Abstract
Herein we describe a platform for degradomic-peptidomic analyses. The human blood peptidome was isolated through application of AC/SEC, which enriched its components by >300-fold. The isolated peptidome components were separated by the long column HRLC providing a peak capacity of ~300 for species having MWs of up to 20 kDa. The separated species were identified by the FT MS/MS-UStags sequencing method. We identified >200 peptidome peptides that originated from 29 protein substrates from the blood plasma of a single healthy person. The peptidome peptides identified had MWs range of 0.5–14 kDa and identifications were achieved with extremely low (near zero) false discovery rates through searching the IPI human protein database (~70,000 entries). Some of the peptidome peptides identified have mutations and modifications such as acetylation, acetylhexosamine, amidation, cysteinylation, didehydro, oxidation, and pyro-glu. The capabilities described enable the global analysis of the peptidome peptides to identify degradome targets such as degradome proteases, proteases inhibitors, and other relevant substrates, the cleavage specificities for the degradation of individual substrates, as well as a potential basis for using the various extents of substrate degradation for diagnostic purposes.
INTRODUCTION
The human degradome (1) contains more than 500 proteases (2) responsible for protein degradation to control protein quality and functions. Aberrant degradation of proteins is associated with pathological states such as tumor progression, invasion, and metastasis (3). Protein degradation products, embracing intracellular and/or intercellular peptides, make up the peptidome. The peptidome has been previously explored for clinical diagnosis purposes (4) due to the expectation that the peptidome is a result of both physiology- and pathology-related proteolytic activities.
Human blood is attractive for diagnostic purposes as it is easily sampled. The use of blood peptidomics for diagnostic purposes, however, is controversial. The major issue is whether the peptidome peptides, the targets of blood peptidomics studies (5–10), is of practical value for the clinical diagnosis of disease due to potential issues with the stability of its components (11), and thus issues associated with sample collection, processing, storage, etc. However, a key fundamental issue is whether the blood has detectable peptidome-degradome that can in fact reflect changes due to the perturbations introduced by specific disease states since most blood peptidome peptides reported (5–10) are products from the degradation of common blood proteins. If such a distinctive degradome does exist, then we believe it should be feasible to develop suitable controls, sample handling and processing, etc. to enable blood degradome peptidomics analyses that could potentially identify more effective targets (including proteases and relevant protease activators and inhibitors, and their substrates) for both diagnostic and therapeutic purposes.
A key step in this direction is the capability for global peptidomic measurements to obtain an unbiased view of the degradome. The most broadly applied MALDI-TOF MS and SELDI-TOF MS platforms, mainly previously used for “top-down” analysis of the blood peptidome (5), provide only limited coverage. For example, to date only ~40 proteins in total [compiled from 27 publications (5–7, 9, and references therein)] have been reported as human blood degradome substrates. LC-MS/MS approaches used for “bottom-up” proteomics analysis (12), have also been applied to study the blood peptidome (13), enabling identification of many blood peptidome peptides. The peptides identified are generally of the same size range as found by bottom-up measurements, and it is difficult to evaluate the confidence of peptides identified as false identification rates were not reported (13).
To date no LC-MS/MS approach optimized for the global analysis of the blood peptidome and its larger peptides has been reported. LC-MS/MS challenges include the larger size of peptidome peptides, and obtaining high LC efficiency for separation of the large peptides. Also problematic is the use of conventional methods for identification of the larger peptides (e.g., with charge states of >+3), and effectiveness remains uncertain for MS/MS identification of peptidome peptides in searching against large protein databases (e.g., the IPI human protein database contains ~70,000 entries) without the use of specific “enzyme rules”, although such concerns are well-recognized (14–16). While only slow progress his being made in the development of “top-down” proteomics approaches (17), peptidomics is more practical due to the intermediate and more modest size range of constituents.
Recently, we described a high-resolution FT MS/MS-based approach for degradome analyses (18) that combined efficient and high-resolution LC (HRLC) separations and FT MS/MS (19) with the UStags method for confident identification of the peptidome peptides (20). In this study we apply AC/SEC approaches for depletion of abundant blood proteins (21) and enrichment of the small to medium size blood plasma peptidome components in conjunction with de novo sequencing for identification of peptidome peptide modifications and mutations (22). We demonstrate that this integrated AC/SEC-HRLC-FT MS/MS-UStags strategy provides efficient isolation and separation of the blood plasma peptidome and confident identification of both small and large peptidome peptides, both with and without modifications and mutations. The present work is a prelude to a broader study of blood plasma for early-stage breast cancer patients providing new degradome insights, and a potential spur for the refinement of clinical sample processing approaches and much broader study scope needed to evaluate the true clinical potential of blood plasma peptidome measurements.
EXPERIMENTAL PROCEDURES
Isolation of the human blood peptidome using AC/SEC
The human plasma used for estimation of the AC/SEC-LC-FT MS/MS-UStags platform was obtained from a single, healthy volunteer (Stanford University School of Medicine, Palo Alto, CA). Approval for the conduct of this study was obtained from the Institutional Review Boards of the Stanford University School of Medicine and the Pacific Northwest National Laboratory in accordance with federal regulations.
