Abstract
Ribosome biogenesis is facilitated by a growing list of assembly cofactors, including helicases, GTPases, chaperones, and other proteins, but the specific functions of many of these assembly cofactors are still unclear. The effect of three assembly cofactors on 30S ribosome assembly was determined in vitro using a previously developed mass spectrometry-based method that monitors the rRNA binding kinetics of ribosomal proteins. The essential GTPase Era caused several late-binding proteins to bind rRNA faster when included in a 30S reconstitution. RimM enabled faster binding of S9 and S19, and inhibited the binding of S12 and S13, perhaps by blocking those proteins' binding sites. RimP caused proteins S5 and S12 to bind dramatically faster. These quantitative kinetic data provide important clues about the roles of these assembly cofactors in the mechanism of 30S biogenesis.
Communication Body
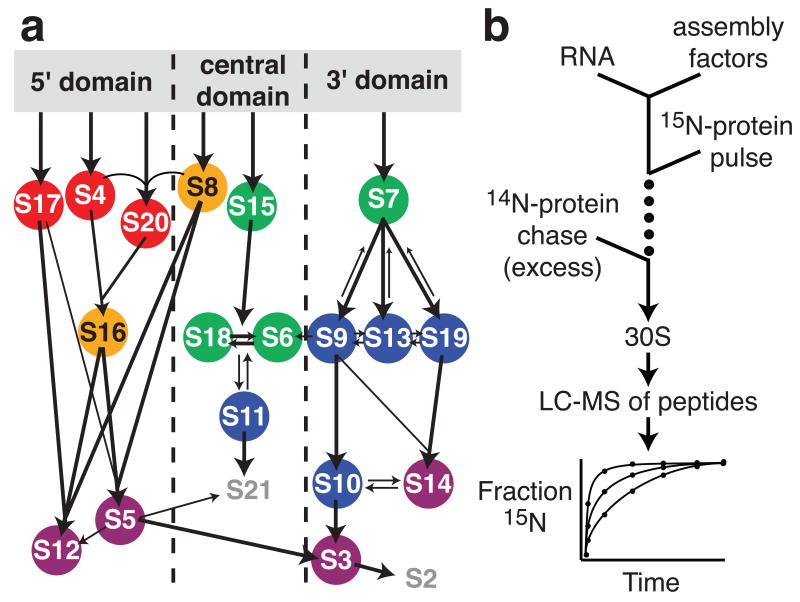
A significant fraction of the mass of rapidly growing bacteria consists of ribosomes,1 which are essential to all living cells. In eukaryotes, ribosome biogenesis is assisted by hundreds of protein cofactors,2,3 whereas the cytoplasmic assembly of ribosomes is facilitated by only approximately twenty cofactors in bacteria.4 In vitro reconstitution studies of E. coli 30S5 and 50S6 ribosomal subunits have provided a framework for understanding the mechanism of ribosome assembly and have demonstrated that protein binding is hierarchical, with some ribosomal proteins (r-proteins) binding directly to the rRNA and others requiring prior binding of one or more proteins.7 For the small subunit, or 30S subunit, these protein-binding dependencies are described by the Nomura equilibrium assembly map (Fig 1a) that was constructed from the results of reconstitution experiments including various combinations of proteins.7 The 30S ribosomal subunit is composed of three discrete structural units8 that can be reconstituted independently in vitro from their RNA and r-protein components: the 5′ domain, the central domain, and the 3′ domain.9-11 Kinetic data shows that 5′ domain r-proteins bind fastest, followed by central domain r-proteins, and 3′ domain r-proteins bind slowest (Fig. 1a).12-14 While the role of many r-proteins in assembly have been investigated in more detail since the assembly map was published,9,15 there are fewer data describing the function of assembly cofactors in vitro.
Figure 1.
