Abstract
Human milk contains large amounts of complex oligosaccharides that putatively modulate the intestinal microbiota of breast-fed infants by acting as decoy binding sites for pathogens and as prebiotics for enrichment of beneficial bacteria. Several bifidobacterial species have been shown to grow well on human milk oligosaccharides. However, little data exists on other bacterial species. In this work we examined 16 bacterial strains belonging to 10 different genera for growth on human milk oligosaccharides. For this propose, we used a chemically-defined medium, ZMB1, which allows vigorous growth of a number gut–related microorganisms in a fashion similar to complex media. Interestingly, Bifidobacterium longum subsp. infantis, Bacteroides fragilis and Bacteroides vulgatus strains were able to metabolize milk oligosaccharides with high efficiency, while Enterococcus, Streptococcus, Veillonella, Eubacterium, Clostridium, and Escherichia coli strains grew less well or not at all. Mass spectrometry-based glycoprofiling of the oligosaccharide consumption behavior revealed a specific preference for fucosylated oligosaccharides by Bifidobacterium longum subsp. infantis and Bacteroides vulgatus. This work expands the current knowledge of human milk oligosaccharides consumption by gut microbes, revealing bacteroides as avid consumer of this substrate. These results provide insight on how human milk oligosaccharides shape the infant intestinal microbiota.
Keywords: human milk oligosaccharides, gut microbiota, chemically-defined medium, ZMB1, MALDI-FTICR-MS, bacteroides
Introduction
Human milk, the sole nourishment for breast-fed infants, is an interesting model of a food shaped by evolution to promote the healthy development of newborns. Of the various components in human milk, oligosaccharides constitute a significant fraction, being the third most abundant molecular species in terms of concentration after lactose and lipids (1). Up to 200 different structures have been defined for human milk oligosaccharides (HMOs) (2). All HMO structures follow the same basic configuration: a lactose core at the reducing end, elongated by N-acetyl-lactosamine units with at least twelve different types of glycosidic bonds, wherein fucose and sialic acid residues are added to terminal positions (3). The linear and branched HMOs vary in size, from three to thirty-two sugars, being mostly fucosylated neutral oligosaccharides (2). Infants digest a minor portion of HMOs present in the breast milk, while a fraction passes undigested through the intestine (4). Different researchers have confirmed that HMOs are resistant to enzymatic hydrolysis from intestinal brush border membrane and pancreatic juices (5, 6). We hypothesize that, given the high concentration of oligosaccharides in human milk, these polymers must have an important role in the well-being of the baby, particularly when considering the evolutionary forces that have shaped the contents of human milk—to ensure the least energy burden on the mother and the greatest survival benefit for the infant (7). One of the functions attributed to HMOs is a role in the development of neonatal intestine. HMOs modulate intestinal cell proliferation and maturation in vitro, suggesting that mucosal barrier of the gut can be affected by these milk components (8). In addition, HMOs are considered a mechanism to protect the newborn against exogenous infections (3). Study of the response of epithelial cells to the presence of of 3’sialyllactose, an acidic HMO, has suggested that the expression of various glycosyltransferases is diminished. Thus, this particular HMO seems able to modify the glycan content of the epithelial cell surface and the receptor sites for some pathogens (9). Moreover, different in vitro studies have demonstrated that HMOs bind and block the infection of pathogenic bacteria to animal cells by acting as receptor analogs to the intestinal cell glycans (10–12). A recent study shows how HMOs can inhibit transfer of HIV-1 to CD4+ lymphocytes (13).
Another hypothesis regarding the bioactive function of HMOs is a role as “prebiotic” (14). Prebiotics are defined as those substances that allow specific changes, both in the composition and/or activity in the gastrointestinal microbiota, conferring benefits upon host health (15). Prebiotics stimulate the growth of the “beneficial” bacteria including bifidobacteria, genus generally predominant in the intestinal microbiota of the breast-bed infants (16). We recently demonstrated that Bifidobacterium longum subsp. infantis ATCC15697 (termed “Bifidobacterium infantis”) grows on HMOs as a sole sugar source while Lactobacillus gasseri, an adult gut isolate, does not (17). To examine this further, we developed a method for quantifying consumption of individual HMOs using matrix-assisted laser desorption/ionization-Fourier transform ion cyclotron resonance mass spectrometry (MALDI-FTICR MS) (2, 18). With this method, we can detect individual neutral oligosaccharides, which represent 90–95% of total HMOs (2). This approach allowed us the determination of the HMO consumption profile of different species belonging to bifidobacterial group (Bifidobacterium breve, Bifidobacterium longum subsp. longum, Bifidobacterium infantis, Bifidobacterium bifidum and Bifidobacterium adolescentis) (19, 20). Comparisons among the selected species and strains revealed significant differences in their consumption profiles. Some of the examined microorganisms show preferences for consuming specific non-fucosylated or non-sialylated oligosaccharide structures, while other strains reveal a broader glycoside consumption profile including fucosylated glycans. Subsequent genomic analysis of a “high consumer” of HMOs, Bifidobacterium infantis ATCC15697, revealed the existence of a 43 kb gene cluster dedicated to HMOs import and processing (7).
