Abstract
Pc2 (Cbx4) is a member of the chromobox family of polycomb proteins, and is a SUMO E3 ligase for the transcriptional corepressor, CtBP1. Here we show that both CtBP1 and Pc2 are phosphorylated by the kinase Akt1, which is activated by growth factor signaling via the PI3-kinase pathway. In the presence of Pc2, phosphorylation of CtBP1 is increased, and this requires interaction of both CtBP1 and Akt1 with Pc2. Pc2 promotes CtBP1 phosphorylation by recruiting Akt1, and in part by preventing de-phosphorylation of activated Akt1. Alteration of the Akt-phosphorylated residue in CtBP1 to a phosphomimetic results in decreased CtBP1 dimerization, but does not prevent interaction with other transcriptional regulators. The phosphomimetic mutant of CtBP1 is expressed at a lower level than the wild type protein, resulting in decreased transcriptional repression. We show that this CtBP1 mutant is targeted for poly-ubiquitylation and is less stable than the wild type protein. Coexpression of Pc2 and Akt1 together results in both phosphorylation and ubiquitylation of CtBP1, thereby targeting CtBP1 for degradation. This work suggests that Pc2 may coordinate multiple enzymatic activities to regulate CtBP1 function.
Keywords: CtBP, polycomb, Akt, Pc2, kinase
CtBP1 (Carboxyl-terminus binding protein) is a transcriptional corepressor, first identified by its ability to bind to the carboxyl-terminus of the adenovirus E1a protein 1; 2. CtBP1 and its paralog, CtBP2, have been shown to interact with transcriptional regulators that contain a short amino-acid motif (PxDLS) 3. Mouse Ctbp1 and Ctbp2 have been shown to have both distinct and overlapping functions during mouse development 4. Homozygous loss of function mutations in both genes results in early embryonic lethality, clearly demonstrating the essential role of these corepressors. In addition to binding PxDLS containing transcriptional regulators, CtBP1 can recruit enzymatic activities involved in the regulation of gene expression, including histone methylation and deacetylation 5, suggesting that CtBP1 is a targeting subunit of a larger transcriptional regulatory complex. The crystal structure of CtBP1 has been solved, and it has been shown to form homodimers. Disruption of CtBP1 dimerization by the introduction of multiple point mutations results in decreased transcriptional repression, suggesting that the functional unit is a dimer 6. In support of a role for CtBP1 dimerization, mutational analyses have suggested that a monomeric CtBP1 can interact with components of the general transcriptional repression complex, but that such mutants fail to repress gene expression due to an inability to assemble the corepressor complex on specific DNA bound transcriptional repressors 7. Recent work has suggested that for CtBP2, one subunit of the dimer interacts with a PxDLS motif containing transcriptional repressor, while the other subunit recruits general transcriptional regulators such as histone deacetylases, via non-PxDLS interactions 8.
Polycomb proteins are a large and diverse group of proteins, with common functions in the stable repression of gene expression 9. Mammalian cells have at least five chromobox (Cbx) proteins, with some similarity to the prototypic Drosophila Pc. Cbx proteins have an amino terminal chromodomain, which can bind methylated lysines on histone H3 10; 11; 12; 13, and a small hydrophobic carboxyl-terminal domain, termed the c-box 14; 15. Cbx4, Cbx7 and Cbx8 have all been shown to associate with the PRC1 polycomb complex 16; 17. Cbx4 (Pc2; Polycomb 2) is also a SUMO E3 ligase, which recruits the SUMO conjugating enzyme, Ubc9 18; 19. Known Pc2 sumoylation substrates include the transcriptional regulators, CtBP1 and SIP1 (Smad-interacting protein 1), the de novo DNA methyl transferase, Dnmt3a, and the homeodomain interaction protein kinase, HIPK2 19; 20; 21; 22. Pc2 can also be phosphorylated by HIPK2, and it has been proposed that HIPK2 and Pc2 regulate each others activity 22.
Signaling via the Akt or protein kinase B pathway inhibits apoptosis and mutations which alter pathway activity are associated with numerous forms of cancer 23. Activation of Akt1 requires the phosphorylation of two residues within Akt1, resulting in a conformational change that allows substrate access to the kinase domain. In Akt1, threonine 308 within the kinase domain is phosphorylated by PDK1 24; 25, and the mTOR/rictor complex has been shown to provide the second activating phosphorylation, on serine 473 26; 27. Although PDK-mediated activation of Akt1 occurs at the plasma membrane, Akt1 is found in the nucleus, and it may shuttle between nucleus and cytoplasm 28; 29; 30. Interestingly, recent work has shown that active nuclear Akt1 can be recruited to PML nuclear bodies, where it is inactivated by phosphatases 31. This may represent a mechanism to limit Akt1 activity within the nucleus. However, numerous nuclear substrates of Akt1 are known 32, and while some may be phosphorylated in the cytoplasm prior to nuclear import, the presence of Akt1 in the nucleus suggests that it may phosphorylate substrates within the nucleus.
We show that Pc2 and CtBP1 are phosphorylated by Akt1, and that Pc2 recruits both Akt1 and CtBP1. Phosphorylation of CtBP1 by Akt1 is stimulated by Pc2, and this appears to be due partly to the protection of Akt1 from inactivation. This provides evidence for a novel level of regulation of the PI-3 kinase/Akt1 pathway within the nucleus, and further suggests that Pc2 acts as a nuclear platform to integrate numerous signaling inputs.
RESULTS
Pc2 promotes Akt1-mediated phosphorylation of CtBP1
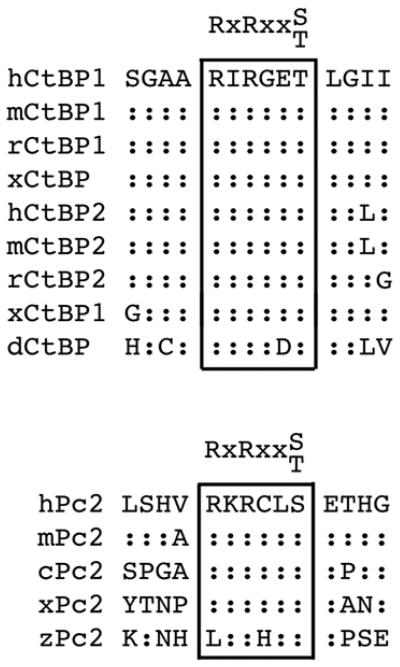
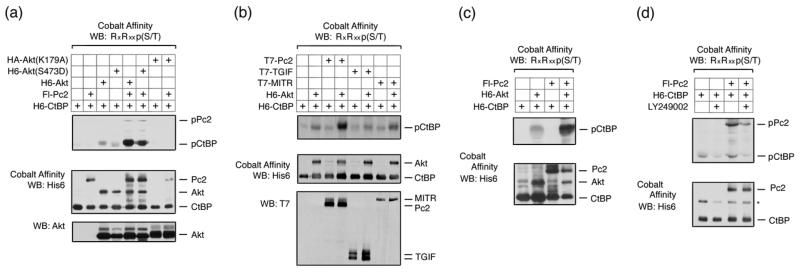
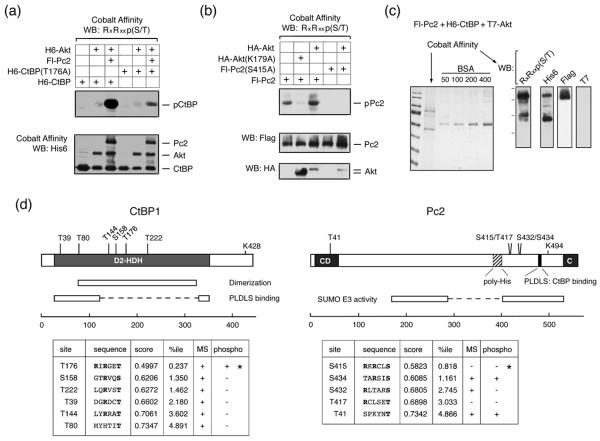
Both CtBP1 and CtBP2, as well as Pc2, contain consensus Akt1 phosphorylation sites, which are conserved among several vertebrate species (Fig 1). We were, therefore, interested to know whether Pc2 or CtBP1 are phosphorylated by Akt1. To test whether Akt1 can phosphorylate CtBP1, we coexpressed six histidine (H6)-tagged CtBP1 together with wild type, inactive (K179A), or constitutively activated (S473D) Akt1 in COS1 cells. Cells were then lysed in 6M guanidine HCl, and H6-tagged proteins were purified via metal affinity and analyzed by western blot with a phospho-specific antibody, which recognizes the phosphorylated Akt1 consensus site. As shown in Figure 2a, coexpression of CtBP1 with either wild type or a constitutively active Akt1 mutant (S473D) resulted in some phosphorylation of CtBP1, whereas no phospho-reactive band was seen with the inactive K179A mutant of Akt1. When Pc2 was coexpressed, the level of CtBP1 phosphorylation was dramatically increased. Some phosphorylation of Pc2 was also detected with wild type or activated Akt1. Note that Pc2 contains an internal poly-histidine stretch, so binds cobalt agarose. To test whether the increased phosphorylation of CtBP1 in the presence of Pc2 and Akt1 was specific to Pc2 we coexpressed CtBP1 and Akt1 with TGIF (thymine/guanine-interacting factor) and MITR (MEF2-interacting transcription repressor), both of which interact with CtBP1 via PLDLS-like motifs 33; 34. In contrast to the effects of Pc2, we observed little or no increase in CtBP1 phosphorylation when we co-expressed either of these CtBP-interacting proteins (Fig 2b). We observed a similar Akt-dependent phosphorylation of CtBP1 in 293T cells that was also increased by coexpression of Pc2 (Fig 2c). To test whether endogenous Akt1 could phosphorylate CtBP1, we transfected 293T cells with CtBP1 alone or together with Pc2 and treated cells with the PI-3 kinase inhibitor, LY249002, which blocks activation of Akt1. As shown in Figure 2d, we observed phosphorylated CtBP1 and Pc2, and the level of phosphorylation of both proteins was decreased in the presence of the inhibitor. Thus, it appears that both Pc2 and CtBP1 can be phosphorylated by Akt1, and that Pc2 can increase the level of CtBP1 phosphorylation.