The plasma sample was first depleted of its 12 abundant proteins using a 12.7 × 79.0 mm IgY12 LC10 AC column (Beckman Coulter, Fullerton, CA) on an Agilent 1100 series HPLC system (Agilent, Palo Alto, CA) according to the manufacturer’s recommendations and previously described method (23). Isolation of peptidome was accomplished using a Superdex 200 10/300 GL SEC column (GE Healthcare, Piscataway, NJ) and an Agilent 1100 series HPLC system equipped with a UV detector. The SEC column was calibrated with various sizes of standard proteins (Sigma-Aldrich, St. Louis, MO), including E. coli beta-galactosidase (464 kDa), rabbit phosphorylase (97 kDa), bovine serum albumin (66.43 kDa), bovine carbonic anhydrase (29 kDa), bovine beta-lactoglobulin (18.4 kDa), and bovine cytochrome c (12.4 kDa). Based on a calibrated elution time, 12.5 mg (protein content) depleted sample (i.e., the AC flow through fraction) was separated and species <20 kDa were collected, denatured for 1 h at 37 °C with 8 M urea, and diluted to 1.3 M urea with 50 mM NH4HCO3 (pH 8.0). The resulting solution was concentrated in Amicon® Ultra-15 filters (3 kDa nominal MW limit, Millipore, Billerica, MA) followed by buffer exchange to 50 mM NH4HCO3 (pH 8.0). The isolated peptidome component content was determined by BCA assay (Pierce, Rockford, IL), which indicated ~600 μg of the denatured <20 kDa species were obtained from a 12.5 mg depleted sample after SEC and cleanup steps.
HRLC-FT MS/MS experiments
HRLC was performed on a 100 cm × 100 μm i.d. LC column containing C4-bonded particles (3- μm, 300 Å size, Sepax Technologies, Inc. Newark, DE) using a 20,000-psi LC system (19). The sample (2.3 μg/μL) was injected onto the LC column, and separated with a gradient generated by replacing mobile phase A (acetonitrile/H2O/acetic acid, 10:90:0.2, v/v/v) with B (acetonitrile/isopropyl alcohol/H2O/acetic acid/trifluoroacetic acid, 60:30:10:0.2:0.1, v/v/v/v/v) in a static mixer at a constant pressure of 13,000 psi.
An LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific, San Jose, CA) was used for analysis of the LC eluent. The heated capillary temperature and spray voltage were held at 300 °C and 2.0 kV, respectively. The FT MS and FT MS/MS data were collected with AGC targets of 1×106 and 3×105, respectively. Spectra were acquired at a 60K resolution, using a survey scan with 400 ≤m/z ≤ 2000 followed by FT MS/MS of the 5 most intense ions from the survey scan (monoisotopic precursor selection not enabled). FT MS/MS employed an isolation window of 3 m/z units and 35% normalized collision energy for CID. Dynamic exclusion was enabled with no repeat counts, using a mass window of ±3 m/z units and a duration cycle of 30 s. FT MS and MS/MS measurements with 1 and 3 micro scans were selected, respectively, for two HRLC-FT MS/MS analyses. Mass calibration was performed according to the method provided by the instrument manufacturer.
Data analysis
Both database searching-UStag and de novo sequencing-UStag methods were employed for FT MS/MS identification of the human blood peptidome peptides from the IPI human protein database, and are described in the Supplementary Notes.
RESULTS
Effectiveness of AC/SEC for isolation of the blood peptidome
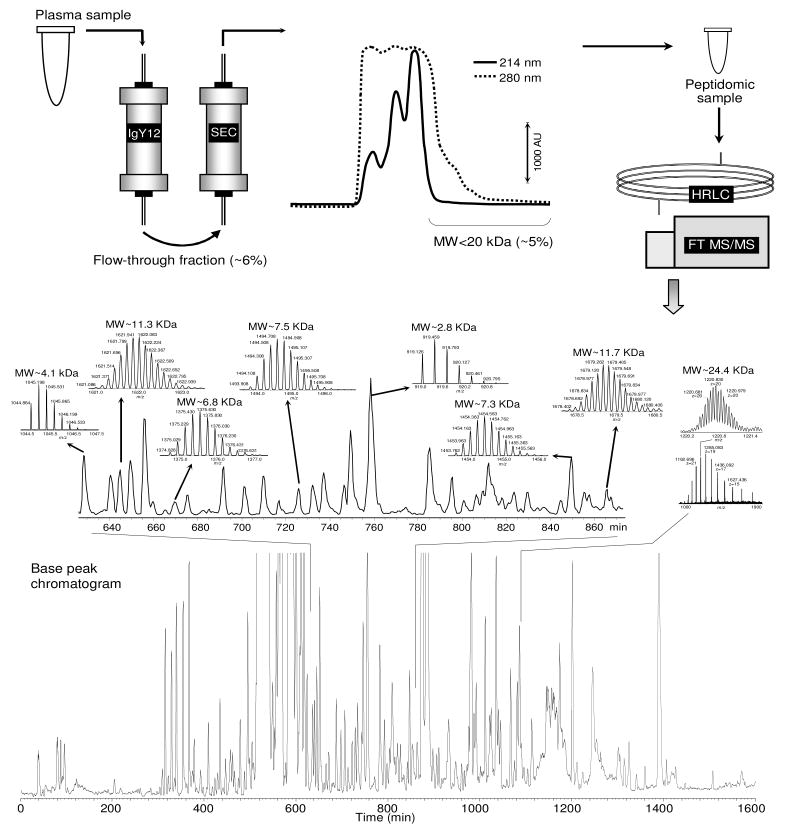
Figure 1 summarizes the isolation/separation performances for blood peptidome components. The combined AC/SEC approach enriched peptidome components ~330-fold [i.e. 1/(6% × 5%)]. The IgY12 AC column removed the 12 highest abundance proteins, which represent 94% of the human blood proteins (23). Subsequent SEC using a 20 kDa-cutoff resulted in isolating ~5% of the protein mass (i.e., 0.6 μg/12.5 μg, see experimental section) from the AC-depleted sample. Peak tailing across the 20 kDa-cutoff (evident at 280 nm) revealed that some components with MWs >20 kDa may still be present in the isolated sample.