A) Nomura assembly map. Arrows represent protein-binding dependencies at equilibrium.7; 42 Colored circles represent protein binding rates at 40°C as determined by pulse-chase.14 Red, 10-14 min-1; orange, 5.4-6 min-1; green, 2.2-3.6 min-1; blue, 0.39-0.84 min-1; purple, 0.14-0.24 min-1. B) Schematic of pulse-chase experiments with factors. Stable-isotope pulse-chase experiments were performed as described previously with minor changes.14 PC/QMS experiments were performed by mixing 16S rRNA with assembly factor before adding a pulse of 15N TP30 and incubating for varying amounts of time, then chasing the assembly reaction with an excess of 14N TP30. A 2× excess of assembly factor (360 pmol) was preincubated with 180 pmol of 16S rRNA for 20 minutes at 40°C in a volume of 555 μl of reconstitution buffer (RB, 25 mM Tris pH 7.5, 20 mM MgCl2, 330 mM KCl, 2 mM DTT). For the experiment with Era, the KCl concentration was 250 mM during the pre-incubation, and then 30 sec before the pulse, 60 μl of 100 μM GTP in a higher KCl buffer was added, bringing the KCl concentration to 330 mM and the GTP concentration to 10 μM. The pulse consisted of 45 μl of 6 μM 15N TP30 in RB, and ranged in length from 10 sec to 40 min. After addition of a chase of 135 μl of 10 μM 14N TP30 in RB, the assembly reaction was incubated a further 40 min at 40°C and then chilled on ice. Sample processing and LC-MS analysis were performed as described previously.14,43 Briefly, the 30S subunits were loaded onto sucrose gradients and purified by ultracentrifugation. The r-proteins were extracted, digested with trypsin, and analysed by LC-MS using an Agilent ESI-TOF. In-house peptide identification software was used which takes advantage of the difference between 14N and 15N peptide peaks for identification purposes.14 Quantitation was performed using an isotope fitting approach called least squares Fourier transform convolution (LS-FTC).43
In the cell, ribosome assembly is very efficient, taking less than 3 minutes during exponential growth.16 The r-proteins bind to the nascent ribosomal RNA transcript as their binding sites become available.17,18 Efficient assembly is facilitated by assembly cofactors including GTPases, helicases, protein-folding chaperones, RNA modification enzymes such as methyltransferases and pseudouridinylases, and other proteins.4,19,20 Many of these factors have been characterized using genetic studies, but little in vitro data regarding their functions is available. One exception is DnaK, the protein-folding chaperone, which has been shown to increase the yield of low-temperature 30S reconstitutions,21-23 although the specific mechanism is unknown.
A number of GTPases have been implicated in ribosome assembly.24 Era (E. coli Ras-like protein) is essential in E. coli and is one of only two GTPases which have been implicated in small subunit assembly.4,25 Cells depleted in Era show inhibition of translation,25 incomplete 16S rRNA processing, and accumulation of 30S and 50S subunits with fewer 70S ribosomes and polysomes.26 Overexpression of era can suppress the cold sensitive phenotype of a mutant of rbfA, a 30S assembly co-factor.26 Era is a 34-kDa two-domain protein with a GTP-binding domain at the N-terminus and a KH RNA-binding domain at the C-terminus,27 and binds the 30S ribosomal subunit between the head and platform, preventing 50S association28 and likely preventing translation initiation.29 The GTPase activity of Era is stimulated by binding to an RNA fragment identical to the 3′ terminus of the 16S rRNA, which is positioned between the head and the platform in the structure.29 This binding suggests that Era may be involved as a chaperone in the rRNA processing performed by RNases during biogenesis.29 Overexpression of the methyltransferase ksgA, which may act as a checkpoint for assembly completion,30 can suppress the slow growth phenotype of cells depleted in Era.31 This suggests that KsgA and Era may function at similar points in assembly, although Era has been implicated in several cellular processes,32 and the phenotype suppression observed may be related to some other function rather than ribosome assembly.33
RimM (ribosome maturation factor M, also called 21K) was first implicated in 30S biogenesis when it was observed that a rimM deletion strain had reduced translational efficiency.34 Deletion strains of rimM also demonstrate slow growth,34 incomplete 16S rRNA processing,35 and accumulation of 30S and 50S subunits with fewer polysomes.36 RimM is a PRC β-barrel protein which binds the 30S ribosomal subunit35,36 and also the r-protein S19.37 Suppressor mutations to rimM mutations also support a function for RimM in the assembly of the 3′ domain.36 Overexpression of assembly co-factor rbfA can suppress the slow growth phenotype of a rimM deletion strain,35 suggesting the two proteins act at a similar point in assembly. The inverse is not true since overexpression of rimM cannot suppress the slow growth phenotype of an rbfA deletion strain.35 In a rimM deletion strain, most RbfA molecules are associated with 30S subunits,36 indicating that RbfA can bind to pre-30S subunits without RimM and suggesting that RimM may be required for the release of RbfA from 30S particles. These data suggest that RbfA acts before RimM in the 30S assembly pathway.
RimP (formerly YhbC or P15a) is a 15-kDa protein that has recently been shown to be important in 30S ribosomal subunit maturation.38 A rimP deletion strain demonstrates slower growth rates, fewer polysomes, an accumulation of free 50S subunits, and an accumulation of unprocessed pre-16S rRNA. In addition, RimP was found to be associated with 30S subunits in cell extracts.38 An ortholog of RimP, SP14.3 from Streptococcus pneumonia, is a two-domain protein, with both domains resembling known RNA-binding proteins.39 Although RimP was found to interact with S5 and S7 in an interaction network study,40 its mechanism of action on maturing 30S subunits is largely unknown.
