Abstract
Background
The rs3803662 SNP in the TOX3/LOC643714 region was identified as a breast cancer susceptibility genetic variant in recent genome-wide association studies (GWAS) of women of European ancestry and has been replicated in other populations of European ancestry. The position of the causal variant tagged by the rs3803662 marker is still unknown. In fact, because the rs3803662 polymorphism is located between the TOX3 and the LOC643714 loci, it is unclear which gene is the one causally related to the risk of breast cancer. Because LD blocks are smaller in populations of African ancestry, fine-mapping in African ancestry samples may be an effective approach to narrowing the position of the causal variant(s) in the TOX3/LOC643714 locus.
Methods
We evaluated a total of 60 tagging SNPs throughout the TOX3/LOC643714 region in a nested case-control study of breast cancer within the Black Women’s Health Study which included 906 cases and 1,111 controls.
Results
No significant association was found for the rs3803662 SNP. However, four other SNPs (rs3104746, rs3112562, rs3104793, and rs8046994), all of them located in the LOC643714 gene, were associated with risk of breast cancer. The strongest association was observed for rs3104746: each copy of the A-rs3104746 allele was associated with a 23% higher risk of breast cancer, OR (95% CI) = 1.23 (1.05–1.44), P = 0.009.
Conclusions
Our results confirm the association observed in GWAS of European ancestry populations.
Impact
The results narrow the locus to a smaller LD block in the LOC643714 gene.
Keywords: Breast cancer, TOX3/LOC643714, fine-mapping, African-Americans
INTRODUCTION
The TOX/LOC643714 locus on chromosome 16 was one of the first breast cancer regions to be identified through genome-wide association study (GWAS) in populations of European and East Asian origin (1). Out of several SNPs associated with the risk of breast cancer, the rs3803662 (a C-to-T transition) was the most strongly correlated with disease; each copy of the T-allele of the rs3803662 SNP was associated with a 20% increase in the risk of breast cancer. Subsequent studies, also in European ancestry populations, showed that the risk conferred by the rs3803662 polymorphism was either restricted to or more strongly associated with estrogen receptor (ER) positive tumors compared to ER-negative cancers (2, 3). Because most of the replication studies have been carried out in populations of European ancestry (2–4), it is unclear whether the same SNP is associated with risk of breast cancer in populations of African origin. In a subgroup of African American women (422 cases and 447 controls) from the Multiethnic Cohort (MEC) study, the T-rs3803662 allele was associated with a lower risk of breast cancer, the opposite direction to the results in the other ethnic groups (2). In addition, a recent analysis of African American women (810 cases and 1784 controls) from the Southern Community Cohort Study (SCCS) and the Nashville Breast Health Study (NBHS), found no significant association between rs3803662 and seven other SNPs in the TOX3 gene and risk of breast cancer (5).