Figure 1.
AC/SEC-HRLC isolation and separation of the peptidome components from the human blood plasma. The plasma elutes through an AC column to deplete the top 12 abundant proteins with a yield of ~6% for the flow-through fraction. Next, the flow-through fraction is separated through a SEC column with a yield of ~5% for species cut off by <20 kDa. Finally, the SEC-isolated components are separated using HRLC with a separation peak capacity of ~300. These features and experimental conditions are further detailed in the text.
Efficiencies of HRLC for separation of the blood peptidome
The isolated peptidome components were separated on a 100-cm packed capillary LC column. The HRLC-FT MS base peak chromatogram (bottom Figure 1) shows an average base peak width of ~5 min (see expanded portion of the chromatogram) for components with MWs of 2-~12 kDa across the separation. Similar peak widths and shapes were also observed for species up to 24 kDa. We estimated a peak capacity of ~300 (i.e., 1600/5), the LC highest peak capacity reported to date for such large peptides, and similar peak capacities were observed for 200–2000 min long separations (data not shown).
Identification of the blood peptidome both small and large peptides
We searched the whole FT MS/MS dataset collected (http://www.ebi.ac.uk/pride/) against the IPI human database (ftp://ftp.ebi.ac.uk/pub/databases/IPI/), and identified 181 unmodified peptides (Supplementary Table) for the tested sample. These unmodified peptidome peptides originate from 24 protein substrates (Table 1). Additionally, we identified 43 peptidome modified peptides (Supplementary Table) with the UNIMOD modification categories (http://www.unimod.org/) including acetylation, acetylhexosamin, amidation, cysteinylation, didehydro, hydroxylation (oxidation), pyro-glu, and mutations (Table 2). The most often observed were oxidation of Arg, Lys, Met, Phe, and Pro residues, and these oxidized proteins are the result of either post-translational modifications (e.g., on Arg, Lys, and Pro) or artifacts (e.g., on Met and Phe). The blood peptidome peptides identified primarily have MWs in the range of 1000–8000 Da (Figure 2A), and most are >3500 Da and also have identified modifications and mutations (see below) that were absent in the previous blood peptidome studies (5,6,13). The much less biased analyses obtained here for various peptide sizes allows more quantitative characterization of the substrate cleavage specificities (Figure 2B), key insights obtainable from degradome analyses (described below).
Table 1.
Human blood degradome substrates corresponding to unmodified peptidome peptides identified using the AC/SEC-HRLC-FT MS/MS-UStags method.*
| Human plasma protein | Reported or new | Number of fragment | L1 | L2/MW | Concentration (μg/mL) |
|---|---|---|---|---|---|
| Alpha-1,2-mannosidase IA | New | 4 | 36 | 653/73 | --- |
| Alpha-1-antichymotrypsin | New | 5 | 40 | 95/11 | ~500 (5) |
| Alpha-1B-glycoprotein | New | 1 | 22 | 326/35 | 150–300 (5) |
| Alpha-2-antiplasmin | Yes | 1 | 30 | 491/55 | 65–70 (7) |
| Alpha-2-HS-glycoprotein | New | 2 | 28 | 367/39 | ~600 (5) |
| Angiotensinogen | New | 1 | 18 | 485/53 | 28–71 (37) |
| Antithrombin III variant | New | 3 | 35 | 259/29 | 120 (38) |
| Apolipoprotein A-IV | Yes | 46 | 67 | 396/45 | 130–250 (5) |
| Ceruloplasmin | New | 3 | 40 | 1065/122 | 200–600 (5) |
| Complement C3 | Yes | 13 | 47 | 1663/187 | 900–1800 (5) |
| Complement C4 | Yes | 4 | 17 | 1774/193 | 640 (7) |
| Complement C9 | New | 1 | 12 | 559/63 | 61±14 (39) |
| Fibrinogen α | Yes | 21 | 56 | 644/69 | 2000–4000 (5) |
| Gelsolin | New | 1 | 29 | 485/52 | 265–367 (40) |
| Inter-alpha-trypsin inhibitor H4 | Yes | 24 | 102 | 644/71 | 400–700 (5) |
| Kininogen-1 (HMW) | Yes | 8 | 53 | 644/72 | 92±15 (41) |
| Kininogen-1 (LMW) | New | 1 | 15 | 427/48 | 89.9 (42) |
| Leucine-rich alpha-2-glycoprotein | New | 1 | 59 | 347/38 | 50 (43) |
| Multimerin-1 | New | 1 | 33 | 531/58 | --- |
| Pigment epithelium-derived factor | New | 26 | 59 | 216/25 | 4.38±0.59 (44) |
| Plasma serine protease inhibitor | New | 1 | 33 | 406/46 | 5.3±1.7 (45) |
| Thymosin beta-4 | New | 2 | 29 | 44/5 | --- |
| Transthyretin | Yes | 8 | 51 | 147/16 | 200–400 (5) |
| Vitamin D-binding protein | New | 3 | 18 | 474/53 | 226–272 (46) |
Reported: published from previous top-down analysis of blood serum and plasma in refs. (5–7); L1: the number of amino acid residues of the longest fragment identified; L2/MW: the number of the protein amino acid residues/molecular weight (kDa); the substrate (protein) concentration information is from the references indicated; ---: data unavailable.