In order to further understand the role of these cofactors in ribosome biogenesis, we examined their effect on the kinetics of 30S ribosomal subunit reconstitution in vitro, using the previously developed pulse-chase monitored by quantitative mass spectrometry (PC/QMS).13,14 This method monitors the protein binding rates of all r-proteins using a pulse of 15N labeled r-proteins followed by a chase of excess 14N protein (Fig. 1b).14 Here, stoichiometric amounts of the assembly cofactors Era, RimM, and RimP were preincubated with the 16S rRNA to allow their association, and protein-binding rates in the presence of the factors were monitored using the PC/QMS method. To our knowledge, these results are the first to demonstrate effects of assembly cofactors on the rates of incorporation of specific proteins during ribosome assembly.
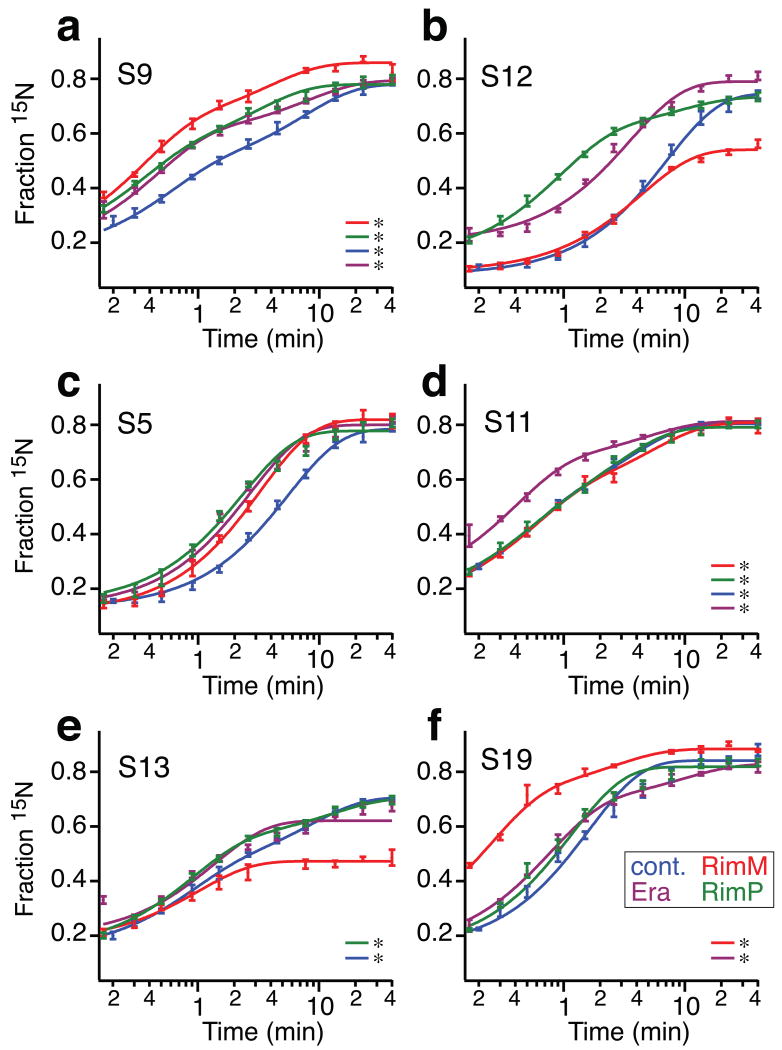
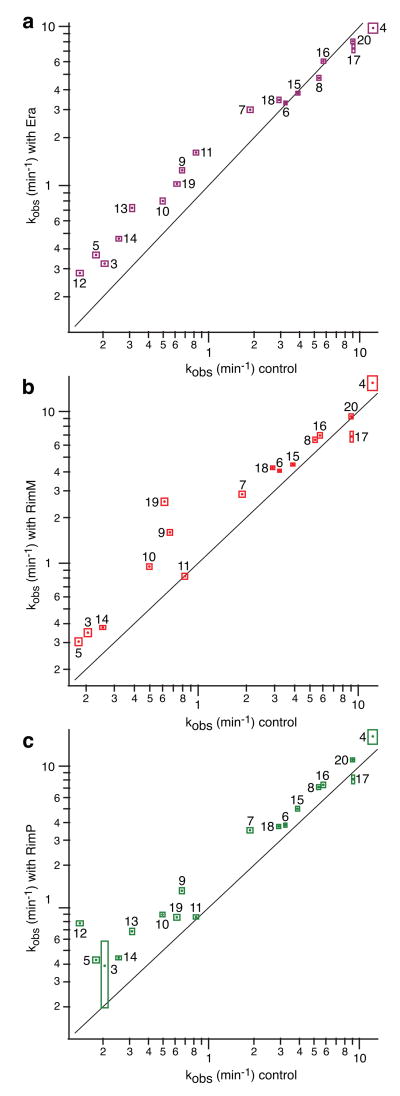
The effects of Era on r-protein binding kinetics were measured by preincubation of Era with the 16S rRNA for 20 min before the start of a PC/QMS kinetic experiment. The nucleotide cofactor GTP was added to the reaction 30 sec before beginning the pulse-chase. Under these conditions, the binding rates of proteins S5, S9, S11, and S12 were accelerated ∼2-fold (Figs. 2a-d). Subtler rate increases were observed for 3′ domain proteins S7, S10, S13, S14, and S19. Overall, the slow-binding proteins bound faster, while the fast-binding proteins were unchanged (Fig. 3a). For several proteins including S9 and S12, the rate increase was more evident in the early time points due to an increase in the rate or the amplitude of the fast kinetic phase, indicating that some pre-30S particles are better substrates for Era than others. The kinetic changes observed for S13 are very subtle, although the calculated rates suggest that S13 binds significantly faster in the presence of Era. The discrepancy between the kinetic curves and the calculated monophasic rate for S13 is due to imperfect fitting of biphasic kinetics with single exponential curves, and was only observed for S13. The fact that Era has several modest effects but no dramatic specific effects suggests that Era does not facilitate the binding of specific proteins, but rather, may facilitate a more global maturation event such as an RNA conformational change. Also, since Era has significant effects on many proteins over a wide range of binding rates, it is possible that Era remains bound to pre-30S subunits throughout several RNA folding and protein binding events.