After birth, the infant gut undergoes a complex process of microbial colonization. Aerobic microbes initially colonize the intestine of the newborn however as oxygen is consumed, the microbiota switches to anaerobic species, composed mainly by bifidobacteria, and to a lesser extent, bacteroides and clostridia (21). Among the components of human milk, oligosaccharides are believed to directly influence the final microbial composition of the infant gut. As described, we have recently demonstrated that specific strains of bifidobacteria can grow vigorously on HMOs. However, the influence of these oligosaccharides on the growth of other bacterial genera present in the gut during the first months of life remained unexplored. In this work, we use a scalable method previously developed by our group (19), examining 16 strains of bacteria belonging to 10 different genera related to gut microbiota. We add the advantage of using a chemically defined media, termed ZMB1, initially designed for the growth of lactococci, enterococci and streptococci (22). We demonstrate that ZMB1, which does not contain any complex component, is a useful minimum medium for growth of a fairly wide array of intestinal microbes in order to test the specificity of HMOs in vitro. Growth curves and detailed glycoprofile analysis of HMOs consumption using MALDI-FTICR MS, evidence that bifidobacteria and bacteroides strains are able to metabolize this substrate, while other strains, such as clostridia, enterococci or Escherichia coli, were not. The results obtained in this work provide an insight in the selectivity of HMOs, showing for first time bacteroides as a species able to consume these sugars.
Materials and Methods
HMOs purification
The oligosaccharide purification was performed according to the method described by Ward et al. (17). Milk samples were provided by the Milk Bank of San Jose, California, and Austin, Texas.
Bacterial strains and growth conditions
Bacteria listed in Table 1 were tested for growth in the presence of HMO. Based in the results obtained by Ward et al. (17), Lactobacillus gasseri UCD235 was included as a control for low growth in HMOs. Seed cultures of all bacteria were prepared as following. Enterococcus faecalis, Streptococcus thermophilus, Escherichia coli. Lactobacillus acidophilus, Lactobacillus gasseri, and Lactococcus lactis strains were incubated overnight while two days of incubation were necessary for Clostriidum perfringens, C. difficile, Eubacterium rectale, Bifidobacterium infantis, Bacteroides fragilis, Bacteroides vulgatus and Veillonela parvula strains. All bacteria were grown in anaerobic conditions at 37°C, using an anaerobic chamber (Coy Laboratory Products, Grass Lake, MI). Reinforced clostridium medium (RCM) was used for growing clostridia, Bacteroides and Eubacterium rectale, M17 supplemented with 2% Glucose for enterococci, streptococci and lactococci growth, MRS for Lactobacillus and Bifidobacterium infantis strains, BHI for Veillonela, and LB for E. coli strains. All complex media was purchased from Becton Dickinson (Franklin Lakes, NJ). Two µl of each resulting overnight culture were used to inoculate 200 µl of various media distributed in the wells of a microplate. Bacterial growths were tested in the complex media LB, MRS, M17 and RCM, as well as in the chemically-defined medium ZMB1, prepared according to the description of Zhang et al. (22). ZMB1 contains 2% glucose as a sole carbon source. After verifying that ZMB1 allowed growth of the various gut bacteria, each strain was grown overnight in the same medium. Two µl of the overnight cultures was added to 200 µl of modified ZMB1 in which the glucose was replaced with 2% HMO and a another 2 µl inoculated into ZMB1 without added sugar. In all the cases the cultures in the wells of the microtiter plates were covered with 40 µl of sterile mineral oil to avoid evaporation. Cell growth was monitored in real time by assessing optical density at 600 nm using a BioTek PowerWave 340 plate reader (BioTek, Winoosky, VT) every 30 min preceded by 15 seconds shaking at variable speed. Two biological replicates (three technical replicates each) were performed for every studied strain and medium. Maximum OD and growth rates were calculated and expressed as the mean of all replicates with the respective standard deviation. These calculations were done with the Bacterial Growth Kinetics Software (F. Breidt, pers. comm.). The maximum OD observed for each strain grown on HMOs, was compared with the maximum OD obtained in the absence of sugar source. This difference in OD (ΔOD) was used as a criterion to evaluate the strain’s ability for growing on milk oligosaccharides.
Table 1.