Table 2.
Modified human blood peptidome peptides and LMW proteins identified using the HRLC-FT MS/MS-UStags method.
| Human plasma protein | Modifications |
|---|---|
| Apolipoprotein A-IV | Pro–oxidation |
| Beta-2-microglobulin | Cys–dehydro; Met–oxidation |
| C1 esterase inhibitor | Pro–oxidation; Met–oxidation |
| Complement C4 | Gly–amidation |
| Complement factor D | Cys–dehydro |
| Fibrinopeptide B | Gln–dehydration |
| Fibrinogen alpha chain | Lys–oxidation; Tyr- oxydation Met–oxidation; Phe–oxidation |
| Inter-alpha-trypsin inhibitor heavy chain H4 | Met–oxidation; Arg–oxidation; Phe–oxidation; Ser–acetylhexosamine; Gln–pyro–glu; Ser–acetylation |
| Kininogen-α (HMW) | Thr–acetylhexosamine |
| Pigment epithelium-derived factor | Met–oxidation |
| Plasma serine protease inhibitor | Arg–oxidation; Pro–oxidation |
| Plasminogen | Pro–oxidation |
| Thymosin beta-4 | Ser–acetylation; Met–oxidation |
| Transthyretin | Cys–cysteinylation |
Figure 2.
The molecular size (MW) distribution of the blood peptidome peptides identified (A) and an example for the substrate cleavage specificity determined for apoA-IV (B). A) The blue and red lines represent the blood plasma peptidome peptides identified in this work and a previous study (ref. 13), respectively. B The sequence encased in red is the signal peptide; the detected peptides are represented by the yellow curves that start from Glu21, and cleavage specificities are presented at P1⊥P1′.
We present examples below of identified blood plasma peptidome larger peptides and their modified forms not reported using previously described methods. Figure 3A shows a ADEFKVKID sequence obtained from an FT MS/MS spectrum (and associated mass uncertainties). The observed sequence is a UStag for apoA-IV (IPI database). The UStag is constructed by y9–y17 (labeled in the figure) counted from apoA-IV Arg201. The UStag prefix, measured from the UStag-containing smallest fragment (y9), agrees with theoretical prediction for the specified protein (i.e., within the mass error tolerance of ≤10 ppm or sequence mass error of ≤0.005 u). Similarly, the UStag suffix, measured from the difference between the precursor and the largest UStag-containing fragment (y17), agrees with that predicted. Agreements for both prefix and suffix of the UStag with the database predictions allow us to assign the spectrum for apo A-IV Val147-Arg201 has having a MW of ~6 kDa, and with charge state (CS, z) of 7 for the peptide dissociated.
Figure 3.
Examples of using the FT MS/MS-UStags method for identification of non-modified (A) and modified (B) peptidome peptides. A) The UStag sequenced from the FT MS/MS spectrum is black color-circled; the UStag-containing smallest fragment (m/z 515.785) is aligned as y8 (m/z 515.786) of the UStag-assigned peptide, and the precursor isotopic peaks agree with those of the assigned peptide. Black peaks and numbers represent measurements and red, predictions of the assigned-peptide sequence. B) The UStag sequenced is black color-circled, and the mass difference measured from the precursor and the UStag-containing largest fragment (m/z 1218.864) predicts the UStag suffix as Ser without modification; the mass difference of the UStag-containing smallest fragment (m/z 1026.276) is determined as the result of UStag prefix Ser-acetylation and Met-oxidation.
