metabolic syndrome.
One of the strengths of the current study was that the study families were recruited from the general population in Beaver Dam and were not ascertained for any clinical characteristics related to diabetes or metabolic syndrome. Thus, it provided us a unique opportunity to investigate the possibility of genetic heterogeneity. The results did provide evidence for genetic heterogeneity since the two subsets of families defined by FHD were linked to several distinct loci. It has been shown that 45% of first-degree relatives of persons with diabetes were insulin resistant compared with only 20% of individuals without FHD (33). Consistently, individuals with parental history of diabetes or FHD had a 2- to 5-fold higher risk of occurrence of metabolic syndrome (28–30), compared to those without FHD. We assume that genes with likely pleiotropic effects are shared between diabetes and metabolic traits, and yet we are aware that diabetes is likely to be an important effect modifier. Therefore, by accounting for the genetic variance due to family propensity to diabetes, we would have more power to identify loci for the factors underlying metabolic traits. Our study also highlighted the importance of stratification by diabetes status in genetic studies on metabolic traits. Further studies are needed to test whether more precise modeling of family propensity to diabetes would improve the ability to localize causative genes underlying the metabolic factors.
Most linkage scans on metabolic traits have been performed in families which were ascertained for specific clinical characteristics, such as diabetes, obesity, hypertension, or cardiovascular diseases. Using this method of family ascertainment could select pedigrees with a stronger genetic component to some of the metabolic-related traits and thus would improve the ability to localize causative genes. However, the results are limited in generalization. Families which are representative of the general population are of great interest for quantitative traits, and linkage studies on such families should provide less biased results. Interestingly, using such families, the Insulin Resistance Atherosclerosis Study Family Study located a locus for metabolic syndrome to chromosome 1q23-q31 (LOD = 2.6) (34), which is the same region showing evidence of linkage with the blood pressure in the current study.
Factor analysis results are contingent on the number and nature of the traits included and the procedures used in the analysis. Previous studies that have used the principal-component factor analysis approach found 2 to 4 distinct factors representing the metabolic syndrome (8–12, 35). However, none of them, including the present study, has been able to replicate each others’ findings. Nevertheless, the chromosome 1q24.3 region found in our study appears to confirm previous linkage findings. This finding is interesting that 1q region, which is essentially identified as type 2 diabetes region (36), failed to show any evidence of linkage with glycemia factor, the factor that accounts for the largest amount of the variability, but showed correspondence with blood pressure factor. Subsequent linkage scans on metabolic syndrome and related quantitative traits, have also detected linkage in the same region but across a broader area of chromosome 1q (10, 34). Collectively, the present and previous studies strongly suggest chromosome 1q may harbor a gene which has pleiotropic effects, and more likely, several genes which have individual or additive effects on components of the metabolic syndrome. This is further supported by the findings that the lamin A/C gene, located on chromosome 1q21, was shown to be associated with the development of dyslipidemia and metabolic syndrome (37).
In the present study, the second highest peak for the blood pressure factor was identified on chromosome 16p12.3-16p12.1. This region contains SCNN1B gene, which is a causative gene for Liddle syndrome, a mendelian form of hypertension (38). Common genetic variants in SCNN1B were shown to affect blood pressure in the general population (39).
We included serum uric acid as a metabolic trait in our analysis. We and others have previously shown that serum uric acid is predictive of incident diabetes and cardiovascular disease (6, 40), and is also an independent determinant of metabolic syndrome (41). In the current study, we found serum uric acid loaded with BMI and HDL cholesterol, which supports the use of serum uric acid as a metabolic syndrome-related trait. The phenotypic correlations among BMI, HDL cholesterol and serum uric acid in the current study (0.23–0.32 in absolute value, Supplementary Table S1) were actually very close to those estimated from the NHLBI Family Heart Study (0.25–0.36 in absolute value), in which significant genetic correlations were also found between serum uric acid and BMI or HDL cholesterol (7).
We note limitations to the current study. First, the major limitation of the present study is the phenotype itself. Our data on metabolic traits were limited due to the study being designed initially as a study of age-related eye disease. However, although the initial focus of the study was eye diseases, the study families were not ascertained based on any eye disease, but rather were a representative sample from the general population, so this should not bias the results of our study. Second, the serum glucose level measured in this study was not fasting. The influence of a non-fasting glucose in the analysis is that addition of variance in serum glucose level due to daily dietary intake would make environmental factors stronger, and thus decrease our study’s power to detect genetic factors. However, this would render results presented here conservative, without an increase in Type I error. Additionally, the use of composite scores of HbA1c and serum glucose from factor analysis, rather than individual traits, would help to reduce the effect of measurement errors. Lastly, we defined FHD as any family member with diabetes. Some of the families are quite extended, with the possibility that in a family with FHD, a portion of the family members are not either first or second degree relatives to individuals with diabetes. This may contribute to increased genetic heterogeneity which would be expected to decrease power but not increase Type I error.