Fig. 1. Pc2 and CtBP have conserved Akt consensus phosphorylation sites.
Alignment of Akt1 consensus sites in CtBP1, CtBP2 and Pc2 are shown. Identities to human sequence are indicated with colons (h: human, m: mouse, r: rat, c: chicken, x: Xenopus laevis, z: zebra fish, d: Drosophila melanogaster).
Fig. 2. Akt1 phosphorylates CtBP1 and Pc2.
(a) COS1 cells were transfected with H6-CtBP1, Flag-Pc2, H6-Akt1, H6-Akt(S473D), or HA-Akt(K179A) expression constructs as indicated. Cells were lysed in 6M guanidine-HCl, proteins purified on cobalt agarose and western blotted with an Akt1 substrate [RxRxxp(S/T)] antibody. Lower panels: 20% of the pulldown was analyzed by western blot using H6 and a portion of the lysate with an Akt1 antibody. (b) COS1 cells were transfected with the indicated expression constructs and analyzed as in panel a. Cobalt affinity purified proteins were analyzed with phospho-substrate and His6 antibodies [upper panels] and part of the lysate was analyzed by direct western blot with a T7 antibody [below]. (c) 293T cells were transfected with H6-CtBP1, H6-Akt1, and Flag-Pc2 expression constructs as indicated, and analyzed as for panel a. (d) 293T cells were transfected with H6-CtBP1 and Flag-Pc2 expression constructs and were left untreated or treated with 50μm LY294002 for 1 hour, and analyzed as for panel a.
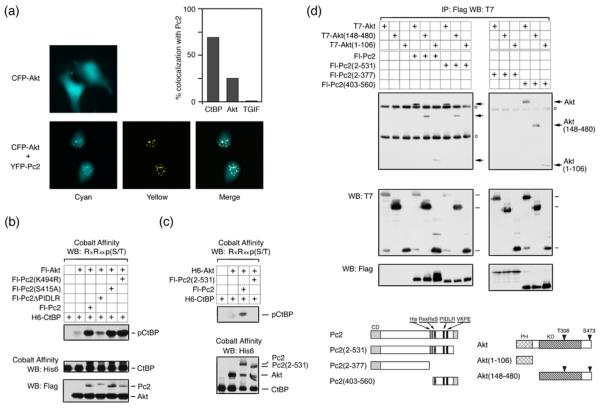
Since Pc2 can recruit CtBP1 to subnuclear polycomb foci 19; 35, we wanted to know whether Akt1 could also be recruited to these foci in the presence of coexpressed Pc2. Coexpression of eYFP-tagged Pc2 with eCFP-Akt1 resulted in colocalization of a proportion of the Akt1 with Pc2 foci (Fig 3a). In contrast, in the absence of coexpressed Pc2, Akt was observed throughout the nucleus and cytoplasm. In cells with clear Pc2 foci, Akt1 and Pc2 colocalized 25% of the time, and CtBP1 colocalized with Pc2 in approximately 70% of Pc2 expressing cells with foci (Fig 3a). In contrast, we never observed colocalization of TGIF (which is not known to interact with Pc2) and Pc2 under these conditions. Coexpression of eYFP-Akt1 with eCFP-CtBP1 did not detectably alter the localization of either protein. However, when Flag-tagged Pc2 was also present, colocalization of both Akt1 and CtBP1 at subnuclear foci was observed (Fig S1). Thus, Pc2 is able to recruit both Akt1 and CtBP1 to subnuclear foci, consistent with the possibility that CtBP1 phosphorylation by Akt1 may occur in the nucleus.
Fig. 3. Interaction of Akt1 and CtBP1 with Pc2 is required for CtBP1 phosphorylation.
(a) COS1 cells were transfected with an eCFP-Akt1 expression vector, with or without eYFP-Pc2. Cells were visualized 24 hours after transfection. % colocalization with Pc2 is shown for Akt1, CtBP1 and TGIF. (b) Lysates from COS1 cells transfected with the indicated expression constructs were analyzed by cobalt affinity purification and western blot, or by direct western blot of the lysate. (c) Transfected COS1 cells were lysed in 6M guanidine-HCl, proteins purified on cobalt agarose, and analyzed by western blot with an Akt1 substrate antibody, as in panel b. (d) COS1 cells were transfected with expression constructs as indicated. Proteins were isolated with anti-Flag agarose and analyzed by western blot with a T7 antibody. Expression in the lysates is shown below. Circles indicate Ig heavy and light chains. The constructs used are shown schematically below. PH: pleckstrin homology domain, KD: kinase domain, CD: chromodomain, His: Pc2 poly-histidine stretch. Numbers indicate amino acids present in each construct.
To begin to test the requirements for Pc2 stimulation of CtBP1 phosphorylation, we tested Akt1 mediated phosphorylation of CtBP1 in the presence of wild type Pc2, or of a mutant form in which the CtBP-interaction motif (PIDLR) had been deleted. The PIDLR mutant of Pc2 was dramatically impaired in its ability to stimulate CtBP1 phosphorylation, whereas mutation of either the single consensus Akt1 phosphorylation site (S415A) or the sumoylation site (K494R) within Pc2 had no effect (Fig 3b). As shown in Figure 3c, the Pc2(2-531) construct was unable to increase the phosphorylation of CtBP1 by Akt1 above that seen with Akt1 alone. One interpretation of this is that a specific interaction of Akt1 with the carboxyl-terminus of Pc2 is required for Pc2 to enhance phosphorylation of CtBP1. However, since this Pc2 mutant is delocalized from polycomb bodies 19; 35, it is also possible that correct Pc2 localization contributes to the increase in CtBP1 phosphorylation by Akt1. We, therefore, next analyzed the interaction of Pc2 with Akt1. A series of Flag-tagged Pc2 expression constructs (Fig 3d) was coexpressed in COS1 cells with T7-tagged Akt1, a deletion mutant lacking the PH domain, or a construct with contains only the PH domain. All three Akt1 constructs coprecipitated with full length Pc2 and with a construct containing only the carboxyl-terminal 158 amino acids [Pc2(403-560)] (Fig 3d). Deletion of this carboxyl-terminal region (construct 2-377) abolished interaction with all Akt1 constructs, whereas, deletion of only the 29 carboxyl-terminal amino acids prevented interaction with the PH domain of Akt1, but did not affect interaction with constructs containing the intact kinase domain. Thus, it appears that Pc2 and Akt1 interact via two domains: The PH domain of Akt1 interacts with the extreme carboxyl-terminus of Pc2, and the kinase domain interacts with a region between amino acids 403 and 531, which contains the phosphorylation site. Together, these results suggest that interaction of both CtBP1 (substrate) and Akt1 (enzyme) with Pc2 are required for the stimulation of Akt1 activity towards CtBP1.