Figure 3B shows the method for identification of modifications of the peptidome peptides. QEKQAGE was sequenced and it is a UStag of thymosin beta-4, a small protein [MW 5 kDa, (24)]. The UStag suffix [105.050 u, determined from the precursor (CS=5), and the largest fragment] agrees with the predicted Ser. The UStag prefix has a mass shift of 58.002 u from that of the database prediction. This mass shift can be explained either by substitutions Ala→Glu and Gly→Asp, or alternatively also by one of 66 combinations of 2 modifications according to UNIMOD (based upon filtering with a mass tolerance of 0.005 u). Observation of y37 and y38+O (highlighted) led to determination of Met-oxidation (15.995 u) and acetylation (42.010 u) that can occur on Lys, Cys, Ser, Thr, Tyr, His, and N-terminal sites (UNIMOD). Observation of y41+O is consistent with Ser-acetylation protein N-terminal processing (25). Additional examples of modified blood peptidome peptide identifications are given in Supplementary Figures 1–5.
Confidence for blood peptidome peptide identifications
Few previous blood peptidome studies to date have reported false identification rates. The present results display extremely low (near zero) false identifications for blood peptidome peptides even using the large IPI human protein database, as determined using a reversed decoy IPI database (see Supplementary Notes). This is largely attributed to the accurate sequencing (and molecular mass determinations) of amino acid residues, as illustrated in Figure 3. Not a single UStag was obtained from the decoy database search, highlighting the extremely low probability of randomly matching UStags. When UStags obtained were used to identify the non-modified peptidome peptides, as shown in Figure 3A, additional criteria for the UStag prefix and suffix masses were applied, which further ensured confident identification. To this point we have not found the UStags approach to provide false identifications for the non-modified peptidome peptides, even from the IPI human database. When the UStags were used to determine modifications and mutations, the proteins were first-pass specified by the UStags approach. Based on the database predictions, the mass shifts for the modified prefix or suffix regions were determined by alignment of the FT MS/MS-measured fragments and the FT MS-measured parent precursors. These mass shifts were assigned as modifications and mutations according to those listed in UNIMOD, and with manual inspection of spectra for determination of the site(s) of modification(s) and/or mutation(s) in the UStag-specified prefix and suffix regions (all FT spectra used for identifications are provided at http://www.ebi.ac.uk/pride/). The error rates associated with possible uncertainties for the exact sites of modifications and mutations arising from the manual aspects of the analysis have not yet been determined.
Direct measurement of the degradome substrate cleavage specificity
Determination of cleavage specificity is a key aspect of the degradome analysis, and a complex procedure has been developed for this purpose (26). In contrast the present approach provides significant simplicity and precision in characterizing substrate cleavage specificity. As exemplified in Figure 2B for apoA-IV, the cleavage specificity is directly accessed by counting the terminal amino acids of the peptidome peptides identified. The Arg201⊥Ser202 is the major cleavage site where further truncations of the long sequences (e.g., 67-residue sequence Arg135-Arg210) start towards both N- and C-terminal directions. We note that the cleavage specificity for individual degradome substrates may be useful for distinguishing healthy subjects from diseased patients, and needs to be further explored.
Identification of the blood plasma peptidome peptides having MWs of >10 kDa
Figure 4 shows an identification of an 11.5-kDa peptidome peptide originated from inter-α-trypsin inhibitor heavy chain H4. The peptide, carrying 12 charges and having a signal/noise ratio of 3.5, can be confidently identified using the method as described for Figure 3A, showing the method effectiveness for identification of >10-kDa peptidome peptides. Identification of such larger peptidome peptides can possibly reveal the roles of functional protein domains (e.g., propeptides, preactivation peptides, etc.) during degradation.
Figure 4.
Identification of an 11.5 kDa peptidome peptide using the method described in this work. The UStag is black-color circled; the black- and red-colored peaks and numbers represent the measurements and the predictions from the assigned-peptide’s sequence, respectively. The peptide identification is completed with the same method as for Figure 3A.
Discovery of protein sequence database errors
Any reference protein database will have errors or differences in protein sequences. We use β2-M as an example to show the potential for revealing protein database errors. UStag PKIVKWD was sequenced for IPI β2-M (IPI00004656.2) and its similar sequence (IPI00796379.1) (see sequences listed at the top of Figure 5). The precursor, however, is 713 Da smaller than the β2-M theoretical sequence, which could not be explained by UNIMOD modifications, nor by sequence truncations as both b and y fragments were observed (see those labeled). We then speculated that the IPI β2-M sequence was possibly in error and inspected IPI β2- M-similar sequence, and found only small b fragments. Alignment of the UStag to IPI β2-M led to determining a mutation Ile99→Met in IPI β2-M-similar sequence, which produces a sequence that agrees with the human B2M gene prediction for β2-M [ref. (27) or SwissProt β2-M]. With Ile99→Met and disulfide Cys25-Cys80 (28), the precursor and most of the fragments were explained (see labeled peaks). The spectrum for precursor measurement (bottom of the figure) additionally indicated the existence of many different β2-M variants evidenced with β2-M y fragments (see the two spectra inserted). β2-M-Fe were speculated to be responsible for the serial peaks based on different types of information including the mass shift of 52.9 Da that agrees with that of Fe3+ →H3+, the reduced MS/MS spectrum quality that would result from the formation of the complex, and the co-existence of Fe with β2-M (29). β2-M has been found to be correlated to the clinical course of disease (30) and involved in cancer (31), however, the relevance of the β2-M different forms remains to be explored.