Kinetic Analysis of Bacterial Growth. Optical Densities were measured at a Wavelength of 600 nm. Growth Rates (hr−1) were calculated using the Bacterial Growth Kinetic Software Package (F. Breidt, pers.comm). Values are reported as a mean (± sd). Bold numbers correspond to obtained values from high HMO consumers.
| Strain | ZMB1 without sugar | ZMB1 (2%Glucose) | Modified ZMB1 (2% HMO) | ΔOD | |
|---|---|---|---|---|---|
| Enterococcus faecalis OG1RF | Max. OD Growth rate |
0.370 (0.070) 0.053 (0.001) |
1.388 (0.054) 0.335 (0.001) |
0.245 (0.001) 0.039 (0.001) |
- |
| Enterococcus faecalis KA117 | Max. OD Growth rate |
0.403 (0.075) 0.079 (0.001) |
1.515 (0.017) 0.230 (0.001) |
0.309 (0.001) 0.027 (0.001) |
- |
| Streptococcus thermophilus MTC330 | Max. OD Growth rate |
0.388 (0.012) 0.028 (0.001) |
1.525 (0.188) 0.404 (0.001) |
0.469 (0.002) 0.026 (0.001) |
0.165 |
| Streptococcus thermophilus MTC360 | Max. OD Growth rate |
0.242 (0.082) 0.496 (0.026) |
1.329 (0.073) 0.330 (0.002) |
0.551 (0.001) 0.059 (0.001) |
0.243 |
| Bifidobacterium infantis ATCC15697 | Max. OD Growth rate |
0.175 (0.156) 0.015 (0.001) |
1.069 (0.062) 0.191 (0.001) |
0.698 (0.295) 0.023 (0.001) |
0.523 |
| Lactococcus lactis IL1403 | Max. OD Growth rate |
0.322 (0.008) 0.014 (0.001) |
1.353 (0.009) 0.253 (0.009) |
0.417 (0.001) 0.054 (0.001) |
0.124 |
| Eubacterium rectale ATCC35183 | Max. OD Growth rate |
0.305 (0.028) 0.047 (0.001) |
1.103 (0.028) 0.275 (0.002) |
0.454 (0.001) 0.052 (0.001) |
0.231 |
| Clostridium perfringens ATCC13124 | Max. OD Growth rate |
0.112 (0.007) 0.096 (0.031) |
0.937 (0.017) 0.147 (0.003) |
0.378 (0.001) 0.025 (0.001) |
0.177 |
| Clostridium perfringens AB1 | Max. OD Growth rate |
0.369 (0.055) 0.031 (0.001) |
1.471 (0.089) 0.117 (0.001) |
0.434 (0.001) 0.029 (0.001) |
0.171 |
| Clostridium difficile AB2 a | Max. OD Growth rate |
0.411 (0.078) 0,011 (0.001) |
1.108 (0.012) 0.184 (0.001) |
- - |
- - |
| Escherichia coli OP50 | Max. OD Growth rate |
0.288 (0.066) 0.073 (0.001) |
1.460 (0.220) 0.231 (0.001) |
0.386 (0.001) 0.056 (0.001) |
0.152 |
| Escherichia coli EC100 | Max. OD Growth rate |
0.319 (0.061) 0.053 (0.001) |
1.095 (0.149) 0.138 (0.001) |
0.383 (0.001) 0.042 (0.001) |
0.028 |
| Lactobacillus acidophilus NCFM | Max. OD Growth rate |
0.222 (0.029) 0.070 (0.001) |
0.815 (0.067) 0.010 (0.001) |
0.517 (0.001) 0.078 (0.001) |
0.290 |
| Bacteroides fragilis ATCC25285 | Max. OD Growth rate |
0.297 (0.001) 0.023 (0.001) |
1.352 (0.001) 0.068 (0.001) |
1.081 (0.001) 0.052 (0.001) |
0.940 |
| Bacteroides vulgatusATCC8482 | Max. OD Growth rate |
0.085 (0.002) 0.002 (0.001) |
1.049 (0.001) 0.629 (0.001) |
0.744 (0.001) 0.028 (0.001) |
0.660 |
| Veillonella parvula ATCC10790 | Max. OD Growth rate |
0.199 (0.001) 0.033 (0.001) |
1.220 (0.001) 0.081 (0.001) |
0.214 (0.001) 0.047 (0.001) |
0.015 |
Oligosaccharide quantitation using deuterium-labeled internal standard method
Bacteria cultures in modified ZMB1 (2% HMO) were collected and centrifuged at 2000 × g for 30 minutes. Supernatants were boiled for 5 minutes and filtered using a MultiScreen 96-well filtration plate (Millipore, Billerica, MA). Remaining oligosaccharides recovered in the supernatants (25 µl), were reduced using 25 µl of 2.0 M sodium borohydride and incubated at 65 °C for 1h. For quantitative analysis deuterated HMOs (50 µl) were added as internal standard. The oligosaccharides were desalted and purified by solid phase extraction, following the method described by LoCascio et al (20).