In summary, with the use of the families ascertained from a homogeneous population in Beaver Dam, we found substantial evidence of genetic heterogeneity by family propensity to diabetes for multiple metabolic factors. The multiple distinct loci found across the genome also suggest complex pathogenesis of the metabolic factors. In addition, our results reconfirm the findings of previous studies which mapped components of metabolic syndrome to the chromosome 1q region.
Supplementary Material
Supplementary Figure S1. Nominal P values of linkage analyses of diabetes as a discrete phenotype in all families (no. of sib-pairs = 1,072) showing coincident linkage with the glycemia factor in the subset of families with history of diabetes (no. of sib-pairs = 519) on chromosome 22.
Supplementary Table S1. Pearson’s correlation coefficients of the seven metabolic traits
Supplementary Table S2. Genome-wide linkage results for the three metabolic factors with a multipoint nominal P value <0.00074
Acknowledgments
This study was supported by National Institutes of Health Grants EY06594 (to R.K. and B.E.K. K.) and EY015286 (to B.E.K.K.), Research to Prevent Blindness Senior Investigator Awards (to R.K. and B.E.K.K.), and in part by the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health. Some of the results of this paper were obtained by using the program package S.A.G.E., which is supported by a U.S. Public Health Service Resource Grant (RR03655) from the National Center for Research Resources. We thank W.H. Linda Kao, Ph.D. (Department of Epidemiology, Johns Hopkins Bloomberg School of Public Health, Baltimore, MD) for her invaluable suggestions.
Footnotes
DISCLOSURE
The authors have no conflict of interest to declare.
References
- 1.Ford ES, Giles WH, Dietz WH. Prevalence of the metabolic syndrome among US adults: findings from the third National Health and Nutrition Examination Survey. JAMA. 2002;287:356–359. doi: 10.1001/jama.287.3.356. [DOI] [PubMed] [Google Scholar]
- 2.Hanson RL, Imperatore G, Bennett PH, Knowler WC. Components of the “metabolic syndrome” and incidence of type 2 diabetes. Diabetes. 2002;51:3120–3127. doi: 10.2337/diabetes.51.10.3120. [DOI] [PubMed] [Google Scholar]
- 3.Kekalainen P, Sarlund H, Pyorala K, Laakso M. Hyperinsulinemia cluster predicts the development of type 2 diabetes independently of family history of diabetes. Diabetes Care. 1999;22:86–92. doi: 10.2337/diacare.22.1.86. [DOI] [PubMed] [Google Scholar]
- 4.Lakka HM, Laaksonen DE, Lakka TA, et al. The metabolic syndrome and total and cardiovascular disease mortality in middle-aged men. JAMA. 2002;288:2709–2716. doi: 10.1001/jama.288.21.2709. [DOI] [PubMed] [Google Scholar]
- 5.Pyorala M, Miettinen H, Halonen P, Laakso M, Pyorala K. Insulin resistance syndrome predicts the risk of coronary heart disease and stroke in healthy middle-aged men: the 22-year follow-up results of the Helsinki Policemen Study. Arterioscler Thromb Vasc Biol. 2000;20:538–544. doi: 10.1161/01.atv.20.2.538. [DOI] [PubMed] [Google Scholar]
- 6.Klein BE, Klein R, Lee KE. Components of the metabolic syndrome and risk of cardiovascular disease and diabetes in beaver dam. Diabetes Care. 2002;25:1790–1794. doi: 10.2337/diacare.25.10.1790. [DOI] [PubMed] [Google Scholar]
- 7.Tang W, Hong Y, Province MA, et al. Familial clustering for features of the metabolic syndrome: the National Heart, Lung, and Blood Institute (NHLBI) Family Heart Study. Diabetes Care. 2006;29:631–636. doi: 10.2337/diacare.29.03.06.dc05-0679. [DOI] [PubMed] [Google Scholar]
- 8.Tang W, Miller MB, Rich SS, et al. Linkage analysis of a composite factor for the multiple metabolic syndrome: the National Heart, Lung, and Blood Institute Family Heart Study. Diabetes. 2003;52:2840–2847. doi: 10.2337/diabetes.52.11.2840. [DOI] [PubMed] [Google Scholar]
- 9.Edwards KL, Hutter CM, Yin WJ, Kim H, Monks SA. Genome-wide Linkage Scan for the Metabolic Syndrome: The GENNID Study. Obesity (Silver Spring) 2008;16:1596–1601. doi: 10.1038/oby.2008.236. [DOI] [PubMed] [Google Scholar]