Figure 5.
Examples of correcting the IPI human database errors and finding complex modifications on the blood β2-M. Top: Two IPI database sequences for β2-M and a similar sequence; the grey-colored sub-sequence is the protein signal peptide. The green-colored sub-sequence HLIWAIR in β2-M does not agree with the measurements from FT MS/MS. The red-colored sub-sequence TLS and mutation Ile→Met for the β2- M-similar sequence were determined experimentally, and the sequence black color-circled is the protein UStag. Middle: The FT MS/MS spectrum provides evidence of potential errors in the β2-M database sequence. Bottom: An FT MS spectrum and two FT MS/MS spectra show the existence of complex modifications and truncations on the β2-M sequence. Further detail provided in the text.
Identification of complex peptidome mutations
Using TTR as an example, we demonstrate the capability for identifying mutations that have not been previously reported for the blood peptidome analysis. TTR can have 73 mutations (32, 33). Three TTR variants (Figure 6, top) were found in the sample, and Cys10-cysteinylation TTR (variant 1) identified has been reported (34). Other two variants (sequences 2 and 3) had similar UStag sequences (black spheres in the sequences) as variant 1. For variant 2, a mass shift of 57.02 u was observed for both of the parent molecular ions (see spectrum) and the b18 and b44 fragments (see MS/MS spectrum 2). We searched the UNIMOD (filtered with a mass range of 57.020 u ± 0.005 u) with consideration of modifications and mutations, and obtained mutations Gly6→Ser that has been previously reported (32) and Ser8→Asn, not reported yet, for explanation of the mass shift prior to Asp18. Using the same method, TTR variant 3 was determined to have additional mutations, i.e., Ala45→Asp, Gly47→Arg, and Ala97→Gly, all of which have been previously reported (32). TTR mutations have been analyzed in study of cardiac and amyloid diseases (32, 33), although the origin of the three TTR sequences identified here remains uncertain.
Figure 6.
Examples showing three TTR variants in a human blood sample. From top to bottom: the 3 TTR variant sequences identified (the signal peptide is grey-colored); the LC-MS base peaks numbered for the corresponding 3 identified sequences and their FT MS measurements (black-, blue-, and brown-color peaks represent normal sequence 1, variant 2, and variant 3, respectively); and 3 FT MS/MS spectra acquired for the z = 12 precursors shown in the FT MS measurements and used to identify the three sequences. Determination of the three variants containing cysteinylation and mutations is detail in the text.
DISCUSSION
The results shown in Figures 2–6 illustrate that the method described in this work enables analysis of blood plasma peptidome peptides (with or without modifications) over a larger molecular size range than previous reports (6, 13). [The peptides reported in ref. (6) have the similar size range as those from ref. (13).] In fact, the large peptides shown in Figures 3–6 were missed by the previous studies (6, 13). The identifications were achieved with a near zero false identification rate, and without requirements for specific peptide termini (“the enzyme rule”). These features, including the capabilities for identification of the modifications and mutations, as well as probing the database errors, enable realization of a global analysis of the blood peptidome for blood degradome study, e.g., the degradome substrate cleavage specificity (Figure 2B) and the degradome substrate selectivity (not shown here).
More than 200 peptidome peptides that originated from 29 degradome substrates were confidently identified from the blood plasma of a single healthy person. In a separate work to be reported later this method has identified ~1000 peptidome peptides from ~70 degradome substrates from blood plasma pooled from the early-state breast cancer patients (not shown here). This analytical coverage should be mainly contributed from the AC/SEC-HRLC isolation and separation efficiency of the strategy that effectively enriched the peptidome by >300 fold and separated the peptidome components with a peak capacity of ~300. C4-bonded stationary phase was selected since the SEC-isolated peptidome contained some large species which were observed from the C4 column, but not from the C18 (3 μm, 300 Å pores) column. Data collected during sample loading indicated that very few species eluted from the long column. Any possible losses of small hydrophilic peptides could be minimized, if necessary, by reducing the organic solvent content in mobile phase A, e.g., from 10% in this study to 5% or lower. The use of other (e.g., C5, C8 or phenyl) bonded particles as the stationary phase may also be an alternative, but needs to be optimized for the peptidome samples to be analyzed.
The blood peptidome peptides were analyzed without subjecting samples to any chemical reactions during processing. Although the strategy increases the difficulty of identifying some native peptidome components, it provides a powerful capability for identifying the structures of naturally occurring peptidome peptides, e.g., sequences linked by disulfides (will be reported elsewhere). This also reduced the chances of reaction side products (35) from interfering with the spectral interpretation (20). A concern associated with not inhibiting reactions is the possible ex vivo proteolysis of the peptides after sampling, which can be solved by addition of protease inhibitors in the samples prior to sample processing or even during blood sampling.