Pc2 maintains phosphorylated active Akt1
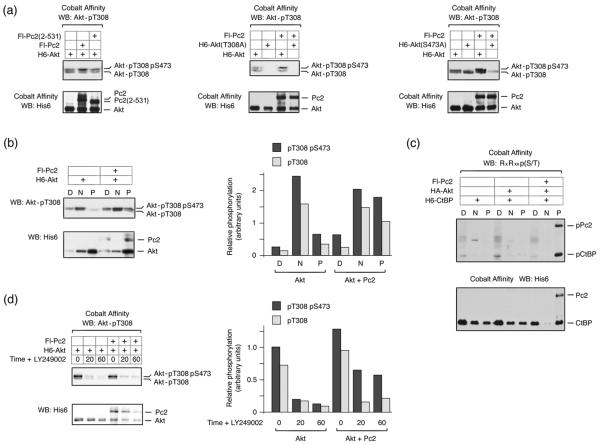
We were next interested to test whether Pc2 promoted CtBP1 phosphorylation by Akt1 simply by bringing together enzyme and substrate, or whether Pc2 might also affect Akt1 activity. Akt1 is activated by phosphorylation at two specific residues, threonine 308 and serine 473, which are phosphorylated by PDK1 and mTOR/rictor respectively. When H6-Akt1 is isolated from COS1 cell lysates and analyzed using a phospho-specific antibody, which recognizes phosphorylated T308, two bands are visible by western blot (Fig 4a). Mutation of threonine 308 to alanine abolishes reactivity with this antibody for both bands, whereas, alteration of serine 473 to alanine results in loss of only the slower migrating isoform (Fig 4a). Thus, the slower migrating isoform represents the fully active doubly phosphorylated Akt1, whereas the faster migrating band recognized by this antibody is phosphorylated only on T308. Interestingly, in the presence of coexpressed Pc2, we noticed a small but reproducible increase in the amount of the slower migrating doubly phosphorylated Akt1, which was not seen with the 2-531 deletion mutant of Pc2 that failed to promote CtBP phosphorylation (Fig 4a). Coexpression of PDK1 with Akt1 resulted in a dramatic increase in both the upper and lower phospho-T308 bands, whereas in the presence of Pc2 it was clearly only the upper band that was increased (Fig S2). It should also be noted that compared to over-expression of PDK1, Pc2 results in a modest increase in activated Akt1.
Fig. 4. Pc2 protects Threonine 308 phosphorylation on Akt1.
(a) COS1 cells were transfected with H6-Akt1, Flag-Pc2, or Pc2(2-531) expression constructs as indicated. Akt1 expression constructs were either wild type, T308A mutant (center) or S473A mutant (right). Proteins were purified on cobalt agarose and western blotted with an Akt1 phospho-T308 antibody, or a His6 antibody. The bands corresponding to Akt1 phosphorylated on T308 or both T308 and S473 are shown. (b) COS1 cells transfected with H6-Akt1 and Flag-Pc2 were partitioned into digitonin soluble [D], NP-40 soluble [N], and pellet [P] fractions, which were analyzed for phosphorylated Akt1 by western blot. Relative phospho-T308 levels, in arbitrary units normalized to total expression of Akt1 are shown to the right. (c) COS1 cells were transfected with the indicated constructs and fractionated as in b. Fractions were western blotted for Pc2 and CtBP with a His6 antibody and for the phosphorylated proteins using the phospho-Akt substrate antibody. (d) 293T cells were transfected with H6-Akt1 and Flag-Pc2, then treated with 50μm LY294002 for the indicated times, and analyzed as in b. Relative phospho-T308 levels of Akt1 are shown to the right.
To more easily visualize the effect of Pc2 on Akt1 phosphorylation, we subjected transfected COS1 cells to differential permeabilization with detergent. Cells were first treated with digitonin to release the soluble cytosolic fraction, then with NP-40 to release soluble nuclear proteins, and finally an insoluble pellet fraction was collected. Analysis of these fractions by western blotting for endogenous components reveals that the small GTPase, Ran, is primarily released in the digitonin fraction and TGIF is present in the NP-40 soluble fraction, consistent with these being cytosolic and nuclear fractions (Fig S3). We also probed for Lamin A and histone H3, and found that both remained exclusively in the pellet fraction, as expected (Fig S3). Although the majority of the Akt1 is present in the insoluble fraction, most of the T308 phosphorylated Akt1 is in the NP-40 soluble fraction (Fig 4b). In the presence of Pc2, there is a clear increase in the amount of doubly phosphorylated Akt1 in the insoluble fraction, consistent with interaction of active Akt1 with Pc2-containing complexes. Additionally, when we performed similar experiments and probed for the presence of phosphorylated CtBP1, we found that the low level of phospho-CtBP seen in the absence of Pc2 was present in the digitonin soluble fraction, whereas, all of the increase in phosphorylated CtBP1 in the presence of Pc2 co-fractionated into the insoluble fraction with Pc2 (Fig 4c). Following removal of the activating stimulus, Akt1 is rapidly inactivated by phosphatases. To determine whether the increase in doubly phosphorylated Akt1 in the presence of Pc2 was due to the inability of phosphatases to inactivate Akt1, we expressed Akt1 alone or together with Pc2 in 293T cells, and treated them with LY249002 for 20 or 60 minutes, to inhibit further Akt1 activation, or left them untreated. In cells expressing Akt1 alone, both the phospho-T308 form and the fully active doubly phosphorylated form of Akt1 were rapidly lost, decreasing to less than 20% within 20 minutes (Fig 4d). In contrast, in cells expressing Pc2 and Akt1, a greater proportion of the total Akt1 remained doubly phosphorylated, even out to 60 minutes. These results suggest that Pc2 interacts with fully active Akt1, phosphorylated at both T308 and S473, and may prevent the access of phosphatases, which normally result in rapid loss of phosphate from threonine 308. Thus, once activated Akt1 enters the nucleus, Pc2 may protect a specific pool of Akt1 from inactivation.
Identification of phosphorylation sites on Pc2 and CtBP1
Since CtBP1 contains a single conserved consensus site for Akt1, we converted the putative phosphorylated residue (T176) to an alanine. The robust phosphorylation of CtBP1 in the presence of both Akt1 and Pc2 was greatly reduced when the T176A mutant of CtBP1 was transfected in place of wild-type (Fig 5a), suggesting that T176 is the major Akt1 phosphorylation site in CtBP1. However, since there is some residual reactivity with the phospho-substrate antibody we cannot rule out the presence of a second site. As with CtBP1, there is a single good match to the Akt consensus phosphorylation site in Pc2, at serine 415. We, therefore, tested phosphorylation of the Pc2 S415A mutant by Akt1. As shown in Figure 5b, Akt-mediated Pc2 phosphorylation was abolished by the presence of the S415A mutation, suggesting that this is indeed the major Akt1 phosphorylation site in Pc2. To confirm that the sites identified in Pc2 and CtBP1 by western blotting were indeed phosphorylated in the presence of Akt1, and to identify other potential phosphorylation sites, we performed an analysis of phosphorylated peptides by mass spectrometry (MS). COS1 cells were cotransfected with His-tagged CtBP1, Flag-tagged Pc2 and T7-tagged Akt1 expression constructs, and CtBP1 and Pc2 were purified under denaturing conditions, using cobalt agarose. A portion of the purified protein was subjected to gel electrophoresis and analyzed by Coomassie blue staining, or western blotting. As shown in Figure 5c, Pc2 and CtBP1 were the major bands isolated, and the phosphorylated forms of both proteins were detectable with a phospho-specific Akt substrate antibody. T7-tagged Akt was not detected in the cobalt agarose purified fraction, as expected. Purified proteins were then digested with either trypsin or chymotrypsin, and modified peptides were identified by MS. For CtBP1, we achieved 81% coverage of the entire protein, including 34 of 45 (76%) serines and threonines (supplemental Tables 1 and 2). Importantly, this included coverage of all six of the closest matches to a consensus Akt phosphorylation site, as predicted by the Scansite motif scanning program (http://scansite.mit.edu/motifscan_seq.phtml) (Fig 5d). However, we detected phosphorylation at only one of these sites in CtBP1 by MS, and this corresponded to phosphorylation of T176, which is present within the highest confidence Akt phosphorylation consensus motif. This suggests that for CtBP1, T176 is at least the major Akt phosphorylation site, and provides confirmation of the identification by western blotting with the phospho-specific antibody.