- 10.Loos RJ, Katzmarzyk PT, Rao DC, et al. Genome-wide linkage scan for the metabolic syndrome in the HERITAGE Family Study. J Clin Endocrinol Metab. 2003;88:5935–5943. doi: 10.1210/jc.2003-030553. [DOI] [PubMed] [Google Scholar]
- 11.Bosse Y, Despres JP, Chagnon YC, et al. Quantitative trait locus on 15q for a metabolic syndrome variable derived from factor analysis. Obesity (Silver Spring) 2007;15:544–550. doi: 10.1038/oby.2007.577. [DOI] [PubMed] [Google Scholar]
- 12.Kraja AT, Hunt SC, Pankow JS, et al. Quantitative trait loci for metabolic syndrome in the Hypertension Genetic Epidemiology Network study. Obes Res. 2005;13:1885–1890. doi: 10.1038/oby.2005.231. [DOI] [PubMed] [Google Scholar]
- 13.Klein R, Klein BE, Linton KL, De Mets DL. The Beaver Dam Eye Study: visual acuity. Ophthalmology. 1991;98:1310–1315. doi: 10.1016/s0161-6420(91)32137-7. [DOI] [PubMed] [Google Scholar]
- 14.Klein AP, Duggal P, Lee KE, Klein R, Bailey-Wilson JE, Klein BE. Confirmation of linkage to ocular refraction on chromosome 22q and identification of a novel linkage region on 1q. Arch Ophthalmol. 2007;125:80–85. doi: 10.1001/archopht.125.1.80. [DOI] [PubMed] [Google Scholar]
- 15.Duggal P, Klein AP, Lee KE, Klein R, Klein BE, Bailey-Wilson JE. Identification of novel genetic loci for intraocular pressure: a genomewide scan of the Beaver Dam Eye Study. Arch Ophthalmol. 2007;125:74–79. doi: 10.1001/archopht.125.1.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.The hypertension detection and follow-up program: Hypertension detection and follow-up program cooperative group. Prev Med. 1976;5:207–215. doi: 10.1016/0091-7435(76)90039-6. [DOI] [PubMed] [Google Scholar]
- 17.Allain CC, Poon LS, Chan CS, Richmond W, Fu PC. Enzymatic determination of total serum cholesterol. Clin Chem. 1974;20:470–475. [PubMed] [Google Scholar]
- 18.Lopes-Virella MF, Stone P, Ellis S, Colwell JA. Cholesterol determination in high-density lipoproteins separated by three different methods. Clin Chem. 1977;23:882–884. [PubMed] [Google Scholar]
- 19.Klenk DC, Hermanson GT, Krohn RI, et al. Determination of glycosylated hemoglobin by affinity chromatography: comparison with colorimetric and ion-exchange methods, and effects of common interferences. Clin Chem. 1982;28:2088–2094. [PubMed] [Google Scholar]
- 20.Fossati P, Prencipe L, Berti G. Use of 3,5-dichloro-2-hydroxybenzenesulfonic acid/4-aminophenazone chromogenic system in direct enzymic assay of uric acid in serum and urine. Clin Chem. 1980;26:227–231. [PubMed] [Google Scholar]
- 21.Klein R, Klein BE, Moss SE, Linton KL The Beaver Dam Eye Study. Retinopathy in adults with newly discovered and previously diagnosed diabetes mellitus. Ophthalmology. 1992;99:58–62. doi: 10.1016/s0161-6420(92)32011-1. [DOI] [PubMed] [Google Scholar]
- 22.Manly BFJ. Factor analysis. In: Manly BFJ, editor. Multivariate statistical methods. London: Chapman & Hall; 1994. pp. 93–106. [Google Scholar]
- 23.Broman KW, Murray JC, Sheffield VC, White RL, Weber JL. Comprehensive human genetic maps: individual and sex-specific variation in recombination. Am J Hum Genet. 1998;63:861–869. doi: 10.1086/302011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Broman KW, Weber JL. Estimation of pairwise relationships in the presence of genotyping errors. Am J Hum Genet. 1998;63:1563–1564. doi: 10.1086/302112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.George VT, Elston RC. Generalized modulus power transformations. Commun Statist -Theory Meth. 1988;17:2933–2952. [Google Scholar]
- 26.Elston RC, Buxbaum S, Jacobs KB, Olson JM. Haseman and Elston revisited. Genet Epidemiol. 2000;19:1–17. doi: 10.1002/1098-2272(200007)19:1<1::AID-GEPI1>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 27.Shete S, Jacobs KB, Elston RC. Adding further power to the Haseman and Elston method for detecting linkage in larger sibships: weighting sums and differences. Hum Hered. 2003;55:79–85. doi: 10.1159/000072312. [DOI] [PubMed] [Google Scholar]