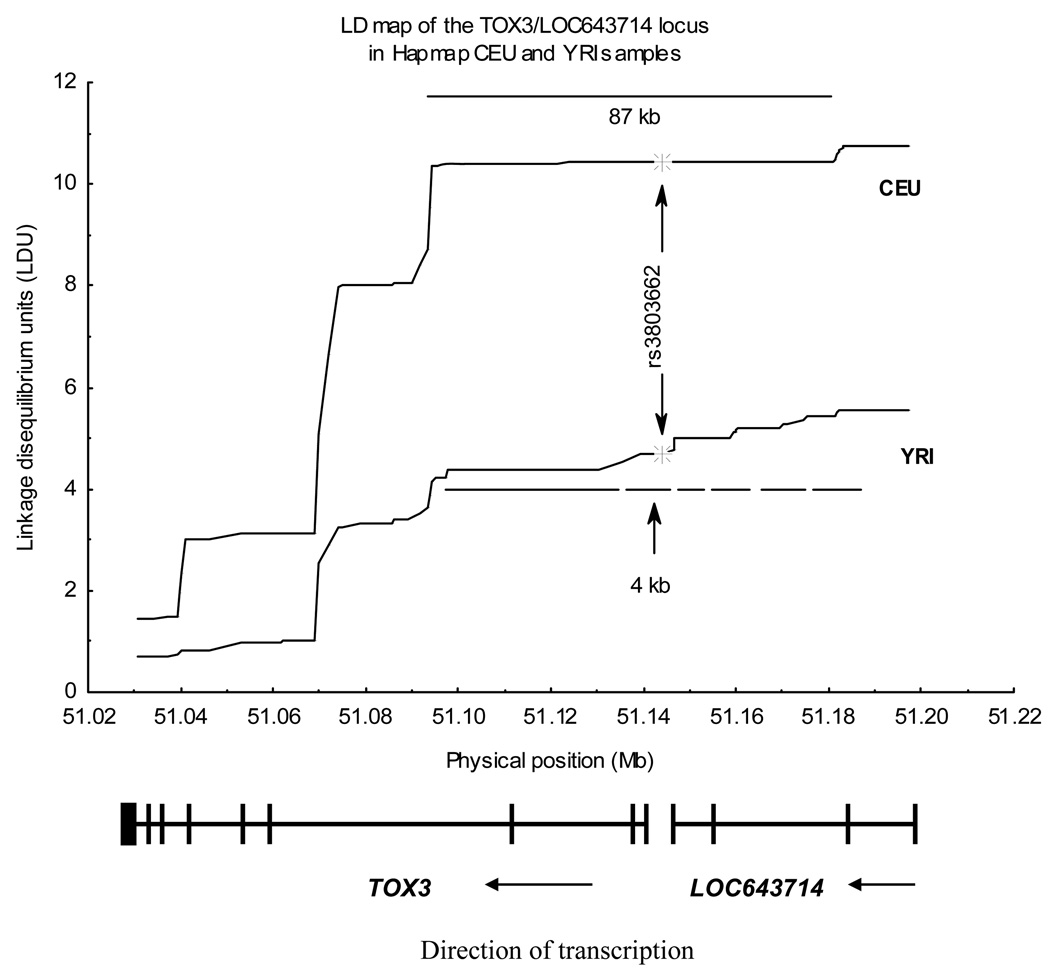
The reasons for the differences in results between African American women and women of European or Asian ancestry remain to be determined. The high allele frequency of the T-rs3803662 allele in African Americans (about 50%) as well as the considerable sample size of the SCCS/NBHS study (810 cases and 1784 controls) makes low statistical power an unlikely explanation. Difference in the linkage disequilibrium (LD) structure in the TOX3/LOC643714 region between populations of European and African origin is a more likely reason (Figure 1). In the HapMap CEU population, the rs3803662 SNP resides in an LD block that is more than 80 kb and covers part of the TOX3 and LOC643714 loci. In the HapMap YRI population, this big LD block is split into smaller blocks and includes some gaps of low LD. In particular, the rs3803662 SNP is located inside a small 4 kb LD block in HapMap Yoruba samples. It is therefore possible that the causal variant(s) tagged by rs3803662 SNP in populations of European ancestry will be tagged by different SNPs in populations of African ancestry. If we assume the same causal polymorphisms in Europeans and Africans, the smaller LD blocks observed in the latter allows a more accurate mapping of the causal variants.
Figure 1. Linkage disequilibrium (LD) structure of the TOX3/LOC643714 region in HapMap CEU and YRI populations.
LD structure was assessed using linkage disequilibrium units (LDU) estimated with the SNPBrowser software (25). LDU are proportional to the decay of LD along the physical distance. Regions of constant (‘flat’) LDU represent LD blocks. Note that the big block (from 51.09 Mb to 51.18 Mb) that covers parts of the TOX3 and LOC643714 genes in HapMap CEU population and it is split into smaller blocks and gaps in the HapMapYRI population.
We conducted a nested case-control study to narrow the position of the causal variant for breast cancer in the TOX3/LOC643714 region. To this end, we performed fine-scale mapping of the entire TOX3/LOC643714 locus including genotyping of the index rs3803662 SNP.
DESIGN AND METHODS
Study population
We conducted a nested-case control study within the ongoing Black Women’s Health Study (BWHS) that has been described elsewhere (6). Briefly, the study began in 1995 when women 21–69 years of age from across the United States completed a 14-page postal health questionnaire. The initial cohort comprises 59,000 women who self-identified as “black” and had a valid address. Follow-up questionnaires are sent every two years. Average follow-up of the baseline cohort through the completed 2-year cycles to date is greater than 80%.