MALDI-FTICR Mass spectrometry analysis
The mass spectra analyses were performed on a HiRes Matrix-assisted laser desorption/ionization-Fourier transform ion cyclotron resonance mass spectrometry instrument (MALDI-FTICR) with an external MADI source, a 355-nm pulsed Nd:YAG laser, a quadrupole ion guide, and a 7.0 Tesla superconducting magnet (IonSpec Corp. Irvine, CA). 2,5-Dihydroxybenzoic acid (DHB) was used as matrix and samples were prepared following the fast evaporation technique. 1 µl of analyte (glycans) was spotted onto a 100-well stainless steel sample plate (Applied Biosystems, Foster City, CA) followed by the addition of 0.2 µl of 0.1 mM NaCl as a dopant and 1 µl of 0.4 M DHB. The spots were allowed to dry under a stream of air before analysis. Fifteen acquisitions were recorded for each replicate. MALDI-FTICR MS analysis was performed in the positive ion mode in the m/z scans range of 220–4500 with 1024K data points and 1 Mhz ADC rate acquired. The ratio of deuterated species to undeuterated species (D/H) and percent of consumption was calculated according to Ninonuevo et al. (20) for the 14 most abundant HMO signals present in the spectra. Standard deviations for the percent consumption values were calculated from the measured mass spectral intensities via error propagation. Student’s t tests (p=0.05) were performed to determine the significance of the difference in consumption values between strains.
Results
Application of ZMB1 as chemically-defined medium for growth of intestinal bacteria
Our previous work to characterize HMO-consumption by bifidobacteria employed a rich medium, MRS (19, 20). However when tested, other lactic acid bacteria showed significant background growth in MRS medium lacking any added sugar, thus making testing for HMO growth problematic since it would be impossible to discern weak, or even moderate, growth on HMO from background growth (see Supplementary Figure 1). To circumvent this problem we employed the newly developed chemically-defined medium ZMB1 (22) which allowed robust growth of all the species examined here. Indeed, to our knowledge, no other chemically-defined medium has been described for use among this many genera. As seen Supplementary Figure 2, much less background growth of S. thermophilius is observed in ZMB1 lacking glucose in contrast to MRS lacking glucose (Supplementary Figure 1). Thus, ZMB1 was ideal for the evaluation of HMO consumption by the range of genera tested herein. Using ZMB1, the maximum OD values obtained with glucose as a sugar source ranged from 0.815, in the case of Lactobacillus acidophilus NCFM, to 1.5 for Enterococcus faecalis 117 (Table 1).
Growth of gut-related microorganisms on HMOs
The various strains were grown in modified ZMB1 with HMOs as sole sugar source. In addition, all the selected strains were grown in ZMB1 media without sugar. Maximum optical densities (ODs), ΔOD (Maximum OD in modified ZMB1 minus maximum OD in ZMB1 without sugar) and growth rates are shown in Table 1. Among all the tested microorganisms, Bacterioides fragilis ATCC25285 and Bacterioides vulgatus ATCC8482 reached the highest cell density on HMOs, with ΔOD > 0.6. We have previously demonstrated the B. infantis ATCC15967 is able to consume milk oligosaccharides (19, 20). Thus, this strain was used as a positive control for consumption. However, the maximum OD reached using modified ZMB1 with 2% HMO (0.698; Table 1) was lower than that observed previously in rich media (modified MRS with 1.6% HMO) (17). Interestingly, the species with higher OD (Bacterioides fragilis, Bacterioides vulgatus and Bifidobacterium infantis) also show the longest lag times among the strains tested (data not shown). Some strains show weak, but noticeable, growth on HMOs including Lb. acidophilus, C. perfringens, Escherichia coli OP50, Eubacterium rectale and S. thermophilus, with ΔODs between 0.1 and 0.29. C. difficile, Enterococcus faecalis, V. parvula and Escherichia coli EC100, did not grow at all in this substrate. It should be noted that all these species were capable of vigorous growth on ZMB1 media containing glucose (to ODs of 0.8–1.5).
Determination of HMO consumption profile using MALDI-FTICR mass spectrometry
In order to determine the specific HMO structures consumed from the species that grew well on HMOs, supernatants from Bacteroides fragilis, Bacteroides vulgatus. and Bifidobacterium infantis cultures were recovered after fermentation and remaining HMOs were purified, reduced and profiled by MALDI-RTICR-MS as previously described by LoCascio et al (19). Consumption of 14 neutral oligosaccharides with a degree of polymerization from 4 to 12 was monitored, representing up to 95% of milk oligosaccharides (2). Table 2 shows the mass to charge ratio (m/z) of the types of sugars analyzed, their composition in hexose, N-acetylhexosamine and fucose residues, degree of polymerization (DP) and the relative concentration of each oligosaccharide in the total HMO pool extracted from multiple samples of human milk, according to the data described by Ninonuevo et al. (2).
Table 2.