Fig. 5. Identification of phosphorylation sites in CtBP1 and Pc2.
(a) COS1 cells were transfected with H6-CtBP1, H6-CtBP(T176A), Flag-Pc2, and H6-Akt1 expression constructs as indicated and phosphorylation was analyzed by western blot with an Akt phospho-substrate antibody. (b) COS1 cells were transfected with Fl-Pc2 or Fl-Pc2(S415A), together with a control vector, HA-Akt1, or HA-Akt(K179A) expression constructs, and analyzed by western blot as in A. (c) A sample used for MS analysis is shown. Pc2 and CtBP1 were purified from transiently transfected COS1 cells expressing both proteins together with T7-Akt1. A portion of the purified proteins was analyzed by Coomassie blue staining (left), or by western blot with the indicated antibodies (right). (d) CtBP1 and Pc2 are shown schematically, together with the positions of the closest matches to consensus Akt phosphorylation sites. The sequences of consensus Akt phosphorylation sites are shown below, together with the score and percentile as predicted by Scansite. A + in the MS column indicates that peptides spanning the site were identified in the MS analysis. A + in the phospho column indicates that phosphorylation was detected at that site by MS. * indicates the two sites which were identified by western blotting with the phospho-substrate antibody.
For Pc2, we obtained 62% coverage of the protein, including 41 out of 58 (71%) serine and threonine residues. Here we identified numerous phosphorylated peptides, including seven phosphorylated serines and three threonines (supplemental Tables 1 and 2). Among the five best matches to an Akt consensus site, peptides containing three were found in the MS analysis, and two of these (S434 and T41) were phosphorylated (Fig 5d). Peptides spanning S415 of Pc2 were not detected in our MS analysis. Including the S415 site identified by western blot, it appears that at least three potential Akt sites can be phosphorylated in Pc2. However, one of these (T41) is a very low confidence match to the consensus, whereas the S434 site identified by MS is the second best match after the S415 site identified by blotting and mutational analysis. Among the sites identified by MS in Pc2, S434 had the highest confidence prediction for known kinase motifs (1.161 percentile for Akt; Fig 5d and supplemental Table 1). The combination of western blotting and mutational data with the MS analysis suggests that Pc2 is phosphorylated by Akt on at least two residues, S415 and S434, whereas CtBP1 appears to be phosphorylated by Akt1 primarily on a single residue, T176.
Mutation of T176 affects CtBP1 dimerization and protein expression
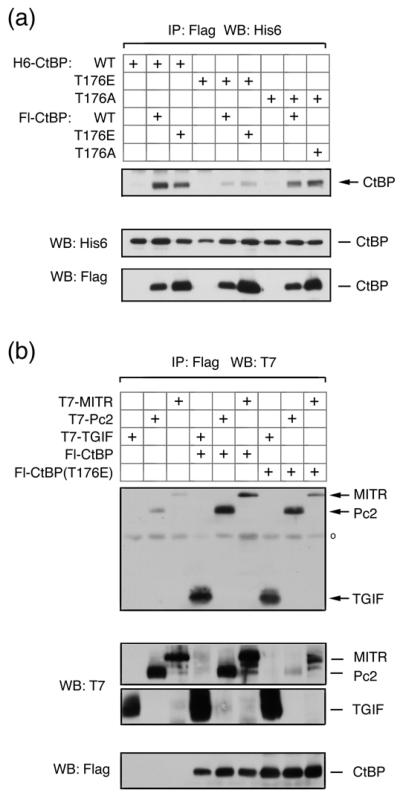
We were next interested to know whether phosphorylation of CtBP1 and Pc2 by Akt1 might affect their function. We tested the effects of mutating both S415 and S434 in Pc2, singly and together, but did not observe any consistent effects on CtBP1 or Akt1 interaction, or on the ability of Pc2 to promote CtBP1 sumoylation (data not shown). We next tested the effects of mutating the single Akt1 site in CtBP1. CtBP1 forms homodimers, and mutations which disrupt dimerization also affect transcriptional repression 6. We first tested the formation of CtBP1 dimers using either wild type or mutant CtBP1 proteins in which T176 had been altered to alanine (T176A) or glutamic acid (T176E), to either prevent or mimic phosphorylation of this site. COS1 cells were transfected with combinations of His6- and Flag-tagged wild type and mutant CtBP1, and proteins were precipitated via the Flag epitope. The T176E mutant was clearly impaired in its ability to dimerize either with wild type or T176E mutant CtBP1, whereas, the T176A mutation had a lesser effect on the ability of CtBP1 to dimerize (Fig 6a). We next tested interaction of mutant CtBP1 with transcriptional regulators which recruit CtBP1 via interaction with PLDLS-like motifs. When protein complexes were isolated from COS1 cells, via the Flag epitope on CtBP1 or the T176E mutant, similar amounts of co-precipitating epitope tagged TGIF1, Pc2 or MITR were present, suggesting that the T176E mutation in CtBP1 does not prevent interaction with PLDLS-containing transcriptional regulators (Fig 6b). Together, these results suggest that alteration of threonine 176 to a negatively charged residue decreases the ability of CtBP1 to dimerize, but has little effect on interaction with proteins containing a PLDLS motif.
Fig. 6. Mimicking Akt1-mediated phosphorylation of CtBP1 decreases dimerization.
(a) COS1 cells were cotransfected with H6- or Flag-tagged CtBP1, CtBP(T176E) or CtBP(T176A) expression constructs, as indicated. Proteins were precipitated with anti-Flag agarose and analyzed by western blot with a H6 antibody. Expression of transfected proteins in the lysates is shown below. (b) COS1 cells were cotransfected with T7 tagged TGIF, Pc2, or MITR, together with a control vector, Flag-CtBP1 or CtBP(T176E) expression constructs. Proteins were precipitated with anti-Flag agarose and analyzed by western blot with a T7 antibody. Expression in the lysates is shown below.
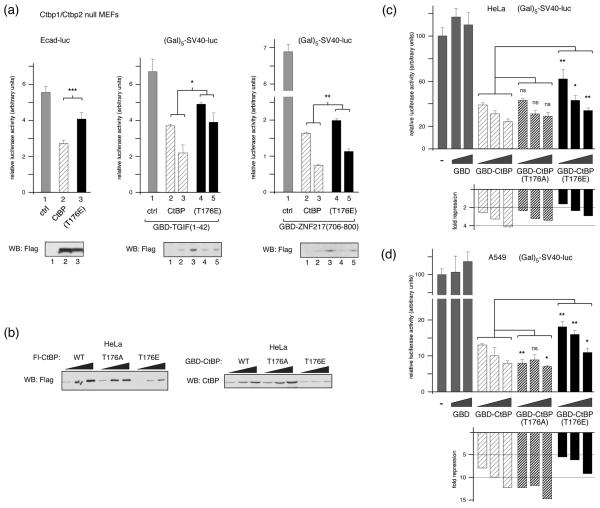
We next analyzed the ability of the T176E mutant form of CtBP1 to repress transcription in mouse embryo fibroblasts (MEFs) lacking both Ctbp1 and Ctbp2 (referred to here as Ctbp1/2 null; 4). The E-cadherin gene is repressed by SIP1, dependent on its ability to recruit CtBP1, and E-cadherin transcriptional reporters are derepressed in Ctpb1/2 null MEFs. As shown in Figure 7a, we observed a decrease in E-cadherin reporter activity in Ctpb1/2 null MEFs, when we co-expressed wild type CtBP1. However, there was significantly less repression when we co-expressed the T176E mutant version of CtBP1. We next analyzed a simpler artificial system for targeting CtBP1, in which CtBP1 is targeted to a reporter with five GaI4 binding sites upstream of the SV40 promoter, via a fusion between the GaI4 DNA binding domain (GBD) and the CtBP recruitment domains of either TGIF or ZNF217. TGIF contains a PLDLS motif which recruits CtBP1 33, whereas CtBPs interact with this region of ZNF217 via an alternate peptide motif (RRTGCPPAL in ZNF217), which binds to a different region of CtBP1 36. These two fusions, therefore, test alternate modes of CtBP1 recruitment. Repression of SV40 promoter activity by both fusions is almost completely dependent on CtBP1 (data not shown). As with the E-cadherin reporter, we observed significantly more repression via both GBD fusions with the wild type CtBP1 than with the T176E mutant (Fig 7a). When we analyzed expression of the Flag-tagged CtBP1 constructs in these experiments by western blotting, we observed a consistently lower level of expression of the T176E mutant than the wild type protein (Fig 7a). This difference may not have been apparent in previous experiments with COS1 cells due to the very high level of expression in these cells. To test whether the lower expression of the T176E in Ctbp1/2 null MEFs was also seen in other cell types, we transfected increasing amounts of Flag-tagged wild type, T176E and T176A CtBP1 into HeLa cells. We also performed a similar comparison using GBD fusions to CtBP1 and the two T176 mutants. As shown in Figure 7b, the T176E mutant was less well expressed than the wild type or T176A, whether tagged with Flag or GBD. We then used the GBD fusions to test the ability of CtBP1 to repress transcription when tethered to a heterologous promoter directly in HeLa cells. As shown in Figure 7c, the T176E mutant repressed significantly less well at each of the three amounts tested. Additionally, we repeated these analyses in A549 cells which have lower expression of endogenous CtBP1 and CtBP2 than HeLa 35, with similar results, although in this case the T176A also appeared to repress slightly better than the wild type protein (Fig 7d). This analysis suggests that the presence of a negative charge at position 176 in CtBP1 reduces its steady state expression level, thereby resulting in decreased transcriptional repression.