Figure 2.
The kinetic effects of Era, RimM, and RimP on in vitro reconstitutions. a) – f) Protein binding progress curves of selected proteins showing an experiment with RimM (red), RimP (green), Era (purple) and a control14 (blue). Curves marked with asterisks are fit to a double exponential based on the results of a statistical f-test, using a 95% confidence interval. Era, RimM and RimP were prepared as follows. Plasmids pSTN022, pMW487 and pSTN021 encoding N-terminally his-tagged Era, RimM and RimP, respectively, expressed from a T7/lac promoter were constructed by PCR amplification of DNA fragments containing the respective structural gene using the following primer pairs: era-pETM10-F (5′ CATGCCATGGGCATCGATAAAAGTTACTGCGG 3′) and era-pETM10-R (5′-CATGCTCGAGTTAAAGATCGTCAACGTAACCGAG-3′), rimM-NcoI-F (5′-GGTCACCATGGGCAAACAACTCACCGCGCAA-3′ and rimM-pETM10-R (5′-CATGCTCGAGTTTAAAAACCAGGATCCCAATCTAC-3′), yhbC-pETM10-F (5′--CATGCCATGGGCTTGTCCACATTAGAGC-3′) and yhbC-pETM10-R (5′-CATGCTCGAGTTAAAAGTGGGGAACCAGGTTCG-3′), and cloning of the DNA fragments, after trimming of their ends with NcoI and XhoI, into NcoI and XhoI digested expression vector pETM10 (http://www.pepcore.embl.de/strains_vectors/vectors/bacterial_expression.html). Plasmid clones verified by DNA sequencing were introduced into strain BL21 (DE3)44 for overproduction of the respective proteins. The resulting strains were grown in LB45 at 37°C to a cell density of approx. 50 Klett units, at which 0.5 mM IPTG was added, to induce expression from the T7/lac promoter on the plasmids, and the temperature shifted to ∼21°C for further incubation overnight. Cells from 1 L of culture were harvested by centrifugation and dissolved in 5 ml of 20 mM Tris-HCl, pH 8.0, containing 0.5 M NaCl, 10% glycerol and 5mM imidazole, then 55 μl of 100 mM phenylmethanesulphonylfluoride (PMSF) and 250 μl of 10 mg/ml lyzosyme were added before freeze/thawing three times in liquid nitrogen/water, followed by 80 min centrifugation (20,800 × g) at 4°C. The obtained supernatants were passed through Ni-NTA columns (Qiagen) by gravity flow, the columns were washed with 5 ml aliquots of 20 mM Tris-HCl, pH 8.0, containing 0.5 M NaCl, 10% glycerol and increasing concentrations of imidazole (5, 20, 50, 100 and 500 mM, respectively). Wash fractions containing the respective proteins were pooled and further purified by FPLC using a HiLoad 16/60 Superdex 200 prep grade column (GE Healthcare Life Sciences) equilibrated in 20 mM Tris-HCl, pH 8.0, containing 0.5 M NaCl and 10% glycerol. Prior to kinetic experiments, the proteins were dialysed into TKMD (25 mM Tris pH 7.5, 20 mM MgCl2, 1 M KCl, 2 mM DTT) for several hours or overnight. Native 30S subunits, 16S rRNA, and total r-proteins (TP30) were prepared from E. coli MRE600 cells as previously described13,14 except that cells were lysed using a Bead Beater (BioSpec).
Figure 3.
First-order calculated rates of the control experiment14 compared with those from an experiment with a) Era, b) RimM, and c) RimP. Bounding boxes show fit errors. Black line is y = x. For S9, the rate increase in the presence of Era occurred mainly in the faster of two kinetic phases, indicating that some pre-30S particles are better substrates for Era than others. The kinetic changes observed for S13 with Era and RimP are very subtle, although the calculated rates suggest that S13 binds significantly faster in the presence of Era.