Masses and Composition of Human Milk Oligosaccharides analyzed. m/z is the mass to charge ratio,
| HMO (m/z) | Hexose | N-Acetylhexosamine | Fucose | DPa | Relative abundance (%)b |
|---|---|---|---|---|---|
| 732.25 | 3 | 1 | - | 4 | 20 |
| 878.31 | 3 | 1 | 1 | 5 | 1 |
| 1024.36 | 3 | 1 | 2 | 6 | <1 |
| 1097.38 | 4 | 2 | - | 6 | 10 |
| 1243.44 | 4 | 2 | 1 | 7 | 25 |
| 1389.50 | 4 | 2 | 2 | 8 | 15 |
| 1462.51 | 5 | 3 | - | 8 | 3 |
| 1535.55 | 4 | 2 | 3 | 9 | <1 |
| 1608.57 | 5 | 3 | 1 | 9 | 7 |
| 1754.63 | 5 | 3 | 2 | 10 | 8 |
| 1827.64 | 6 | 4 | - | 10 | 2 |
| 1900.69 | 5 | 3 | 3 | 11 | 5 |
| 1973.70 | 6 | 4 | 1 | 11 | 2.5 |
| 2119.76 | 6 | 4 | 2 | 12 | 3 |
DP=Degree of Polymerization,
as measured in Ninonuevo et al. (2)
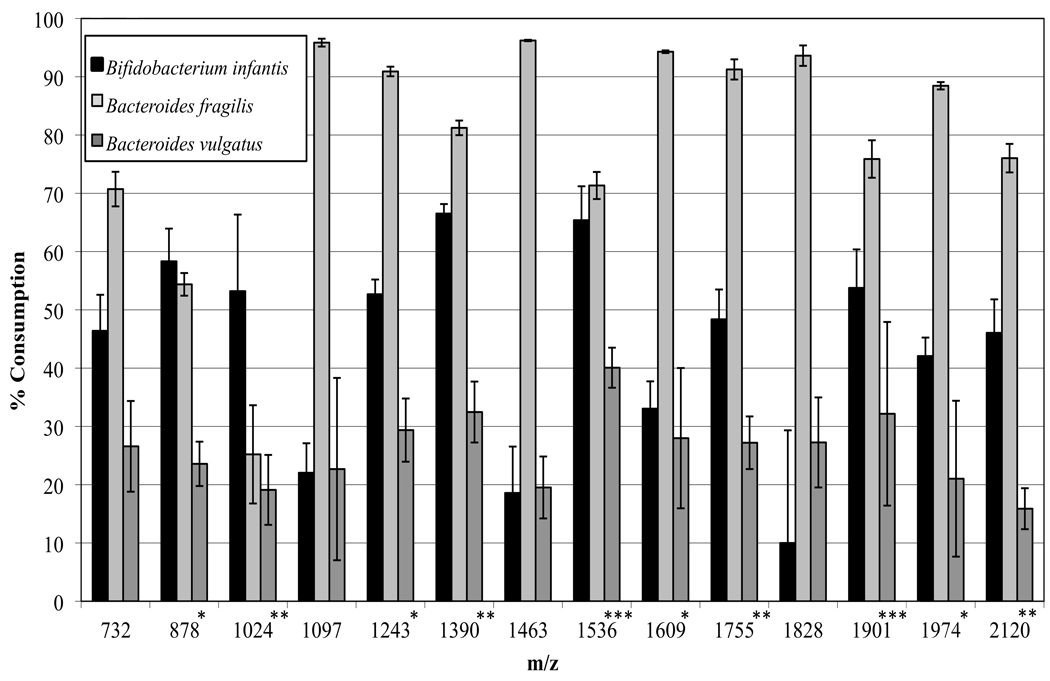
Glycoprofiling of Bifidobacterium infantis ATCC15697 confirmed the high metabolic capacity for HMOs of this strain. The consumption profile appeared different to that described in previous work where modified MRS (1.6 % HMO) was used as growth medium (20). When grown in modified ZMB1 with 2% HMO, Bifidobacterium infantis ATCC15697 show significant consumption of highly fucosylated structures with a high degree of polymerization (DP), a phenotype not witnessed previously in the MRS grown cells (20). Interestingly, the relative consumption of oligosaccharides with two (m/z 1024.36, 1389.50, 1754.63) and three (m/z 1535.55 and 1900.69) fucose residues was higher than the corresponding monofucosylated (m/z 878.31, 1243.44, 1608.57, 1973.70) or non-fucosylated oligosaccharides (m/z 732.25, 1097.97, 1462.52) (Figure 1). In addition, Bifidobacterium infantis did not completely consume glycans with DP ≤ 7 (m/z 732.25, 878.31, 1097.39, 1243.44 and 1389.50), which constitute almost 70% of the total HMO pool. Thus, in this culture medium, the total consumption of HMOs from the initial pool ranges from 45 to 65%. The data contrast with the complete utilization of these small polymers when Bifidobacterium infantis is grown in modified MRS (20). Moreover, Bifidobacterium infantis manifested a strong ability to catabolize specific oligosaccharides with DP>7. These results reinforce the concept that Bifidobacterium infantis ATCC15697 has the ability to consume the wide range of structures present in the HMO pool.
Figure 1.
Human milk oligosaccharide consumption profiles of Bifidobacterium infantis, Bacteroides fragilis and Bacteroides vulgatus in modified ZMB1 (2% HMO), as determined using MALDI-FTICR MS. Asterisks represents number of fucose residues on the specific oligosaccharide depicted by the mass/charge ratio (m/z).