Fig. 7. Decreased expression and transcriptional repression by CtBP(T176E).
(a) Ctbp1/2 double null MEFs were transfected with an E-cadherin promoter-based Luciferase reporter and expression vectors encoding Flag-CtBP1 or CtBP(T176E) [left], or with a (Gal)5-SV40-Luciferase reporter, an expression vector encoding a Gal4 DNA binding domain (GBD) fusion to amino acids 1-42 of TGIF [center], or the (Gal)5-SV40-Luciferase reporter and a GBD fusion to amino-acids 706-800 of ZNF217 [right], with increasing amounts of Flag-CtBP1 or CtBP(T176E). Luciferase activity is presented as mean + s.d. of triplicate transfections. CtBP1 expression was assayed by western blot with a Flag antibody (below). (b) Relative expression in HeLa cells of increasing amounts of transfected Flag- or GBD-tagged CtBP1, or the T176A and T176E mutants, was analyzed by western blot (triangles represent increasing amounts of transfected CtBP1 expression construct; 100, 200 and 400ng per 35mm dish). HeLa cells (c), or A549 lung epithelial cells (d) were transfected with the (Gal)5-SV40-Luciferase reporter and an expression vector encoding the GBD, or a GBD fusion to wild type or T176A, or T176E CtBP1 as indicated. Luciferase activity is presented as mean + s.d. of triplicate transfections (upper panels) and as fold-repression relative to the control transfection (below). Triangles represent increasing amounts of transfected GBD expression construct; 3, 9 and 27ng per 35mm well (or 0, 9 and 27ng for the GBD). Significance testing was by student's T test. ***: p < 0.001, **: p < 0.01, *: p < 0.05, ns: not significant.
Phosphorylation of T176 induces ubiquitylation and degradation of CtBP1
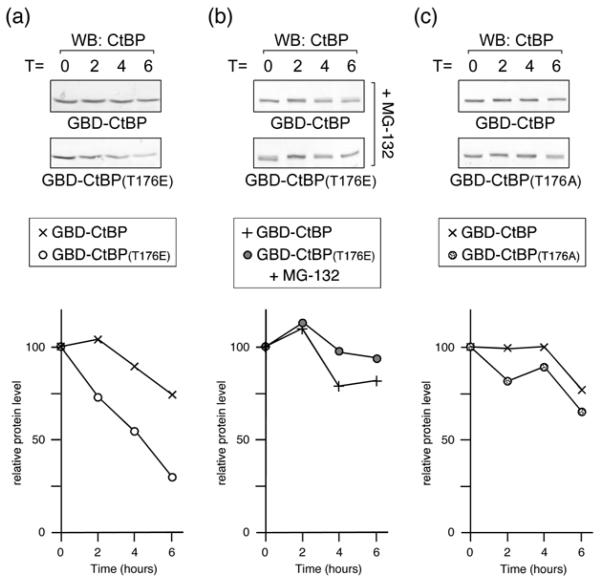
To test whether the decreased expression of T176E CtBP1 was due to a change in protein stability, we analyzed the half-life of GBD-CtBP1 and the T176E mutant. HeLa cells were transfected with wild type or T176E GBD-CtBP1 and treated with cycloheximide for 2-6 hours, to prevent further protein synthesis. As shown in Figure 8a, the wild type protein was relatively stable, whereas, the T176E mutant had a clearly reduced half-life. When we repeated this experiment but pretreated the cells with MG-132 for 30 minutes prior to the addition of cycloheximide, to inhibit the proteasome, we effectively abolished the difference in half-lives between the wild type and T176E mutant CtBP1 (Fig 8b). In contrast to the T176E mutation, alteration of T176 to alanine did not dramatically reduce the half-life of CtBP1 (Fig. 8c). Thus, it appears that conversion of threonine 176 to an acidic residue results in a proteasome-dependent decrease in CtBP1 stability.
Fig. 8. Half-life analysis of CtBP1.
(a) HeLa cells were transfected with GBD-CtBP1 or the T176E mutant, and 40 hours later incubated with cycloheximide. Cells were lysed at the indicated times (in hours) after cycloheximide addition, and western blotted with a CtBP1 antibody. The relative amount of protein for wild type and T176E mutant is plotted below. (b) Cells were treated and analyzed as in a, except that 30 minutes prior to the addition of cycloheximide, MG-132 was added to inhibit the proteasome. (c) HeLa cells were transfected with GBD-CtBP1 or the T176A mutant, and analyzed as in a.
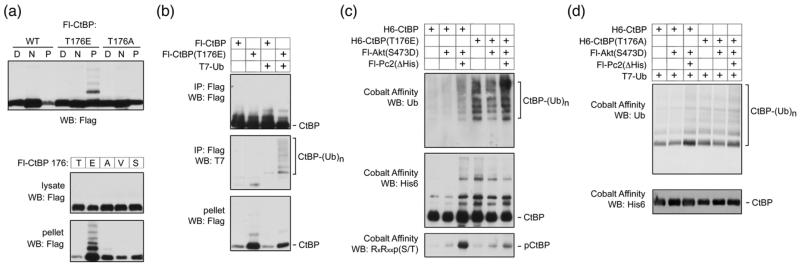
In some of our western blotting analyses we noticed a minor shifted band associated with the T176E mutant, which was too small to be sumoylated CtBP1. To analyze this more carefully, we transfected COS1 cells with Flag tagged wild type, T176E or T176A CtBP1, and fractionated cells into digitonin-soluble, NP-40-soluble and pellet fractions. As shown in Figure 9a, a slower migrating band was clearly visible in the insoluble pellet fraction with the T176E mutant, but was not seen for the wild type or T176A. To confirm that it was specifically introduction of a negative charge that induced this slower migrating form, we compared NP-40 lysate and pellet fractions from cells expressing wild type CtBP1, and forms in which T176 had been altered to E, A, V or S residues (Fig 9a). Again, only the T176E resulted in a slower migrating band present in the insoluble fraction, although in this case, it appeared that a ladder of slower migrating forms was detectable, consistent with poly-ubiquitylation. To test whether the T176E mutant could be ubiquitylated, we coexpressed wild type or mutant CtBP1 in COS1 cells with a T7-tagged ubiquitin construct, immunoprecipitated the Flag-CtBP1 and blotted with an antibody that recognizes the T7 epitope. As shown in Figure 9b, a ladder of slower migrating forms of CtBP1 was clearly present in the Flag precipitate from CtBP(T176E) expressing cells, but was barely detectable with the wild type protein. Thus, it appears that the T176E mutant of CtBP1 can become ubiquitylated, consistent with its reduced stability.
Fig. 9. Ubiquitylation of CtBP1.
(a) COS1 cells were transfected with Flag tagged wild type or mutant CtBP1 as indicated and partitioned into digitonin soluble [D], NP-40 soluble [N], and pellet [P] fractions, which were analyzed by Flag western blot. Lower panels show NP-40 lysate and pellet fractions from COS1 cells expressing Flag-CtBP1, in which residue 176 was either wild type [T], or altered to one of the other indicated amino acids, were analyzed by western blot. (b) COS1 cells were transfected with Flag tagged CtBP1 or the T176E mutant, together with a T7-tagged ubiquitin expression construct, as indicated. Flag immunoprecipitates, or the pellet fraction were analyzed by Flag and T7 western blot as indicated. (c) COS1 cells were transfected with the indicated expression constructs, lysed in guanidine HCl and His-tagged CtBP1 was purified by metal affinity [note the Pc2 construct used here lacks the poly-histidine stretch, so does not bind cobalt agarose]. Bound CtBP1 was analyzed by western blot with antibodies for endogenously expressed ubiquitin, H6-CtBP1 and phosphorylation at T176. (d) COS1 cells were transfected with the indicated expression constructs, lysed in guanidine HCl and CtBP was isolated on cobalt agarose. Bound His-tagged CtBP was detected with a His6 antibody (lower panel) and ubiquitylated CtBP was detected with a T7 antibody (upper panel).