A preliminary application of this strategy to the blood plasma peptidome of the early-stage breast cancer patients and matched healthy controls has shown striking differences, including the degradation performances of the protease system activity, the substrate selectivity, and the cleavage specificity for individual substrates. Instead of searching for individual peptidome peptides as the targets for investigation of biomarkers, these data suggest the possible utility of degradome features constructed through the unbiased, global analysis of the blood plasma peptidome for diagnostic purposes. The separation and methodology for identification described here is the basis for our future studies in which large numbers samples of clinical applications will be able to be rapidly screened using the accurate mass and time tag strategy (36).
Supplementary Material
Acknowledgments
Portions of this research were supported by the NIH National Center for Research Resources (RR18522) and the U.S. Department of Energy Office of Biological and Environmental Research (DOE/BER). Work was performed in the Environmental Molecular Science Laboratory, a DOE/BER national scientific user facility located on the campus of Pacific Northwest National Laboratory (PNNL) in Richland, Washington. PNNL is a multi-program national laboratory operated by Battelle Memorial Institute for the DOE under contract DE-AC05-76RLO-1830.
Abbreviations
- FT MS/MS
Fourier transform tandem mass spectrometry
- UStags
unique sequence tags
- HRLC
high-resolution LC
- AC
affinity chromatography
- SEC
size exclusion chromatography
- LMW
low molecular weight
- MALDI-TOF
matrix-assisted laser desorption/ionization time of flight
- SELDI
surface-enhanced laser desorption/ionization
- LMW
low molecular weight
References
- 1.Lópezín-Otín C, Overall CM. Nat Rev Mol Cell Biol. 2002;3:509–519. doi: 10.1038/nrm858. [DOI] [PubMed] [Google Scholar]
- 2.Puente XS, Sánchez LM, Overall CM, Lópezín-Otín C. Nat Rev Genet. 2003;4:544–558. doi: 10.1038/nrg1111. [DOI] [PubMed] [Google Scholar]
- 3.Koblinski JE, Ahram M, Sloane BF. Clin Chim Acta. 2000;291:113–135. doi: 10.1016/s0009-8981(99)00224-7. [DOI] [PubMed] [Google Scholar]
- 4.Petricoin EF, Belluco C, Araujo RP, Liotta LA. Nat Rev Cancer. 2006;6:961–967. doi: 10.1038/nrc2011. [DOI] [PubMed] [Google Scholar]
- 5.Hortin GL. Clin Chem. 2006;52:1223–1237. doi: 10.1373/clinchem.2006.069252. [DOI] [PubMed] [Google Scholar]
- 6.Villanueva J, et al. J Clin Invest. 2006;116:271–284. doi: 10.1172/JCI26022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koomem JM, Li D, Xiao LC, Liu TC, Coombes KR, Abbruzzese J, Kobayashi R. J Proteome Res. 2005;4:972–981. doi: 10.1021/pr050046x. [DOI] [PubMed] [Google Scholar]
- 8.Villanueva J, Philip J, DeNoyer L, Tempst P. Nat Protoc. 2007;2:588–602. doi: 10.1038/nprot.2007.57. [DOI] [PubMed] [Google Scholar]
- 9.Villanueva J, Nazarian A, Lawlor K, Yi SS, Robbins RJ, Tempst P. Mol Cell Proteomics. 2008;7:509–518. doi: 10.1074/mcp.M700397-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Z, et al. Cancer Res. 2004;64:5882–5890. doi: 10.1158/0008-5472.CAN-04-0746. [DOI] [PubMed] [Google Scholar]
- 11.Yi J, Kim C, Gelfand CA. J Proteome Res. 2006;6:1768–1781. doi: 10.1021/pr060550h. [DOI] [PubMed] [Google Scholar]
- 12.Washburn MP, Wolters D, Yates JR. Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 13.Bakun M, et al. Proteomics Clin Appl. 2009;3:932–946. doi: 10.1002/prca.200800111. [DOI] [PubMed] [Google Scholar]
- 14.The minimun information about a proteomics experiment (MIAPE) http://www.nature.com/nbt/consult/index.html.