We used medical records and cancer registry data to confirm self-reported cases of breast cancer, as well as to gain information on tumor characteristics such as estrogen and progesterone receptor status. We have obtained records or registry data for 1,151 breast cancer cases reported on the BWHS questionnaires, of which 99.4% were confirmed. Self-reported cases that were not confirmed have been excluded.
DNA samples were obtained from BWHS participants by the mouthwash-swish method (7), with all samples stored in freezers at −80°C. Approximately 50% of participants, 27,800 women, provided a sample. Women who provided samples were slightly older than women who did not, but the two groups were similar with regard to educational level, geographic region of residence, body mass index, and family history of breast cancer.
The present study includes all cases of breast cancer who provided a DNA sample and were diagnosed through the end of the 2007 follow-up cycle. Controls were selected from among BWHS participants with DNA samples who were free of breast cancer at the end of the 2007 follow-up period. Controls were matched to cases approximately 1:1 on year of birth (±1-year), and geographical region of residence (Northeast, South, Midwest, and West).
The study protocol was approved by the Institutional Review Board of Boston University.
Selection of tag SNPs and ancestral informative markers
We downloaded SNPs covering the entire TOX3/LOC643714 locus from the HapMap Yoruba (YRI) database (8). We used the Tagger (9) software implemented in Haploview (10) version 4.1 to select the set of common haplotype tagging SNPs with a minor allele frequency (MAF) ≥ 5% and r2 ≥ 0.8. The rs3803662 SNP was forced into the set. We selected 68 tagging SNPs along the TOX3/LOC643714 locus.
We also selected 30 ancestral informative markers (AIMs) to estimate and control for population stratification due to European admixture. The 30 AIMs were selected from a list of validated SNPs in which the top 30 AIMs had allele frequency differences between Africans and Europeans of at least 0.75 (11). We used a Bayesian approach as implemented in the Admixmap software (12, 13) to estimate individual admixture proportions. Eighty-one controls included in this breast cancer study had also been genotyped for a set of 1,536 AIMs used for admixture mapping analyses in a case-control study of systemic lupus erythematosus in the BWHS. The correlation (r = 0.87) between the two measures of percent European admixture was highly significant, p<0.0001, confirming the validity of our small set of AIMs. Because Admixmap requires the specification of the ancestral allele frequencies, we also used the Structure software version 2.2 (14, 15), which does not require ancestral allele frequencies, to identify hidden population stratification beyond the one due to European admixture.
Genotyping and quality control
DNA was isolated from mouthwash swish samples from breast cancer cases and controls at the Boston University Molecular Core Genetics Laboratory using the QIAAMP DNA Mini Kit (Qiagen). Whole genome amplification was performed with the Qiagen RePLI-g Kits using the method of multiple displacement amplification. Amplified samples underwent purification and PicoGreen quantification at the Broad Institute Center for Genotyping and Analysis (Cambridge, MA) before being plated for genotyping.
Genotyping was carried out at the Broad Institute Center for Genotyping and Analysis using the Sequenom MassArray iPLEX technology. Ninety-eight blinded duplicate samples were included to assess reproducibility of the genotypes. An average reproducibility of 99% was obtained among the blinded duplicates. All SNPs with calling rate < 90% or a deviation from Hardy-Weinberg equilibrium in the control sample at p < 0.001 were excluded. We also excluded samples with calling rates < 80%. We first genotyped 41 tagging SNPs chosen for the TOX3 gene among 769 cases and 833 controls. After the addition of breast cancer cases and controls identified after the first genotyping plates were made, we genotyped 27 tagging SNPs along the LOC643714 locus in 865 cases and 1073 controls. We successfully genotyped 35 of the TOX3 variants, 25 of the LOC643714 variants and 29 of the AIMs. Mean call rate in the final data set for both SNPs and samples was 98.4%.
Data analysis
We used PLINK (16) software version 1.06 to calculate summary statistics for the genotype data. We tested for association with breast cancer using the Cochran-Armitage trend test of an additive genetic model with 10,000 permutations to calculate empirical p-values. We used PROC LOGISTIC of the SAS statistical software version 9.1.3 (SAS Institute Inc., Cary, NC, USA) to estimate odds ratios (ORs) and 95% confidence intervals (95% CI) for the SNPs significant at the nominal p-value of 0.05. We adjusted the ORs for age, geographical region of residence (Northeast, South, Midwest, West), place of birth (US, foreign country), and European admixture proportion. We used a general genetic model with 2 degrees of freedom because it does not assume any particular inheritance risk pattern. Also, we estimated per-allele ORs if a linear trend was evident.