Bacteroides fragilis ATCC2585 show the highest HMOs consumption rates from all bacteria analyzed, ranging from 25 to 90% (Figure 1), nearly depleting all structures from the HMO pool with high degree of polymerization. Oligosaccharides with lower DP (m/z 732.25, 878.32, 1024), corresponding to tetra, penta and hexasaccharides, were partially metabolized from 40% to 70%. Unlike Bifidobacterium infantis, Bacteroides fragilis exhibited a glycan utilization pattern where the relative utilization of non-fucosylated glycans (m/z values 735.21, 1097.38, 1462.51 and 1827.64) was higher than the consumption of fucosylated oligosaccharides. Indeed, the relative consumption of fucosylated glycans by Bacteroides fragilis decreased as the number fucose residues increased on the HMO structure.
Bacteroides vulgatus ATCC8482 exhibited a moderate growth phenotype on HMOs (ΔOD of 0.66), and the consumption of individual glycans ranges from 16 to 40%. For this strain, no selectivity was observed regarding the degree of polymerization, and the higher consumption rates were those found for highly fucosylated species (m/z value of 1535 and 1901). Bacteroides vulgatus can partially consume all available HMO species, however with noticeably lower efficiency than B. fragilis (Figure 1). Differences between these two Bacteroides strains were also witnessed in the growth kinetics. Bacteroides fragilis exhibited a growth rate of 0.052 per hour in HMOs while Bacteriodes vulgatus had a rate of 0.028 hr−1. Oligosaccharide utilization by Bacteroides vulgatus is quite similar to the level of consumption observed by LoCascio et al (19) for bifidobacteria classified as low-HMO consumers.
Discussion
While breast milk constitutes the dominant nutritional source for newborns and infants during the first months of life, the bioactive effects of various milk components are poorly understood. HMOs constitute one of the most abundant molecules provided by the mother to the newborn through lactation. The high concentration of bifidobacteria in feces from breast-fed infant and the high concentration of HMOs in mother’s breast milk have suggested a “prebiotic” role of these components, which are thought to promote a predominantly bifidobacterial microbiota in the infant gut (16). In vitro analyses has shown that several bifidobacterial species can grow on HMOs, and the genome of Bifidobacterium infantis revealed clusters of glycosidases and oligosaccharide transporters, likely linked to HMOs utilization by this phylotype (7, 19, 20). However, there has been little examination of consumption of HMOs by other bacterial species, including those commonly found in the infant gut.
When comparing sugar consumption by many different species, an optimum situation is to use a common medium in which one can manipulate the sugar source. Unfortunately the requirements of intestinal microbes are diverse and, as a consequence, rich (complex) medium components such as beef or yeast extract often are added to the “common” growth medium. However, as shown in Supplemental Figure 1, complex components can bias the results by allowing significant background growth even in the absence of sugar. For that reason, we suggest that a chemically-defined medium that supports good cell growth is a better choice for the study of HMOs consumption by the selected microbes. ZMB1 medium, originally designed for Lactococcus lactis consists of known quantities of trace elements and vitamins, as well as defined nitrogen and carbon sources. The data presented in Table 1 show that ZMB1 generates high cell density in all the strains evaluated, with minor background growth in same medium lacking a sugar source.
This work demonstrates that HMOs consumption is not an exclusive property of specific strains of bifidobacteria. Two species of Bacteroides are able to metabolize free glycans from breast milk, growing to moderate or high cell densities. Bacteroides is known for its incredible ability to ferment an extend assortment of plant polysaccharides (23, 24), and some species, such as B. fragilis, for its capability of consume host-derived glycoconjugates found in the mucus gel layer from the gastrointestinal cells (25). Intestinal glycans are similar to the structures of HMOs, containing N-acetylglucosamine, galactose, fucose, sialic acid and N-acetylgalactosamine (25). Thus, as most of Bacteroides can consume host-glycans, it is perhaps not surprising that they can also consume HMOs. Overall, Bacteroides fragilis ATCC25285 was the most efficient in the metabolism of HMOs, and its glycan consumption pattern exhibits a trend for preferential degradation of non-fucosylated HMO species and longer oligosaccharides. Bacteroides vulgatus ATCC8482 exhibited a lower consumption of these sugars than Bacteroides fragilis. It is interesting to note that Bacteroides vulgatus ATCC8482 has been revealed as moderate consumer of host-glycans and polysaccharides, which could be extended to its ability to consume HMOs (26). The potential ability for consuming HMOs by Bacteroides was suggested by Bjursell et al. (23), after a transcriptomic-based in silico reconstruction of Bacteroides thetaiotaomicron carbohydrate metabolism, using data obtained in the study of gnotobiotic suckling mice.
As we have previously demonstrated (19, 20), our results indicate that Bifidobacterium infantis metabolizes HMOs. However, the use of the chemically-defined media ZMB1 reveals that Bifidobacterium infantis can consume the fucosylated glycans, ability not observed when B. infantis was grown on rich media (MRS). In addition, Bifidobacterium infantis growth on modified ZMB1 indicated consumption for glycans of all sizes similar to what that witnessed previously on MRS using limiting levels of HMOs (19).