- 15.Mischak H, et al. Proteomic Clin Appl. 2007;1:148–156. doi: 10.1002/prca.200600771. [DOI] [PubMed] [Google Scholar]
- 16.Carr S, et al. Mol Cell Proteomics. 2004;3:531–533. doi: 10.1074/mcp.T400006-MCP200. [DOI] [PubMed] [Google Scholar]
- 17.Cravatt BF, Simon GM, Yates JR., III Nature. 2007;450:991–1000. doi: 10.1038/nature06525. [DOI] [PubMed] [Google Scholar]
- 18.Shen Y, Hixson KK, Tolić N, Camp DG, Purvine SO, Moore RJ, Smith RD. Anal Chem. 2008;80:5819–5828. doi: 10.1021/ac800077w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen Y, Zhang R, Moore RJ, Kim J, Metz TO, Hixson KK, Zhao Z, Livesay EC, Udseth HR, Smith RD. Anal Chem. 2005;77:3090–3100. doi: 10.1021/ac0483062. [DOI] [PubMed] [Google Scholar]
- 20.Shen Y, Tolić N, Hixson KK, Purvine SO, Pasă-Tolić L, Qian WJ, Adkins JN, Moore RJ, Smith RD. Anal Chem. 2008;80:1871–1882. doi: 10.1021/ac702328x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anderson NL, Anderson NG. Mol Cell Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 22.Shen Y, Tolić N, Hixson KK, Purvine SO, Anderson GA, Smith RD. Anal Chem. 2008;80:7742–7754. doi: 10.1021/ac801123p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu T, Qian WJ, Mottaz HM, Gritsenko MA, Norbeck AD, Moore RJ, Purvine SO, Camp DG, II, Smith RD. Mol Cell Proteomics. 2006;5:2167–2174. doi: 10.1074/mcp.T600039-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hannappel E. Ann N Y Acad Sci. 2007;1112:21–37. doi: 10.1196/annals.1415.018. [DOI] [PubMed] [Google Scholar]
- 25.Li X, Chang YH. Proc Natl Acad Sci USA. 1995;92:12357–12361. doi: 10.1073/pnas.92.26.12357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schilling O, Overall CM. Nat Biotech. 2008;26:685–694. doi: 10.1038/nbt1408. [DOI] [PubMed] [Google Scholar]
- 27.Güssow D, Rein R, Ginjaar I, Hochstenbach F, Seemann G, Kottman A, Ploegh HL. J Immunol. 1987;139:3132–3138. [PubMed] [Google Scholar]
- 28.Isenman DE, Painter RH, Dorrington KJ. Proc Nat Acad Sci USA. 1975;72:548–552. doi: 10.1073/pnas.72.2.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miranda CJ, Makui H, Andrews NC, Santos MM. Blood. 2004;103:2847–2849. doi: 10.1182/blood-2003-09-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Traut M, Haufe CC, Eismann U, Deppisch RM, Stein G, Wolf G. Blood Purif. 2007;25:432–440. doi: 10.1159/000110069. [DOI] [PubMed] [Google Scholar]
- 31.Nissen MH, Bjerrum OJ, Plesner T, Wilken M, Rørth M. Clin Exp Immunol. 1987;67:425–432. [PMC free article] [PubMed] [Google Scholar]
- 32.Hou X, Aguilar MI, Small DH. FEBS J. 2007;274:1637–1650. doi: 10.1111/j.1742-4658.2007.05712.x. [DOI] [PubMed] [Google Scholar]
- 33.Hesse A, Altland K, Linke RP, Almeida MR, Saraiva MJM, Steinmetz A, Maisch B. Br Heart J. 1993;70:111–115. doi: 10.1136/hrt.70.2.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kingsbury JS, Laue TM, Klimtchuk ES, Théberge R, Costello CE, Connors LH. J Biol Chem. 2008;283:11887–11896. doi: 10.1074/jbc.M709638200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boja ES, Fales HM. Anal Chem. 2001;73:3576–3582. doi: 10.1021/ac0103423. [DOI] [PubMed] [Google Scholar]
- 36.Shen Y, Strittmatter EF, Zhang R, Metz TO, Moore RJ, Li F, Udseth HR, Smith RD. Anal Chem. 2005;77:7763–7773. doi: 10.1021/ac051257o. [DOI] [PubMed] [Google Scholar]
- 37.Katsurada A, Hagiwara Y, Miyashita K, Satou R, Miyata K, Ohashi N, Navar LG, Kobori H. Am J Physiol Renal Physiol. 2007;293:956–960. doi: 10.1152/ajprenal.00090.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Conard J, Brossatd F, Lie Larsen M, Samama M, Abildgaadr U. Haemostasis. 1983;13:363–368. doi: 10.1159/000214823. [DOI] [PubMed] [Google Scholar]
- 39.Greenstein JD, Peake PW, Charlesworth JA. Clin Exp Immunol. 1996;104:160–166. doi: 10.1046/j.1365-2249.1996.d01-636.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith DB, Janmey PA, Sherwood JA, Howard RJ, Lind SE. Blood. 1988;72:214–218. [PubMed] [Google Scholar]
- 41.Kleniewski J. Thromb Haemost. 1979;42:1045–1055. [PubMed] [Google Scholar]
- 42.Hilgenfeldt U, Puschner T, Riester U, Finsterie J, Hilgenfeldt J, Ritz E. Am J Physiol. 1998;275:F88–93. doi: 10.1152/ajprenal.1998.275.1.F88. [DOI] [PubMed] [Google Scholar]
- 43.Weivoda S, et al. J Immunol Methods. 2008;336:22–29. doi: 10.1016/j.jim.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ogata N, et al. J Clin Endocrinol Metab. 2007;92:1176–1179. doi: 10.1210/jc.2006-2249. [DOI] [PubMed] [Google Scholar]
- 45.Laurell M, Christensson A, Abrahamsson PA, Stenflo J, Lilja H. J Clin Invest. 1992;89:1094–1101. doi: 10.1172/JCI115689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lauridsen AL, Vestergaard P, Nexo E. Clin Chem. 2001;47:753–756. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.