We next tested whether we could detect endogenous ubiquitin on CtBP1 and whether this was affected by coexpression of Akt1 or Pc2. COS1 cells were cotransfected with six histidine tagged wild type or T176E mutant CtBP1, together with expression constructs encoding constitutively active Akt or a mutant of Pc2 which lacks the internal histidine stretch. CtBP1 was purified on cobalt agarose, and western blotted with antibodies to detect the His6 epitope, or ubiquitin. As shown in Figure 9c, CtBP(T176E) was robustly ubiquitylated independent of the coexpression of Akt or Pc2. However, we also observed an increase in ubiquitylation of the wild type CtBP1, when both Akt and Pc2 were both coexpressed. In contrast, expression of Akt alone did not induce either CtBP1 ubiquitylation or significant phosphorylation of CtBP1 (Fig 9c). To test whether the Akt1 and Pc2 mediated increase in CtBP1 ubiquitylation was dependent on phosphorylation of T176, we compared the increase in ubiquitylation of wild type and T176A CtBP1. To better visualize low levels of attached poly-ubiquitin chains, we co-expressed a T7-tagged ubiquitin with His6-tagged CtBP1 and purified proteins under denaturing conditions to increase solubility of the poly-ubiquitylated CtBP1. As shown in Figure 9d, we observed an increase in the amount of poly-ubiquitylated wild type CtBP1 on coexpression of both Akt1 and Pc2 together, whereas, this increase was not apparent with the T176A mutant. Together, this data suggests that in response to Akt-mediated phosphorylation, CtBP1 is ubiquitylated and degraded, and that this effect is promoted by Pc2.
DISCUSSION
Here we demonstrate that Pc2 and Akt1 can promote the phosphorylation of CtBP1 on a single residue within an Akt consensus site. This phosphorylation event promotes ubiquitylation of CtBP1, and we suggest that this decreases the half-life of CtBP1 thereby decreasing transcriptional repression (Fig S4).
CtBP1 is known to be regulated by numerous post-translational modifications, including sumoylation, phosphorylation and ubiquitylation. Sumoylation has been suggested to increase transcriptional repression by CtBP, in part by promoting nuclear localization 37. CtBP1 has been shown to be phosphorylated by Pak1 and HIPK2 at different residues, with both resulting in decreased transcriptional repression by CtBP1 38; 39. HIPK2-mediated phosphorylation of CtBP1 appears to down-regulate function by inducing CtBP1 degradation, whereas Pak1 was suggested to effect sub-cellular redistribution of CtBP1. Interestingly, Pak1 phosphorylates S158 of CtBP1, which is the second best match to an Akt consensus site 38. However, we did not detect phosphorylation of this residue by our MS analysis, suggesting that it is not a target for both kinases. Our analysis suggests that phosphorylation of CtBP1 is a negative regulatory input, with similarities to the regulation by HIPK2; both Akt1- and HIPK2-mediated phosphorylation target CtBP1 for ubiquitin-mediated degradation. CtBP1 phosphorylation by Akt1 appears to both decrease dimerization and induce ubiquitylation. However, we do not as yet know whether the ubiquitylation of CtBP1 is simply a result of decreased dimer formation, or whether it is an independent event. Other mutations which affect CtBP1 dimerization have been shown to maintain at least some interactions with CtBP1 partner proteins, and do not appear to target the protein for ubiquitylation 6; 7. Thus decreasing dimerization and increasing ubiquitylation may represent two independent modes of regulation of CtBP1 function. CtBP1 is a component of a large corepressor complex 5, and based on its ability to interact with a wide array of PxDLS containing transcription factors 3; 40, CtBP1 is likely to represent a key targeting component of such complexes. In this context, it is perhaps not surprising that CtBP1 is targeted by numerous regulatory inputs, since this allows for the modulation of general corepressor recruitment to a large number of transcription factors.
Akt1 activation occurs at the plasma membrane, but once activated, Akt1 can signal at other locations within the cell. Akt1 is found in both the nucleus and cytoplasm, and a nuclear export signal (NES) has been identified in Akt1, suggesting the protein can shuttle between nuclear and cytosolic compartments 28; 29; 30. Here we show that two proteins with nuclear functions, CtBP1 and Pc2, are phosphorylated by Akt1. Although it is possible that they are phosphorylated in the cytoplasm and then translocate to the nucleus, at least for Pc2 we think this is unlikely, since it stably associates with sub-nuclear foci, termed polycomb bodies. Additionally, we show that in the presence of Pc2, Akt1 can localize to these foci. Since Pc2 expression stimulated Akt1-mediated CtBP1 phosphorylation, dependent on interaction of Pc2 with CtBP1, it is likely that phosphorylation of CtBP1 also occurs in the nucleus, at least when in complex with Pc2. However, we cannot rule out the possibility that CtBP1 can also be phosphorylated in the cytoplasm by Akt1. The stimulation of CtBP1 phosphorylation by Akt1 in the presence of Pc2 appears to require interaction of all three proteins, suggesting that Pc2 acts as a scaffold which brings together enzyme and substrate. This recruitment of Akt1 has clear parallels to the SUMO E3 activity of Pc2 towards CtBP1. As with E3 activity, it appears that the effect of Pc2 on Akt1 consists of more than simple co-recruitment of enzyme and substrate. Threonine 308 of Akt1 is normally subject to rapid dephosphorylation, thereby inactivating Akt1 41. However, we show that a pool of Akt1 in the nucleus is protected by Pc2 from dephosphorylation. It appears to be specifically the doubly phosphorylated fully active form of Akt1 that is protected by Pc2, suggesting that Pc2 is preserving a sub-nuclear pool of active Akt1. As yet we have been unable to distinguish between the requirement for a specific Pc2-Akt1 interaction, involving the PH domain and the C-box of Pc2, and localization to polycomb bodies which prevents dephosphorylation of Akt1. We do not know whether the active Akt1 preserved by Pc2 can phosphorylate proteins other than Pc2 and CtBP1, but it remains an intriguing possibility that Akt1 may be released from Pc2 allowing it to act on other nuclear proteins. The effect of Pc2 on CtBP1 phosphorylation appears to be relatively specific, since co-expression of two other PLDLS containing CtBP interacting proteins did not increase CtBP1 phosphorylation at T176. Thus it appears that Pc2 can recruit at least two enzymatic activities (phosphorylation and sumoylation) that modify CtBP1. This may suggest that, in addition to its role in repression of gene expression, Pc2 can act as a platform to recruit protein modifying activities within the nucleus. We have as yet been unable to show that sumoylation regulates Akt1 activity, or of the ability of Pc2 to stimulate CtBP1 phosphorylation, but this remains an intriguing possibility.
Recent work has shown that active Akt1 in the nucleus can be recruited to PML nuclear domains, where it is inactivated by phosphatases 31. Here we show that Pc2 recruits Akt1 to distinct nuclear domains, resulting in protection of the active enzyme rather than inactivation. Pc2 and PML may perform opposing functions in the regulation of nuclear Akt1 function. Once active Akt1 enters the nucleus, it may be subject to differing regulation by these proteins, depending on the specific regulatory inputs and the relative levels of PML and Pc2 within the nucleus. Pc2 interacts with several protein modifying enzymes, including Ubc9, the kinases Akt1 and HIPK2, and proteins involved in histone modification, such as Ring1 and SUV39H1 15. Thus Pc2 may represent a platform which recruits multiple enzymatic activities to specific sub-nuclear domains. Pc2 can be incorporated into the PRC1 polycomb complex, and recent evidence suggests that differing versions of this complex may exist, based in part on their inclusion of different Cbx proteins 16. This may represent a mechanism to recruit different complements of enzymatic activities to PRC1 complexes.