The effects of RimM on r-protein binding kinetics were also measured using PC/QMS with 16S rRNA that was preincubated with RimM for 20 min. Most slow-binding proteins, including 3′ domain proteins S10 and S3, were modestly (1.5-fold) accelerated, but 3′ domain proteins S9 and S19 showed a larger, 2-4 fold acceleration (Figs 2a and f). The stimulatory effects on these 3′ domain proteins are consistent with literature suggesting that RimM binds to the 3′ domain.36 Surprisingly, proteins S12 and S13 displayed a significant decrease in binding extent (Figs. 2b and e). Given that S13 and S19 are close to each other in the 30S subunit structure, and RimM binds S19, it is possible that the lowered binding extent of S13 is due to a physical blocking of the binding site while RimM is bound to S19 on the pre-30S particle. The lowered binding extent of S12 is more surprising, since evidence suggests RimM binds to the 3′ domain, and S12 binds to the 5′ domain. Interestingly, S12 and S13 are the only two proteins that bind on the 50S interaction face of the 30S subunit, and their reduced binding here may suggest that RimM binds on the intersubunit interface. It is also possible that the slight excess of RimM included in the experiment enabled non-specific association that prevented stable association of S12 and S13. However, it is clear that any non-specific association that occurred did not produce further non-native effects since RimM had no effect on many of the fastest-binding proteins (Fig. 3b). Given the known efficiency of ribosome biogenesis in vivo,16 it seems unlikely that RimM stalls assembly by preventing the association of ribosomal proteins. It is possible that in vivo, the release of RimM from the ribosome is facilitated by another ribosome maturation factor. The in vitro experiments described here included only one factor at a time, and so the release of RimM may have been delayed, preventing S13 from binding.
Finally, the effects of RimP on r-protein binding kinetics were measured in a reconstitution reaction starting with 16S rRNA that was preincubated with RimP for 20 min prior to the pulse-chase experiment. The binding rates of most slower-binding proteins were increased, particularly 5′ domain proteins S5 and S12, which bound 2-fold and 6-fold faster, respectively (Figs. 2b and c). The effect of RimP on S12 was the largest kinetic effect observed in this study. Almost all of the 3′ domain proteins bound faster, with S9 showing a 2-fold acceleration (Fig. 2a), and S3, S7, S10, S13, and S14 displaying subtler rate increases. The binding of S12 showed an additional kinetic phase not present in the control experiment, suggesting that some portion of the pre-30S molecules were competent to bind S12 faster than others. Many proteins exhibited no kinetic changes (Figs. 2d and 3c), indicating that these effects are specific. Since RimP accelerates mainly the slowest binding proteins S5 and S12, it is likely that RimP is involved in the last steps of protein binding in vivo. The role of RimP in ribosome biogenesis is only beginning to be characterized, and these data constitute a significant contribution towards the understanding of RimP's function.
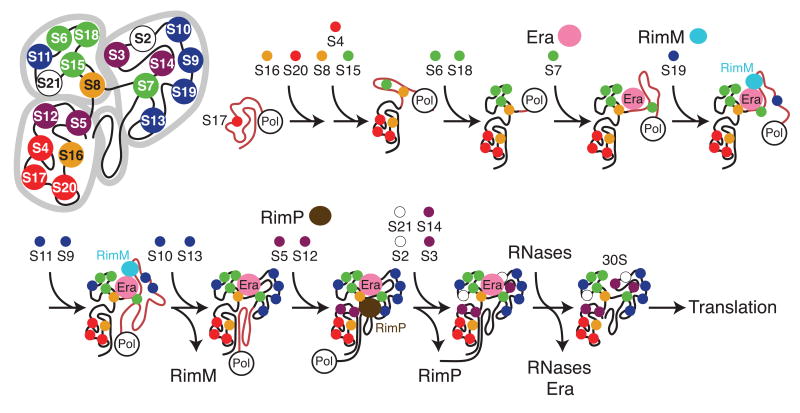
It is noteworthy that in each of these experiments, only a subset of r-proteins showed kinetic changes, and it is likely that the observed effects are specific and relevant to the function of Era, RimM, and RimP. A preliminary experiment to measure the effects of the cofactors in combination indicated that their effects were additive and not synergistic (data not shown). These data, in combination with current knowledge, suggest a potential cotranscriptional model for 30S biogenesis. Figure 4 shows a cartoon model of co-transcriptional ribosome biogenesis with RimM, RimP, and Era facilitating assembly. The order of protein addition shown is based on the in vitro binding rates of r-proteins,13,14 the in vivo order of addition,41 and the binding sites of r-proteins.15 In the model, Era remains bound to the pre-30S particle throughout several stages of assembly due to its subtle global effect on protein binding. This model also reflects structural data29 suggesting that Era may remain bound to the pre-30S particle until rRNA processing is complete. The r-protein binding kinetics presented here neither support nor contradict the hypothesis that Era chaperones the processing of rRNA termini, because mature rRNA was used in these experiments, and a model where Era dissociates after or during the last steps of r-protein binding is consistent with this data. RimM facilitates the assembly of the 3′ domain in the model, and then dissociates prior to the binding of S12 and S13, which is consistent with the hypothesis that RimM does not prevent the association of those proteins in vivo. In the model, RimP associates with the pre-30S particle during the late stages of protein binding in order to facilitate the binding of several late binding proteins in the 5′ and 3′ domains. RimP may dissociate after one of the last steps of protein binding, or it may remain bound to the pre-30S particle until the final stages of rRNA processing.