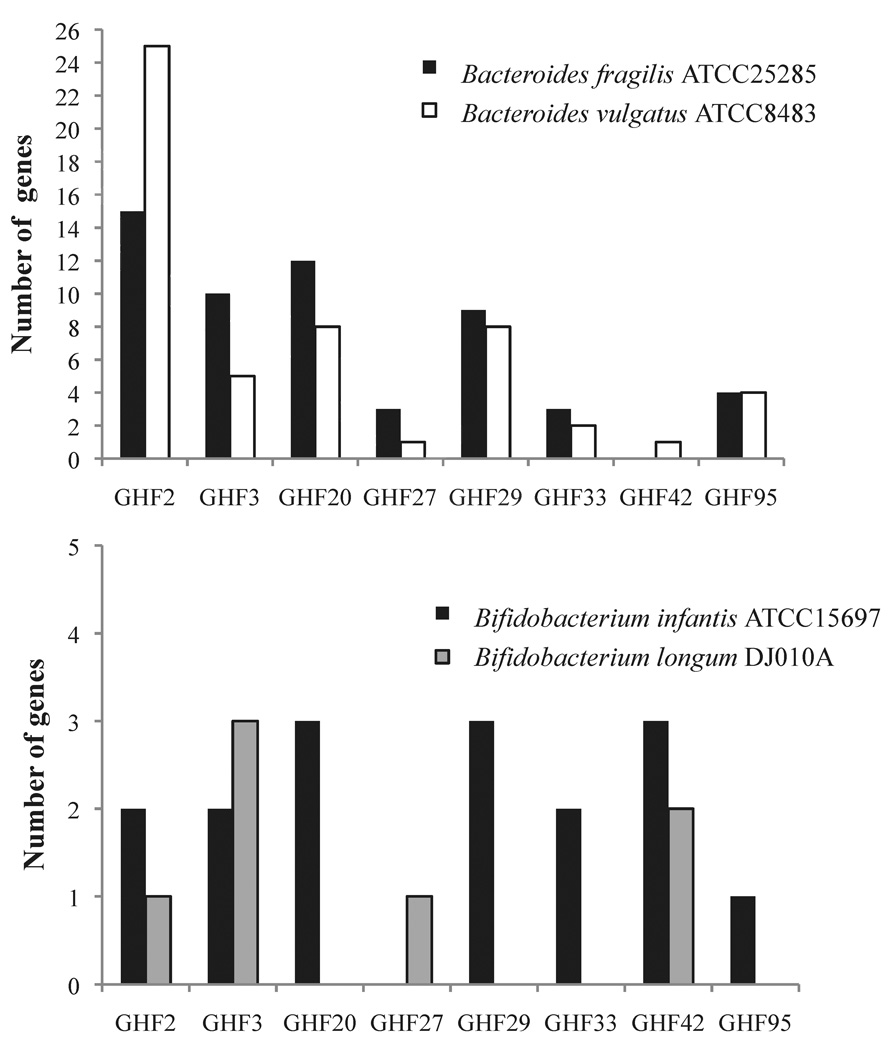
We hypothesize that the consumption differences between the studied strains are related to the specificity of the enzymes involved in cleaving and importing the complex oligosaccharides. Genomic comparisons of Bifidobacterium infantis ATCC15697, Bacteroides fragilis ATCC25285, Bacteroides vulgatus ATCC8482 and the low HMO-consumer Bifidobacterium longum DJO10A (20) show differences in the number of genes related to glycoside hydrolases families (GHFs) (Figure 2). A closer examination of the genomes using CAZy database (27) reveals that Bacteroides fragilis ATCC25285 and Bacteroides vulgatus ATCC8482 have differences in the number of genes that encode glycoside hydrolases genes, with 126 and 159 respectively. The fact that Bacteroides vulgatus genome encodes larger number of cleaving enzymes suggests that differences found in the growth curves and HMOs consumption analysis are derived from cleavage specificities. Differences between these two Bacteroides strains were previously found in the analysis of their host-glycan consumption (26). The genome of Bifidobacterium infantis ATCC332 encodes 42 glycosidehydrolases, which is far lower than the number of carbohydrate cleaving enzymes found in the genomes of the Bacteroides species used in this work. However, the genome of Bifidobacterium infantis encloses specific loci seemingly designed for HMOs import and consumption (7), thus explaining the HMOs growth phenotype of this strain. Conversely, the low HMO consumer Bifidobacterium longum DJO10A (20) does not have any fucosidase (GHF29, GHF95) or sialidases (GHF33), which are directly related to the consumption of specific types of HMOs.
Figure 2.
Annotated genes coding for some carbohydrate active enzymes in the genomes of Bacteroides fragilis ATCC25285, Bacteroides vulgatus ATCC8483, Bifidobacterium infantis ATCC15697 and Bifidobacterium longum DJO10A, as it is described at the CAZy database (http://www.cazy.org) (27). The glycoside hydrolase families (GHF) shown are potentially related to the degradation of HMOs, and they include the following activities: GH2, α-galactosidase; GH3, β-N-Acetylgalactosaminidase; GH20, β-hexosaminidase; GH27, α-N-acetylgalactosaminidase; GH29, α-L-fucosidase; GH33, sialidase; GH42, β-galactosidase; GH95, α-1,2-L-fucosidase.