We used the conditional haplotype method (17, 18) to determine whether the identified significant SNPs represent independent signals or were tagging the same causal variant. This method stratifies by haplotypic background and tests the null hypothesis that one or more SNPs have no independent haplotypic effect once we condition for such background. We conditioned on the top SNP under the trend model to test for independent effects of the other significant SNPs.
Odds ratios (ORs) of haplotypes of significant SNPs were estimated using an expectation substitution approach (19, 20) that estimates the probabilities of all possible haplotype configurations of each individual in the sample, conditional on their genotype and case-control status. Haplotypes with an estimated frequency of < 5% were pooled in one single group and the most common haplotype was used as the reference haplotype.
RESULTS
Table 1 shows characteristics of breast cancer cases and controls. No significant differences were observed in the percentage of European admixture between the groups (19.2% in cases vs. 19.3% in controls). The Admixmap and Structure software gave very similar results, with a correlation of 98.9% between the two estimates of European admixture proportions (Supplementary Figure 1).
Table 1.
General characteristics of breast cancer cases and controls in the Black Women’s Health Study*
| Characteristic | Cases (N = 906) |
Controls (N = 1,111) |
|---|---|---|
| Mean age at baseline 1995 (SD) | 48 (10) | 48 (10) |
| Region of residence % | ||
| Northeast | 26.7 | 25.6 |
| South | 31.0 | 30.8 |
| Midwest | 24.6 | 25.6 |
| West | 17.6 | 17.9 |
| U.S. born % | 93.2 | 94.1 |
| First degree family history of breast cancer % | 14.1 | 8.5 |
| Mean European ancestry % (SD) | 19.2 (10.7) | 19.3 (10.3) |
Total number of subjects includes individuals genotyped for the TOX3 gene and individuals genotyped for the LOC643714 gene.
Because additional population stratification may be present beyond that due to European admixture, we estimated the likelihood of the observed AIM genotypes under different numbers of subpopulation groups. The observed data is best explained by 2 different subpopulations (African and European). However, our results also suggest the presence of a third or even a fourth subpopulation in the BWHS population (Supplementary Figure 2). To assess the impact of this additional population stratification in our odds ratio estimates, we conducted all logistic analyses under different scenarios of 2, 3, and 4 subpopulations. Because no major differences were observed among these three different scenarios, we present the results only under the scenario of 2 subpopulations.