In summary, we show that Pc2 can recruit both CtBP1 and Akt1, resulting in the stimulation of CtBP1 phosphorylation by Akt1. Our data support a model in which, once phosphorylated at T176, CtBP is subject to ubiquitylation and proteasome-mediated degradation (Fig S4). This work suggests a novel mechanism by which Akt1 activity is regulated within the nucleus, and demonstrates a further level of regulation of CtBP1 function.
Materials and Methods
Cell Culture
Ctbp1/2 double null mouse embryonic fibroblasts (MEFs) were a gift of J. Hildebrandt, and were cultured in DMEM with 10% FBS. COS1, HeLa and A549 cells were cultured in DMEM with 10% bovine growth serum. 293T cells were maintained in DMEM with 5% FBS. COS1 and 293T were transfected with LipofectAMINE. MEFs were transfected with LipofectAMINE 2000. HeLa and A549 were transfected with Exgen.
Plasmids
Akt1, Pc2, and CtBP1 constructs were expressed from modified pCMV5 plasmids with amino-terminal T7, Flag, His6, or HA tags. T7-ubiquitin was expressed from pCMV5, with an amino-terminal T7 tag, as a processed ubiquitin, terminating in the di-glycine motif. Fluorescent protein fusions were expressed from pCS2 with amino-terminal fusions to eCFP or eYFP. CtBP1 constructs used in luciferase assays were expressed from pCS2. The (Gal)5-SV40-luc reporter and Gbd-TGIF(1-42) are as described 33. The E-cadherin luciferase reporter contains a region from the mouse E-cadherin promoter (−178 to +91) in pGL3. GBD-CtBP1 expression construct were created in pM (Clontech), with an amino-terminal GBD fusion. Mutations were introduced by standard PCR techniques and verified by sequencing.
Cobalt Affinity Purification and Cell Fractionation
Cells were lysed in 6M guanidine-HCl, 50mM NaH2PO4 (pH 8.0), 10mM Tris-HCl pH 8.0, 100mM NaCl. His6 tagged proteins were bound to Talon Resin (Clontech) at room temperature. Resin was washed 3 times for 30 min with 8M Urea, 50mM NaH2PO4 pH 7.0, 100mM NaCl. Transfected COS1 cells were permeabilized as described in 19.
Coimmunoprecipitation and Western blotting
COS1 cells were lysed in PBS with 1% NP-40 (PBSN) and protease inhibitors (Roche). After centrifugation, lysates were immunoprecipitated with Flag-agarose (Sigma) for 2h at 4°C. Beads were washed 3 times with PBS-N and proteins were separated by SDS-PAGE and transferred to Immobilon-P (Millipore). Proteins were visualized using ECL (Amersham). Antibodies for tagged proteins were against Flag M2 (Sigma), T7 and His6 (Novagen), and HA (12CA5; UVA lymphocyte culture center). Antibodies against Akt1, Akt1 phosphorylated on T308 and a phosphorylated Akt substrate site (RxRxxpS/T) were from Cell Signaling. The anti-CtBP1 antibody was from BD Biosciences, and the anti ubiquitin antibody from Cell Signaling. Blots using phospho-antibodies were blocked in TBS, 0.2% TWEEN-20, 1% BSA, with phosphatase inhibitors. For quantification, blots were incubated with a rabbit Akt1 phospho-T308 antibody and an Alexa goat anti-rabbit secondary antibody, and quantified using a Licor Odyssey infrared imager. For fractionation, transfected COS-1 cells were permeabilized with Transport Buffer (TB: 20mM HEPES, pH 7.4, 110mM K Acetate, 2mM Mg Acetate, 0.5mM EGTA) with 0.005% digitonin, and protease and phosphatase inhibitors, for 5 minutes. The cytosol fraction was removed and cells were washed twice with TB, prior to solubilization in TB with 1% NP40. NP40 soluble and pellet fractions were separated by centrifugation.
Luciferase Assays
Cells were lysed in passive lysis buffer 48 hours after transfection. Where shown, one-third of the lysate was subjected to western blotting with a CtBP1 antibody, and two-thirds were assayed for luciferase activity with an Berthold LB953 luminometer. A Renilla luciferase reporter (phCMV-RL; Promega) was included in all transfections as a control. Significance was tested by Student's T test.
Live Cell Imaging
COS1 cells were plated onto 4-well chamber slides and transfected with eYFP and eCFP fusions using Fugene 6 (Roche). After 24 hr, cells were imaged with a Zeiss Axiovert 135T inverted fluorescence microscope with YFP and CFP filter sets. Images were converted to 8 bit .tif files using Openlab and manipulated in Photoshop 9.
Mass spectrometry
COS1 cells were transfected with CtBP1, Pc2 and Akt expression constructs, lysed directly in 6M guanidine HCl, and the CtBP1 and Pc2 purified on cobalt agarose. Samples were washed six times in 8M urea buffer as above. 10% of each sample was removed and analyzed by western blotting, and the remaining samples were transferred to 50 mM ammonium bicarbonate digested overnight at 37 °C and evaporated to 15 μL for MS analysis. The LC-MS system consisted of a Finnigan LTQ-FT mass spectrometer system with a Protana nanospray ion source interfaced to a self-packed 8 cm × 75 um id Phenomenex Jupiter 10 um C18 reversed-phase capillary column. 0.5-5 μL volumes of the extract were injected and the peptides eluted from the column by an acetonitrile/0.1 M acetic acid gradient at a flow rate of 0.25 μL/min. The nanospray ion source was operated at 2.8 kV. The digest was analyzed using the double play capability of the instrument acquiring full scan mass spectra to determine peptide molecular weights and product ion spectra to determine amino acid sequence in sequential scans. This mode of analysis produces approximately 1200 CAD spectra of ions ranging in abundance over several orders of magnitude. Data were analyzed by database searching using the Sequest search algorithm against Human IPI and CtBP1 and Pc2 protein sequences.
Supplementary Material
Acknowledgements
We thank M.W. Mayo and B.M. Paschal for advice and reagents, and B.A. Hemmings for providing plasmids, and J. Hildebrand for the Ctbp1/2 double null MEFs. This work was supported by NIH grant, HD39926, to D.W.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chinnadurai G. CtBP, an unconventional transcriptional corepressor in development and oncogenesis. Mol Cell. 2002;9:213–24. doi: 10.1016/s1097-2765(02)00443-4. [DOI] [PubMed] [Google Scholar]
- 2.Schaeper U, Subramanian T, Lim L, Boyd JM, Chinnadurai G. Interaction between a cellular protein that binds to the C-terminal region of adenovirus E1A (CtBP) and a novel cellular protein is disrupted by E1A through a conserved PLDLS motif. J Biol Chem. 1998;273:8549–52. doi: 10.1074/jbc.273.15.8549. [DOI] [PubMed] [Google Scholar]
- 3.Turner J, Crossley M. Cloning and characterization of mCtBP2, a co- repressor that associates with basic Kruppel-like factor and other mammalian transcriptional regulators. Embo J. 1998;17:5129–40. doi: 10.1093/emboj/17.17.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hildebrand JD, Soriano P. Overlapping and unique roles for C- terminal binding protein 1 (CtBP1) and CtBP2 during mouse development. Mol Cell Biol. 2002;22:5296–307. doi: 10.1128/MCB.22.15.5296-5307.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi Y, Sawada J, Sui G, Affar el B, Whetstine JR, Lan F, Ogawa H, Luke MP, Nakatani Y. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature. 2003;422:735–8. doi: 10.1038/nature01550. [DOI] [PubMed] [Google Scholar]
- 6.Kumar V, Carlson JE, Ohgi KA, Edwards TA, Rose DW, Escalante CR, Rosenfeld MG, Aggarwal AK. Transcription corepressor CtBP is an NAD(+)-regulated dehydrogenase. Mol Cell. 2002;10:857–69. doi: 10.1016/s1097-2765(02)00650-0. [DOI] [PubMed] [Google Scholar]