Figure 4.
Proposed cotranscriptional model of in vivo 30S ribosome assembly and the point at which factors RimM, RimP, and Era are required. The cartoon 30S is made up of 3 major domains: the 5′ domain (lower left), the central domain (upper left) and the 3′ major domain (upper right). The 3′ minor domain is in the center. Proteins are colored as in Fig. 1. Proteins bind to the nascent, unfolded rRNA (pink) as soon as their binding sites become available. Era is involved throughout several steps of assembly, while RimM facilitates 3′ domain assembly, RimP acts during the later stages of protein binding, and RNAses mature the rRNA termini.
These quantitative kinetic data demonstrate that PC/QMS is a powerful tool for investigating the role of assembly cofactors in vitro. In these experiments, each individual assembly factor caused significant and specific kinetic changes in ribosome assembly. The effects on specific proteins observed in these in vitro experiments provide important clues to the mechanism of these proteins. Consistent with what is known about the role of these cofactors in ribosome biogenesis, these results suggest that Era remains bound to the 30S subunit throughout several stages of assembly, while RimM acts during 3′ domain assembly, and RimP acts during the late stages of r-protein binding. These are the first results to demonstrate specific effects of assembly cofactors on r-protein binding, and these data represent a significant first step towards proposing a complete mechanistic model for 30S ribosome biogenesis.
Supplementary Material
Acknowledgments
This work was supported by a grant from the NIH (R37-GM-53757) to J.R.W, and by the Skaggs Institute for Chemical Biology. P.M.W. was supported by The Carl Trygger Foundation. The authors thank Prof. Gary Siuzdak and Dr. Sunia Trauger of the Scripps Center for Mass Spectrometry for advice and access to instrumentation. Gunter Stier is gratefully acknowledged for providing plasmid pETM10, Dr. Michael T. Sykes for mass spectrometry data analysis programs, and Dr. Zahra Shajani for critical comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bremer H, Dennis PP. Modulation of Chemical Composition and Other Parameters of the Cell by Growth Rate. In: Neidhardt FC, editor. Escherichia Coli and Salmonella: Cellular and Molecular Biology. 2nd. American Society for Microbiology; Washington, DC: 1996. pp. 1553–1569. [Google Scholar]
- 2.Dez C, Tollervey D. Ribosome synthesis meets the cell cycle. Curr Opin Microbiol. 2004;7:631–7. doi: 10.1016/j.mib.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 3.Henras AK, Soudet J, Gerus M, Lebaron S, Caizergues-Ferrer M, Mougin A, Henry Y. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol Life Sci. 2008;65:2334–59. doi: 10.1007/s00018-008-8027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilson DN, Nierhaus KH. The weird and wonderful world of bacterial ribosome regulation. Crit Rev Biochem Mol Biol. 2007;42:187–219. doi: 10.1080/10409230701360843. [DOI] [PubMed] [Google Scholar]
- 5.Traub P, Nomura M. Structure and Function of E. coli Ribosomes, V. Reconstitution of Functionally Active 30S Ribosomal Particles from RNA and Proteins. Proc Natl Acad Sci USA. 1968;59:777–784. doi: 10.1073/pnas.59.3.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nierhaus KH, Dohme F. Total reconstitution of functionally active 50S ribosomal subunits from Escherichia coli. Proc Natl Acad Sci U S A. 1974;71:4713–7. doi: 10.1073/pnas.71.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Held W, Ballou B, Mizushima S, Nomura M. Assembly Mapping of 30S Ribosomal Proteins from Escherichia Coli: Further Studies. J Biol Chem. 1974;249:3103–3111. [PubMed] [Google Scholar]
- 8.Schuwirth B, Borovinskaya M, Hau C, Zhang W, Vila-Sanjurjo A, Holton J, Cate J. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–34. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 9.Recht M, Williamson J. RNA tertiary structure and cooperative assembly of a large ribonucleoprotein complex. J Mol Biol. 2004;344:395–407. doi: 10.1016/j.jmb.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 10.Samaha R, O'Brien B, O'Brien T, Noller H. Independent in vitro assembly of a ribonucleoprotein particle containing the 3′ domain of 16S rRNA. Proc Natl Acad Sci U S A. 1994;91:7884–8. doi: 10.1073/pnas.91.17.7884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weitzmann C, Cunningham P, Nurse K, Ofengand J. Chemical evidence for domain assembly of the Escherichia coli 30S ribosome. FASEB J. 1993;7:177–80. doi: 10.1096/fasebj.7.1.7916699. [DOI] [PubMed] [Google Scholar]