- 28.Hunt KJ, Heiss G, Sholinsky PD, Province MA. Familial history of metabolic disorders and the multiple metabolic syndrome: the NHLBI family heart study. Genet Epidemiol. 2000;19:395–409. doi: 10.1002/1098-2272(200012)19:4<395::AID-GEPI10>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 29.Liese AD, Mayer-Davis EJ, Tyroler HA, et al. Familial components of the multiple metabolic syndrome: the ARIC study. Diabetologia. 1997;40:963–970. doi: 10.1007/s001250050775. [DOI] [PubMed] [Google Scholar]
- 30.Rodriguez-Moran M, Guerrero-Romero F. The parental phenotype of diabetes, but not of essential hypertension, is linked to the development of metabolic syndrome in Mexican individuals. Acta Diabetol. 2001;38:87–91. doi: 10.1007/s005920170019. [DOI] [PubMed] [Google Scholar]
- 31.Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- 32.Risch N, Giuffra L. Model misspecification and multipoint linkage analysis. Hum Hered. 1992;42:77–92. doi: 10.1159/000154047. [DOI] [PubMed] [Google Scholar]
- 33.Groop L, Forsblom C, Lehtovirta M, et al. Metabolic consequences of a family history of NIDDM (the Botnia study): evidence for sex-specific parental effects. Diabetes. 1996;45:1585–1593. doi: 10.2337/diab.45.11.1585. [DOI] [PubMed] [Google Scholar]
- 34.Langefeld CD, Wagenknecht LE, Rotter JI, et al. Linkage of the metabolic syndrome to 1q23-q31 in Hispanic families: the Insulin Resistance Atherosclerosis Study Family Study. Diabetes. 2004;53:1170–1174. doi: 10.2337/diabetes.53.4.1170. [DOI] [PubMed] [Google Scholar]
- 35.Edwards KL, Burchfiel CM, Sharp DS, et al. Factors of the insulin resistance syndrome in nondiabetic and diabetic elderly Japanese-American men. Am J Epidemiol. 1998;147:441–7. doi: 10.1093/oxfordjournals.aje.a009469. [DOI] [PubMed] [Google Scholar]
- 36.McCarthy MI. Growing evidence for diabetes susceptibility genes from genome scan data. Curr Diab Rep. 2003;3:159–167. doi: 10.1007/s11892-003-0040-y. [DOI] [PubMed] [Google Scholar]
- 37.Steinle NI, Kazlauskaite R, Imumorin IG, et al. Variation in the lamin A/C gene: associations with metabolic syndrome. Arterioscler Thromb Vasc Biol. 2004;24:1708–1713. doi: 10.1161/01.ATV.0000136384.53705.c9. [DOI] [PubMed] [Google Scholar]
- 38.Shimkets RA, Warnock DG, Bositis CM, et al. Liddle’s syndrome: heritable human hypertension caused by mutations in the beta subunit of the epithelial sodium channel. Cell. 1994;79:407–414. doi: 10.1016/0092-8674(94)90250-x. [DOI] [PubMed] [Google Scholar]
- 39.Tobin MD, Tomaszewski M, Braund PS, et al. Common variants in genes underlying monogenic hypertension and hypotension and blood pressure in the general population. Hypertension. 2008;51:1658–1664. doi: 10.1161/HYPERTENSIONAHA.108.112664. [DOI] [PubMed] [Google Scholar]
- 40.Boyko EJ, de Courten M, Zimmet PZ, Chitson P, Tuomilehto J, Alberti KG. Features of the metabolic syndrome predict higher risk of diabetes and impaired glucose tolerance: a prospective study in Mauritius. Diabetes Care. 2000;23:1242–1248. doi: 10.2337/diacare.23.9.1242. [DOI] [PubMed] [Google Scholar]
- 41.Onat A, Uyarel H, Hergenc G, et al. Serum uric acid is a determinant of metabolic syndrome in a population-based study. Am J Hypertens. 2006;19:1055–1062. doi: 10.1016/j.amjhyper.2006.02.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1. Nominal P values of linkage analyses of diabetes as a discrete phenotype in all families (no. of sib-pairs = 1,072) showing coincident linkage with the glycemia factor in the subset of families with history of diabetes (no. of sib-pairs = 519) on chromosome 22.
Supplementary Table S1. Pearson’s correlation coefficients of the seven metabolic traits
Supplementary Table S2. Genome-wide linkage results for the three metabolic factors with a multipoint nominal P value <0.00074