- 7.Kuppuswamy M, Vijayalingam S, Zhao LJ, Zhou Y, Subramanian T, Ryerse J, Chinnadurai G. Role of the PLDLS-binding cleft region of CtBP1 in recruitment of core and auxiliary components of the corepressor complex. Mol Cell Biol. 2008;28:269–81. doi: 10.1128/MCB.01077-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao LJ, Kuppuswamy M, Vijayalingam S, Chinnadurai G. Interaction of ZEB and histone deacetylase with the PLDLS-binding cleft region of monomeric C-terminal binding protein 2. BMC Mol Biol. 2009;10:89. doi: 10.1186/1471-2199-10-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–45. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 10.Fischle W, Wang Y, Jacobs SA, Kim Y, Allis CD, Khorasanizadeh S. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 2003;17:1870–81. doi: 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacobs SA, Khorasanizadeh S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science. 2002;295:2080–3. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- 12.Min J, Zhang Y, Xu RM. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev. 2003;17:1823–8. doi: 10.1101/gad.269603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nielsen PR, Nietlispach D, Mott HR, Callaghan J, Bannister A, Kouzarides T, Murzin AG, Murzina NV, Laue ED. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature. 2002;416:103–7. doi: 10.1038/nature722. [DOI] [PubMed] [Google Scholar]
- 14.Satijn DP, Olson DJ, van der Vlag J, Hamer KM, Lambrechts C, Masselink H, Gunster MJ, Sewalt RG, van Driel R, Otte AP. Interference with the expression of a novel human polycomb protein, hPc2, results in cellular transformation and apoptosis. Mol Cell Biol. 1997;17:6076–86. doi: 10.1128/mcb.17.10.6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wotton D, Merrill JC. Pc2 and SUMOylation. Biochem Soc Trans. 2007;35:1401–4. doi: 10.1042/BST0351401. [DOI] [PubMed] [Google Scholar]
- 16.Dietrich N, Bracken AP, Trinh E, Schjerling CK, Koseki H, Rappsilber J, Helin K, Hansen KH. Bypass of senescence by the polycomb group protein CBX8 through direct binding to the INK4A-ARF locus. Embo J. 2007;26:1637–48. doi: 10.1038/sj.emboj.7601632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levine SS, Weiss A, Erdjument-Bromage H, Shao Z, Tempst P, Kingston RE. The core of the polycomb repressive complex is compositionally and functionally conserved in flies and humans. Mol Cell Biol. 2002;22:6070–8. doi: 10.1128/MCB.22.17.6070-6078.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kagey MH, Melhuish TA, Powers SE, Wotton D. Multiple activities contribute to Pc2 E3 function. Embo J. 2005;24:108–19. doi: 10.1038/sj.emboj.7600506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kagey MH, Melhuish TA, Wotton D. The polycomb protein Pc2 is a SUMO E3. Cell. 2003;113:127–37. doi: 10.1016/s0092-8674(03)00159-4. [DOI] [PubMed] [Google Scholar]
- 20.Li B, Zhou J, Liu P, Hu J, Jin H, Shimono Y, Takahashi M, Xu G. Polycomb protein Cbx4 promotes SUMO modification of de novo DNA methyltransferase Dnmt3a. Biochem J. 2007 doi: 10.1042/BJ20061873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Long J, Zuo D, Park M. Pc2-mediated sumoylation of Smad- interacting protein 1 attenuates transcriptional repression of E-cadherin. J Biol Chem. 2005;280:35477–89. doi: 10.1074/jbc.M504477200. [DOI] [PubMed] [Google Scholar]
- 22.Roscic A, Moller A, Calzado MA, Renner F, Wimmer VC, Gresko E, Ludi KS, Schmitz ML. Phosphorylation-dependent control of Pc2 SUMO E3 ligase activity by its substrate protein HIPK2. Mol Cell. 2006;24:77–89. doi: 10.1016/j.molcel.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 23.Altomare DA, Testa JR. Perturbations of the AKT signaling pathway in human cancer. Oncogene. 2005;24:7455–64. doi: 10.1038/sj.onc.1209085. [DOI] [PubMed] [Google Scholar]
- 24.Alessi DR, James SR, Downes CP, Holmes AB, Gaffney PR, Reese CB, Cohen P. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Balpha. Curr Biol. 1997;7:261–9. doi: 10.1016/s0960-9822(06)00122-9. [DOI] [PubMed] [Google Scholar]
- 25.Stephens L, Anderson K, Stokoe D, Erdjument-Bromage H, Painter GF, Holmes AB, Gaffney PR, Reese CB, McCormick F, Tempst P, Coadwell J, Hawkins PT. Protein kinase B kinases that mediate phosphatidylinositol 3,4,5-trisphosphate-dependent activation of protein kinase B. Science. 1998;279:710–4. doi: 10.1126/science.279.5351.710. [DOI] [PubMed] [Google Scholar]
- 26.Hresko RC, Mueckler M. mTOR.RICTOR is the Ser473 kinase for Akt/protein kinase B in 3T3-L1 adipocytes. J Biol Chem. 2005;280:40406–16. doi: 10.1074/jbc.M508361200. [DOI] [PubMed] [Google Scholar]
- 27.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 28.Andjelkovic M, Alessi DR, Meier R, Fernandez A, Lamb NJ, Frech M, Cron P, Cohen P, Lucocq JM, Hemmings BA. Role of translocation in the activation and function of protein kinase B. J Biol Chem. 1997;272:31515–24. doi: 10.1074/jbc.272.50.31515. [DOI] [PubMed] [Google Scholar]
- 29.Meier R, Alessi DR, Cron P, Andjelkovic M, Hemmings BA. Mitogenic activation, phosphorylation, and nuclear translocation of protein kinase Bbeta. J Biol Chem. 1997;272:30491–7. doi: 10.1074/jbc.272.48.30491. [DOI] [PubMed] [Google Scholar]
- 30.Saji M, Vasko V, Kada F, Allbritton EH, Burman KD, Ringel MD. Akt1 contains a functional leucine-rich nuclear export sequence. Biochem Biophys Res Commun. 2005;332:167–73. doi: 10.1016/j.bbrc.2005.04.109. [DOI] [PubMed] [Google Scholar]
- 31.Trotman LC, Alimonti A, Scaglioni PP, Koutcher JA, Cordon-Cardo C, Pandolfi PP. Identification of a tumour suppressor network opposing nuclear Akt function. Nature. 2006;441:523–7. doi: 10.1038/nature04809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–74. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Melhuish TA, Wotton D. The interaction of C-terminal binding protein with the Smad corepressor TG-interacting factor is disrupted by a holoprosencephaly mutation in TGIF. J Biol Chem. 2000 doi: 10.1074/jbc.C000416200. [DOI] [PubMed] [Google Scholar]
- 34.Zhang CL, McKinsey TA, Lu JR, Olson EN. Association of COOH-terminal-binding protein (CtBP) and MEF2- interacting transcription repressor (MITR) contributes to transcriptional repression of the MEF2 transcription factor. J Biol Chem. 2001;276:35–9. doi: 10.1074/jbc.M007364200. [DOI] [PubMed] [Google Scholar]
- 35.Sewalt RG, Gunster MJ, van der Vlag J, Satijn DP, Otte AP. C-Terminal binding protein is a transcriptional repressor that interacts with a specific class of vertebrate Polycomb proteins. Mol Cell Biol. 1999;19:777–87. doi: 10.1128/mcb.19.1.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quinlan KG, Nardini M, Verger A, Francescato P, Yaswen P, Corda D, Bolognesi M, Crossley M. Specific recognition of ZNF217 and other zinc finger proteins at a surface groove of C-terminal binding proteins. Mol Cell Biol. 2006;26:8159–72. doi: 10.1128/MCB.00680-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin X, Sun B, Liang M, Liang YY, Gast A, Hildebrand J, Brunicardi FC, Melchior F, Feng XH. Opposed regulation of corepressor CtBP by SUMOylation and PDZ binding. Mol Cell. 2003;11:1389–96. doi: 10.1016/s1097-2765(03)00175-8. [DOI] [PubMed] [Google Scholar]
- 38.Barnes CJ, Vadlamudi RK, Mishra SK, Jacobson RH, Li F, Kumar R. Functional inactivation of a transcriptional corepressor by a signaling kinase. Nat Struct Biol. 2003;10:622–8. doi: 10.1038/nsb957. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Q, Yoshimatsu Y, Hildebrand J, Frisch SM, Goodman RH. Homeodomain interacting protein kinase 2 promotes apoptosis by downregulating the transcriptional corepressor CtBP. Cell. 2003;115:177–86. doi: 10.1016/s0092-8674(03)00802-x. [DOI] [PubMed] [Google Scholar]
- 40.Chinnadurai G. Transcriptional regulation by C-terminal binding proteins. Int J Biochem Cell Biol. 2007;39:1593–607. doi: 10.1016/j.biocel.2007.01.025. [DOI] [PubMed] [Google Scholar]
- 41.Millward TA, Zolnierowicz S, Hemmings BA. Regulation of protein kinase cascades by protein phosphatase 2A. Trends Biochem Sci. 1999;24:186–91. doi: 10.1016/s0968-0004(99)01375-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.