The rs3803662 polymorphism was not significantly associated with risk of breast cancer overall (Table 2), or with particular subtypes of tumors defined by ER and PR status. Per allele ORs (95% CI) were 1.00 (0.81–1.25) for ER-positive tumors and 0.98 (0.76–1.26) for ER-negative breast cancer cases.
Table 2.
Odds ratios (ORs) and 95% confidence intervals (CIs) for the previously reported rs3803662 SNP and four newly identified significant SNPs in the LOC643714 gene
| SNP | High-risk allele frequency (%) |
Heterozygous OR (95% CI) |
Homozygous OR (95% CI) |
PerAllele OR (95% CI) |
P for trend | Dominant OR (95% CI) |
P for dominant | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Cases (N) |
Controls (N) |
Adj† | Perm‡ | Adj† | Perm‡ | |||||
| rs3803662_T* | 49.9 (753) |
50.9 (825) |
0.92 (0.72–1.18) | 0.92 (0.69–1.22) | 0.96 (0.83–1.10) | 0.56 | 1.0 | 0.92 (0.73–1.16) | 0.47 | 1.0 |
| rs3104746_A | 22.9 (857) |
19.6 (1,067) |
1.28 (1.05–1.55) | 1.38 (0.89–2.13) | 1.23 (1.05–1.44) | 0.009 | 0.20 | 1.29 (1.07–1.55) | 0.008 | 0.16 |
| rs3112562_G | 50.6 (849) |
47.0 (1,056) |
1.34 (1.07–1.67) | 1.36 (1.04–1.78) | 1.17 (1.02–1.34) | 0.020 | 0.30 | 1.34 (1.09–1.66) | 0.006 | 0.12 |
| rs3104793_C | 60.0 (861) |
57.0 (1,066) |
1.40 (1.08–1.81) | 1.39 (1.05–1.83) | 1.14 (1.00–1.30) | 0.050 | 0.23 | 1.39 (1.09–1.78) | 0.009 | 0.05 |
| rs8046994_T | 36.6 (846) |
33.3 (1,053) |
1.25 (1.03–1.52) | 1.25 (0.92–1.69) | 1.16 (1.01–1.33) | 0.035 | 0.16 | 1.25 (1.04–1.51) | 0.017 | 0.09 |
The T-allele of the rs3803662 was defined as the “high-risk” allele to be consistent with previous reports
Adjusted for age, geographical region of residence, place of birth, and European admixture
Permutation 100,000 times
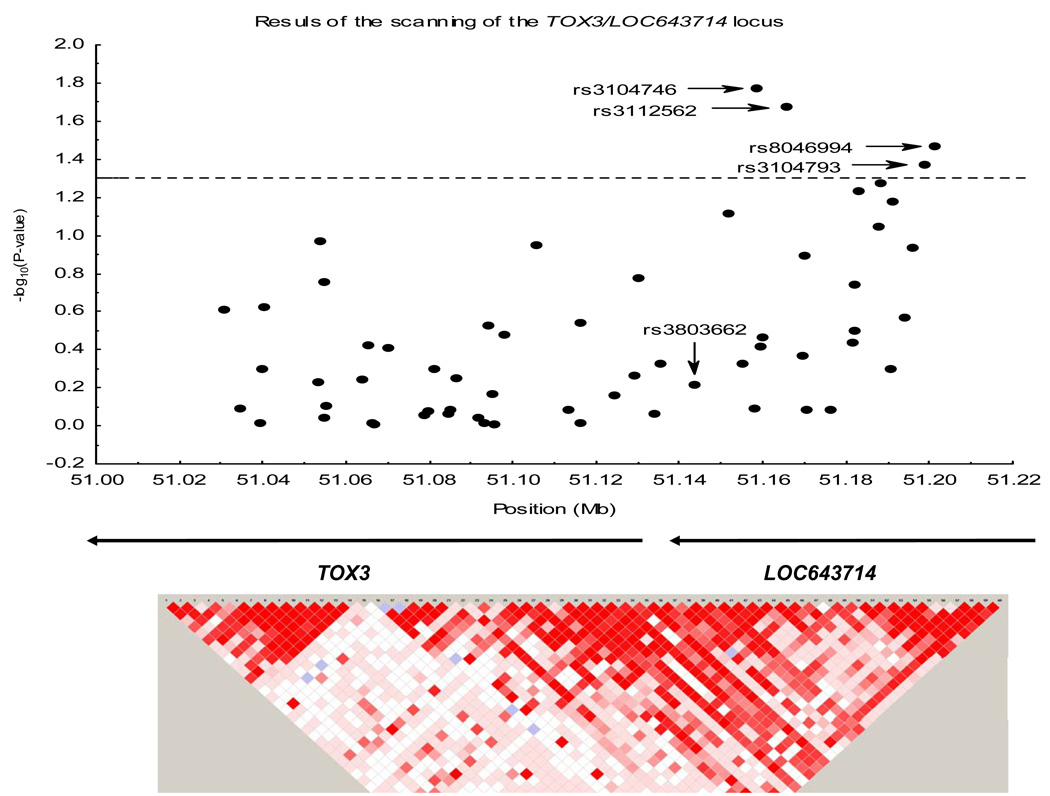
Of the SNPs scanned in the TOX3/LOC643714 region (Supplementary Table 1), four SNPs, all of them in the LOC643714 locus, were associated with breast cancer at the nominal alpha = 0.05 level of significance (Figure 2). Two of the SNPs (rs3104746 and rs3112562) were located inside intron 2 of LOC643714, and the other two SNPs (rs3104793 and rs8046994) were located in the 5’ region of LOC643714 (Supplementary Figure 3). The per allele odds ratios (95% CI) were 1.23 (1.05–1.44) for the A-allele of rs3104746, 1.17 (1.02–1.34) for the G-allele of rs3112562, 1.14 (1.00–1.30) for the C-allele of rs3104793, and 1.16 (1.01–1.33) for T-allele of rs8046994 (Table 2). The similarity of the ORs for heterozygous and homozygous subjects suggests that risk of disease follows a dominant model. For all four SNPs the risk of disease was stronger under the dominant model compared to the dose-response model, although comparison of the log-likelihoods of both genetic models did not allow us to discriminate between them (data not shown). We observed similar ORs for different subtypes of tumors defined by ER and PR status (data not shown).