- 12.Powers T, Daubresse G, Noller H. Dynamics of in vitro assembly of 16 S rRNA into 30 S ribosomal subunits. J Mol Biol. 1993;232:362–74. doi: 10.1006/jmbi.1993.1396. [DOI] [PubMed] [Google Scholar]
- 13.Talkington M, Siuzdak G, Williamson J. An assembly landscape for the 30S ribosomal subunit. Nature. 2005;438:628–32. doi: 10.1038/nature04261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bunner AE, Trauger SA, Siuzdak G, Williamson JR. Quantitative ESI-TOF Analysis of Macromolecular Assembly Kinetics. Anal Chem. 2008;80:9379–9386. doi: 10.1021/ac8020505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stern S, Powers T, Changchien LM, Noller HF. RNA-protein interactions in 30S ribosomal subunits: folding and function of 16S rRNA. Science. 1989;244:783–90. doi: 10.1126/science.2658053. [DOI] [PubMed] [Google Scholar]
- 16.Champney WS. Kinetics of ribosome synthesis during a nutritional shift-up in Escherischia coli K-12. Mol Gen Genet. 1977;152:259–66. doi: 10.1007/BF00693079. [DOI] [PubMed] [Google Scholar]
- 17.Kaczanowkska M, Rydén-Aulin M. Ribosome Biogenesis and the Translation Process in. Escherichia coli Microbiol Mol Biol R. 2007;71:477–494. doi: 10.1128/MMBR.00013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Narvaez CC, Schaup HW. In vivo transcriptionally coupled assembly of Escherichia coli ribosomal subunits. J Mol Biol. 1979;134:1–22. doi: 10.1016/0022-2836(79)90411-x. [DOI] [PubMed] [Google Scholar]
- 19.Connolly K, Culver G. Deconstructing ribosome construction. Trends Biochem Sci. 2009;34:256–63. doi: 10.1016/j.tibs.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ofengand J, Rudd KE. Bacterial, Archeal, and Organellar rRNA Pseudouridines and Methylated Nucleosides and Their Enzymes. In: Garret RA, Douthwaite SR, Liljas A, Matheson AT, Moore PB, Noller HF, editors. The Ribosome: Structure, Function, Antibiotics, and Cellular Interactions. ASM Press; Washington, D.C: 2000. pp. 175–189. [Google Scholar]
- 21.Maki JA, Schnobrich DJ, Culver GM. The DnaK chaperone system facilitates 30S ribosomal subunit assembly. Mol Cell. 2002;10:129–38. doi: 10.1016/s1097-2765(02)00562-2. [DOI] [PubMed] [Google Scholar]
- 22.Maki JA, Southworth DR, Culver GM. Demonstration of the role of the DnaK chaperone system in assembly of 30S ribosomal subunits using a purified in vitro system. RNA. 2003;9:1418–21. doi: 10.1261/rna.5139703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alix JH, Nierhaus KH. DnaK-facilitated ribosome assembly in Escherichia coli revisited. RNA. 2003;9:787–93. doi: 10.1261/rna.5360203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown ED. Conserved P-loop GTPases of unknown function in bacteria: an emerging and vital ensemble in bacterial physiology. Biochem Cell Biol. 2005;83:738–46. doi: 10.1139/o05-162. [DOI] [PubMed] [Google Scholar]
- 25.Sayed A, Matsuyama S, Inouye M. Era, an essential Escherichia coli small G-protein, binds to the 30S ribosomal subunit. Biochem Biophys Res Commun. 1999;264:51–4. doi: 10.1006/bbrc.1999.1471. [DOI] [PubMed] [Google Scholar]
- 26.Inoue K, Alsina J, Chen J, Inouye M. Suppression of defective ribosome assembly in a rbfA deletion mutant by overexpression of Era, an essential GTPase in Escherichia coli. Mol Microbiol. 2003;48:1005–16. doi: 10.1046/j.1365-2958.2003.03475.x. [DOI] [PubMed] [Google Scholar]
- 27.Chen X, Court DL, Ji X. Crystal structure of ERA: a GTPase-dependent cell cycle regulator containing an RNA binding motif. Proc Natl Acad Sci U S A. 1999;96:8396–401. doi: 10.1073/pnas.96.15.8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharma MR, Barat C, Wilson DN, Booth TM, Kawazoe M, Hori-Takemoto C, Shirouzu M, Yokoyama S, Fucini P, Agrawal RK. Interaction of Era with the 30S ribosomal subunit implications for 30S subunit assembly. Mol Cell. 2005;18:319–29. doi: 10.1016/j.molcel.2005.03.028. [DOI] [PubMed] [Google Scholar]