Figure 2. Scatterplot and LD map of the genotyped tagging SNPs along the TOX3/LOC643714 region.
The four tagging SNPs associated with risk of breast cancer in the BWHS population are located in the LOC643714 locus. The position of the previously reported rs3803662 is also indicated. The dotted line indicates the threshold of significance for alpha = 0.05.
These four SNPs tended to be correlated with each other as measured by the D’ and r2 values, and correlation with the rs3803662 SNP was lower (Table 3), suggesting that the four SNPs may be tagging a single causal variant not tagged by the rs3803662 SNP in African Americans. The conditional haplotype method supports the notion of a single causal variant tagged by these four SNPs. After conditioning by the top SNP, rs3104746 SNP, no other SNP was significantly associated with disease: rs3112562 (P = 0.19), rs3104793 (P = 0.51), and rs8046994 (P = 0.50). Haplotypic odds ratios show haplotypes carrying the A-allele of the rs3104746 SNP tended to be associated with risk of breast cancer (Table 4). In particular, the rs3104746-A/rs3112562-G/rs3104793-C/rs8046994-T haplotype was more frequent in cases compared to controls (15.8% vs. 13.0%) and was associated with a 36% increase in the risk of breast cancer, OR (95% CI) = 1.36 (1.10–1.67). We note that the frequency of the A-allele of the rs3104746 SNP in HapMap CEU samples is 4.2%, compared to 25.4% in HapMap Yoruba samples and 19.6% in the BWHS control population, and has an r2 = 0.10 with rs3803662; therefore, the rs3104746 polymorphism may not be a good tagger of the causal variant in populations of European ancestry.
Table 3.
D’ and r2 values in the BWHS among the previously reported 3803662 SNP, and the four newly identified rs3104746, rs3112562, rs3104793, and rs8046994 SNPs in the LOC643714 gene*
| SNPs | rs3803662 | rs3104746 | rs3112562 | rs3104793 | rs8046994 |
|---|---|---|---|---|---|
| rs3803662 | 0.547 | 0.488 | 0.324 | 0.312 | |
| rs3104746 | 0.085 | 0.997 | 0.858 | 0.511 | |
| rs3112562 | 0.229 | 0.284 | 0.891 | 0.452 | |
| rs3104793 | 0.075 | 0.141 | 0.532 | 0.998 | |
| rs8046994 | 0.053 | 0.131 | 0.116 | 0.381 |
D’ values are above the diagonal; r2 values are below the diagonal
Table 4.
Odds ratios (ORs) and 95% confidence intervals (CIs) of haplotypes of the four newly identified significant SNPs in the LOC643714 gene
| rs3104746 | rs3112562 | rs3104793 | rs8046994 | Haplotype* frequency (%) | OR (95% CI) | P-value† | |
|---|---|---|---|---|---|---|---|
| Cases (N = 821) |
Controls (N = 1,026) |
||||||
| T | C | T | C | 37.8 | 40.9 | 1.00 (reference) | 0.05 |
| T | C | C | T | 9.7 | 9.9 | 1.07 (0.84–1.37) | 0.80 |
| T | G | C | C | 15.3 | 15.8 | 1.09 (0.89–1.33) | 0.68 |
| T | G | C | T | 11.0 | 10.5 | 1.18 (0.93–1.50) | 0.58 |
| A | G | C | C | 5.9 | 5.5 | 1.21 (0.88–1.67) | 0.54 |
| A | G | C | T | 15.8 | 13.0 | 1.36 (1.10–1.67) | 0.01 |
Only haplotypes with frequency ≥ 5% are shown
P-value for difference in haplotype frequencies between cases and controls
The rs3104746 and rs3112562 SNPs are in the same 87 kb LD block of the HapMap CEU sample that contains the rs3803662 SNP. The rs3104793 and rs8046994 SNPs are located in an adjacent 24 kb LD block in the same HapMap CEU sample. We used permutation analysis to evaluate the significance of our results adjusting for multiple comparisons within each LD block (27 SNPs in the large block and 10 SNPs in the small block). Both the trend and dominant models were assessed, with 100,000 permutations within each LD block for each of the genetic models. For the trend model, permutated P-values were 0.20 for rs3104746, 0.30 for rs3112562, 0.23 for rs3104792, and 0.16 for rs8046994. For the dominant model, permutated P-values were 0.16 for rs3104746, 0.12 for rs3112562, 0.05 for rs3104793, and 0.09 for rs8046994.
DISCUSSION
In the present study we confirm the previous finding from GWAS of European ancestry populations of an association of breast cancer risk with a locus in the TOX3/LOC643714 region. The SNP associated with breast cancer in previous reports tagged a wide LD region. Our results point to a narrower region of association, located entirely within the LOC643714 gene.
The rs3803662 SNP, associated with breast cancer in European and Asian ancestry populations, was not associated with risk of breast cancer in the BWHS population. Further, no evidence of association was found with any of the subtypes of breast cancer defined by ER and PR status. In African American women from the Multiethnic Cohort study, the T-allele was associated with lower risk of breast cancer, an association opposite in direction to the results from other ethnic groups (2). However, in a recent study by Zheng et al. (5) in African American women of the Southern Community Cohort Study and the Nashville Breast Health Study, there was no significant association between the rs3803662 SNP and risk of breast cancer, in concordance with our present results. It is noteworthy that the study by Zheng et al. (5) also included 7 SNPs in the TOX3 gene that are in high LD (r2 ≥ 0.8) with rs3803662 in Europeans, and none of these polymorphisms was associated with risk of breast cancer. However, such a small number of SNPs is not enough to cover the genetic variation across the TOX3/LOC643714 region in African Americans. These two studies suggest that the causal variant in the TOX3/LOC643714 locus is not tagged by the rs3803662 SNP in African Americans. Our results suggest that causal variant(s) are not located in the TOX3 gene but rather in the LOC643714 locus. Consistent with our results, an ongoing fine-mapping excluded the coding region of the TOX3 gene using 2,270 breast cancer cases and 2,280 controls in an European population from the United Kingdom and narrowed the associated region from the 5’ end of the TOX3 gene through the 3’ end of the LOC643714 locus (21). At the time of this writing, no function is known for the LOC643714 gene. According to Entrez Nucleotide and based on computational analysis the LOC643714 locus codes for a small mRNA of 1028 base-pairs that would be translated into a hypothetical protein of 55 amino acids (22).
Although we used ancestral informative markers to estimate European admixture proportions, residual confounding due to population stratification might still be present. While we cannot completely rule out the presence of residual confounding, we think its effects, if any, on the present results are negligible. First, the selected AIMs provided us with estimates of European ancestry highly correlated with estimates using 1,536 AIMs distributed throughout the genome. Also, the present estimates of European admixture are similar to those reported from other African American populations (11, 23, 24). Control for European admixture removed the major source of confounding due to population stratification. Although more subtle population stratification may still exist, the extra adjustment for a third or even a fourth subpopulation as identified by the Structure software did not materially change our odds ratio estimates. We also note that we adjusted for geographical region of residence and birthplace, therefore reducing potential confounding due to population stratification.
In summary, our present results provide evidence that the TOX3/LOC643714 locus may contribute to the genetic susceptibility of breast cancer in African ancestry populations. The newly identified genetic variants located in the LOC643714 gene may be tagging the same causal variant. These findings help to narrow the localization of the causal variant(s) in the TOX3/LOC643714 region.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Black Women’s Health Study participants for their continuing participation in this research effort. We also thank Dr. Clint Baldwin of Boston University for advice on genotyping and interpretation of genotype data.
FUNDING
This work was supported by grants R01CA058420 and R01CA098663 from the National Cancer Institute, Division of Cancer Control and Population Science (http://www.cancercontrol.cancer.gov). The Broad Institute Center for Genotyping and Analysis is supported by grant U54 RR020278 from the National Center for Research Resources (http://www.broadinstitute.org/sections/science/projects/broad/ncrr-center-genotyping-analysis).