- 29.Tu C, Zhou X, Tropea JE, Austin BP, Waugh DS, Court DL, Ji X. Structure of ERA in complex with the 3′ end of 16S rRNA: Implications for ribosome biogenesis. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0904032106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Connolly K, Rife JP, Culver G. Mechanistic insight into the ribosome biogenesis functions of the ancient protein KsgA. Mol Microbiol. 2008;70:1062–75. doi: 10.1111/j.1365-2958.2008.06485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu Q, Inouye M. The gene for 16S rRNA methyltransferase (ksgA) functions as a multicopy suppressor for a cold-sensitive mutant of era, an essential RAS-like GTP-binding protein in Escherichia coli. J Bacteriol. 1998;180:5243–6. doi: 10.1128/jb.180.19.5243-5246.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Britton RA, Powell BS, Dasgupta S, Sun Q, Margolin W, Lupski JR, Court DL. Cell cycle arrest in Era GTPase mutants: a potential growth rate-regulated checkpoint in Escherichia coli. Mol Microbiol. 1998;27:739–50. doi: 10.1046/j.1365-2958.1998.00719.x. [DOI] [PubMed] [Google Scholar]
- 33.Inoue K, Basu S, Inouye M. Dissection of 16S rRNA methyltransferase (KsgA) function in Escherichia coli. J Bacteriol. 2007;189:8510–8. doi: 10.1128/JB.01259-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bylund GO, Persson BC, Lundberg LA, Wikstrom PM. A novel ribosome-associated protein is important for efficient translation in Escherichia coli. J Bacteriol. 1997;179:4567–74. doi: 10.1128/jb.179.14.4567-4574.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bylund GO, Wipemo LC, Lundberg LA, Wikström PM. RimM and RbfA are essential for efficient processing of 16S rRNA in Escherichia coli. J Bacteriol. 1998;180:73–82. doi: 10.1128/jb.180.1.73-82.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lövgren JM, Bylund GO, Srivastava MK, Lundberg LA, Persson OP, Wingsle G, Wikström PM. The PRC-barrel domain of the ribosome maturation protein RimM mediates binding to ribosomal protein S19 in the 30S ribosomal subunits. RNA. 2004;10:1798–812. doi: 10.1261/rna.7720204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suzuki S, Tatsuguchi A, Matsumoto E, Kawazoe M, Kaminishi T, Shirouzu M, Muto Y, Takemoto C, Yokoyama S. Structural characterization of the ribosome maturation protein, RimM. J Bacteriol. 2007;189:6397–406. doi: 10.1128/JB.00024-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nord S, Bylund GO, Lövgren JM, Wikström PM. The RimP protein is important for maturation of the 30S ribosomal subunit. J Mol Biol. 2009;386:742–53. doi: 10.1016/j.jmb.2008.12.076. [DOI] [PubMed] [Google Scholar]
- 39.Yu L, Gunasekera AH, Mack J, Olejniczak ET, Chovan LE, Ruan X, Towne DL, Lerner CG, Fesik SW. Solution structure and function of a conserved protein SP14.3 encoded by an essential Streptococcus pneumoniae gene. J Mol Biol. 2001;311:593–604. doi: 10.1006/jmbi.2001.4894. [DOI] [PubMed] [Google Scholar]
- 40.Butland G, Peregrin-Alvarez JM, Li J, Yang W, Yang X, Canadien V, Starostine A, Richards D, Beattie B, Krogan N, Davey M, Parkinson J, Greenblatt J, Emili A. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature. 2005;433:531–7. doi: 10.1038/nature03239. [DOI] [PubMed] [Google Scholar]
- 41.Pichon J, Marvaldi J, Marchis-Mouren G. The In Vivo Order of Protein Addition in the Course of Escherichia coli 30 S and 50 S Subunit Biogenesis. J Mol Biol. 1975;96:125–137. doi: 10.1016/0022-2836(75)90186-2. [DOI] [PubMed] [Google Scholar]
- 42.Grondek J, Culver G. Assembly of the 30S ribosomal subunit: positioning ribosomal protein S13 in the S7 assembly branch. RNA. 2004;10:1861–6. doi: 10.1261/rna.7130504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sperling E, Bunner AE, Sykes MT, Williamson JR. Quantitative Analysis of Isotope Distributions In Proteomic Mass Spectrometry Using Least-Squares Fourier Transform Convolution. Anal Chem. 2008;80:4906–4917. doi: 10.1021/ac800080v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Studier FW, Moffatt BA. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–30. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 45.Bertani G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol. 1951;62:293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.