REFERENCES
- 1.Easton DF, Pooley KA, Dunning AM, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stacey SN, Manolescu A, Sulem P, et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2007;39:865–869. doi: 10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- 3.Garcia-Closas M, Hall P, Nevanlinna H, et al. Heterogeneity of breast cancer associations with five susceptibility loci by clinical and pathological characteristics. PLoS Genet. 2008;4:e1000054. doi: 10.1371/journal.pgen.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Antoniou AC, Spurdle AB, Sinilnikova OM, et al. Common breast cancer-predisposition alleles are associated with breast cancer risk in BRCA1 and BRCA2 mutation carriers. Am J Hum Genet. 2008;82:937–948. doi: 10.1016/j.ajhg.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng W, Cai Q, Signorello LB, et al. Evaluation of 11 Breast Cancer Susceptibility Loci in African-American Women. Cancer Epidemiol Biomarkers Prev. 2009 doi: 10.1158/1055-9965.EPI-09-0624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosenberg L, Adams-Campbell L, Palmer JR. The Black Women's Health Study: a follow-up study for causes and preventions of illness. J Am Med Womens Assoc. 1995;50:56–58. [PubMed] [Google Scholar]
- 7.Cozier YC, Palmer JR, Rosenberg L. Comparison of methods for collection of DNA samples by mail in the Black Women's Health Study. Ann Epidemiol. 2004;14:117–122. doi: 10.1016/S1047-2797(03)00132-7. [DOI] [PubMed] [Google Scholar]
- 8.The International HapMap Consortium. The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 9.de Bakker PI, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 10.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 11.Smith MW, Patterson N, Lautenberger JA, et al. A high-density admixture map for disease gene discovery in african americans. Am J Hum Genet. 2004;74:1001–1013. doi: 10.1086/420856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McKeigue PM, Carpenter JR, Parra EJ, Shriver MD. Estimation of admixture and detection of linkage in admixed populations by a Bayesian approach: application to African-American populations. Ann Hum Genet. 2000;64:171–186. doi: 10.1017/S0003480000008022. [DOI] [PubMed] [Google Scholar]
- 13.Hoggart CJ, Parra EJ, Shriver MD, et al. Control of confounding of genetic associations in stratified populations. Am J Hum Genet. 2003;72:1492–1504. doi: 10.1086/375613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valdes AM, Thomson G. Detecting disease-predisposing variants: the haplotype method. Am J Hum Genet. 1997;60:703–716. [PMC free article] [PubMed] [Google Scholar]
- 18.Valdes AM, McWeeney S, Thomson G. HLA class II DR-DQ amino acids and insulin-dependent diabetes mellitus: application of the haplotype method. Am J Hum Genet. 1997;60:717–728. [PMC free article] [PubMed] [Google Scholar]
- 19.Zaykin DV, Westfall PH, Young SS, Karnoub MA, Wagner MJ, Ehm MG. Testing association of statistically inferred haplotypes with discrete and continuous traits in samples of unrelated individuals. Hum Hered. 2002;53:79–91. doi: 10.1159/000057986. [DOI] [PubMed] [Google Scholar]
- 20.Stram DO, Leigh Pearce C, Bretsky P, et al. Modeling and E-M estimation of haplotype-specific relative risks from genotype data for a case-control study of unrelated individuals. Hum Hered. 2003;55:179–190. doi: 10.1159/000073202. [DOI] [PubMed] [Google Scholar]
- 21.Ahmed S, Maranian M, Gregory CS, et al. From association to cause: fine mapping of the TNRC9 gene region, a novel susceptibility locus identified in the first genome-wide association study for breast cancer. Breast Cancer Research. 2008;10:S26. [Google Scholar]
- 22.Entrez Nucleotide. National Library of Medicine (US) National Center for Biotechnology Information. http://www.ncbi.nlm.nih.gov/sites/entrez.
- 23.Halder I, Yang BZ, Kranzler HR, Stein MB, Shriver MD, Gelernter J. Measurement of admixture proportions and description of admixture structure in different U.S. populations. Hum Mutat. 2009;30:1299–1309. doi: 10.1002/humu.21045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reiner AP, Ziv E, Lind DL, et al. Population structure, admixture, and aging-related phenotypes in African American adults: the Cardiovascular Health Study. Am J Hum Genet. 2005;76:463–477. doi: 10.1086/428654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De La Vega FM, Isaac HI, Scafe CR. A tool for selecting SNPs for association studies based on observed linkage disequilibrium patterns. Pac Symp Biocomput. 2006:487–498. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.