Studies of the influence of breast-feeding in the intestinal microbiota of newborns and infants reveal that breast-fed subjects develop a microbiota rich in bifidobacteria (28, 29). Other anaerobes such as clostridia and Bacteroides are present in lower concentration, and facultative anaerobes such as enterococci or E. coli are even less numerous (30). In contrast to breast-fed infants, formula-fed infants are often colonized by a more diverse microbiota (29, 31). Among the all the components in human milk, such as proteins, lactose, or nucleotides, HMOs are the only ones which have been proved to play an important role in the stimulation of the growth of specific bacterial species (32). In this work, the clostridia, eubacteria, enterococci and Escherichia coli strains were revealed as non-HMO consumers, reinforcing the concept that HMOs could enhance the growth of specific groups of bacteria in the gut. Such selectively also helps explain why enterococci, often the initial colonizers of the newborn gut, are commonly replaced by bifidobacteria, within the first few weeks of breast-feeding (16). Even though a set of 18 isolates represents a small subset of the gut microbiome, in vitro analysis of the growth of different intestinal species in HMOs, combined with an in-depth analysis of oligosaccharide consumption profiles, constitutes the first and necessary step toward understanding the role of this breast milk component. The results obtained provide a basis for future in vivo studies, in order to understand the shifts in infant intestinal microbiota and a possible competition between Bacteroides and bifidobacterial populations during lactation.
Supplementary Material
Acknowledgements
We are grateful to Dr. Fred Breidt at the USDA–ARS for the Bacterial Growth Kinetics Software package.
This publication was made possible in part by grant support from the University of California Discovery Grant Program, the California Dairy Research Foundation, USDA NRI-CSREES Award 2008-35200-18776 and NIH (HD059127).
Footnotes
Supporting Information Available: Supporting information includes growth curves as well as kinetic parameters of the species in complex media. This material is available free of charge via the Internet at http://pubs.acs.org.
Literature cited
- 1.Kunz C, Rudloff S, Baier W, Klein N, Strobel S. Oligosaccharides in human milk: Structural, functional, and metabolic aspects. Annu. Rev.Nutr. 2000;20:699–722. doi: 10.1146/annurev.nutr.20.1.699. [DOI] [PubMed] [Google Scholar]
- 2.Ninonuevo MR, Park Y, Yin HF, Zhang JH, Ward RE, Clowers BH, German JB, Freeman SL, Killeen K, Grimm R, Lebrilla CB. A strategy for annotating the human milk glycome. J. Agric.Food Chem. 2006;54:7471–7480. doi: 10.1021/jf0615810. [DOI] [PubMed] [Google Scholar]
- 3.Newburg DS, Ruiz-Palacios GM, Morrow AL. Human milk glycans protect infants against enteric pathogens. Annu. Rev.Nutr. 2005;25:37–58. doi: 10.1146/annurev.nutr.25.050304.092553. [DOI] [PubMed] [Google Scholar]
- 4.Chaturvedi P, Warren CD, Buescher CR, Pickering LK, Newburg DS. Bioactive Components of Human Milk. Vol. 501. New York: Kluwer Academic/Plenum Publ; 2001. Survival of human milk oligosaccharides in the intestine of infants; pp. 315–323. [DOI] [PubMed] [Google Scholar]
- 5.Engfer MB, Stahl B, Finke B, Sawatzki G, Daniel H. Human milk oligosaccharides are resistant to enzymatic hydrolysis in the upper gastrointestinal tract. Am. J. Clin. Nutr. 2000;71:1589–1596. doi: 10.1093/ajcn/71.6.1589. [DOI] [PubMed] [Google Scholar]
- 6.Gnoth MJ, Kunz C, Kinne-Saffran E, Rudloff S. Human milk oligosaccharides are minimally digested in vitro. J. Nutr. 2000;130:3014–3020. doi: 10.1093/jn/130.12.3014. [DOI] [PubMed] [Google Scholar]
- 7.Sela DA, Chapman J, Adeuya A, Kim JH, Chen F, Whitehead TR, Lapidus A, Rokhsar DS, Lebrilla CB, German JB, Price NP, Richardson PM, Mills DA. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc. Natl. Acad. Sci. USA. 2008;105:18964–18969. doi: 10.1073/pnas.0809584105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuntz S, Rudloff S, Kunz C. Oligosaccharides from human milk influence growth-related characteristics of intestinally transformed and non-transformed intestinal cells. Br. J. Nutr. 2008;99:462–471. doi: 10.1017/S0007114507824068. [DOI] [PubMed] [Google Scholar]
- 9.Angeloni S, Ridet JL, Kusy N, Gao H, Crevoisier F, Guinchard S, Kochhar S, Sigrist H, Sprenger N. Glycoprofiling with micro-arrays of glycoconjugates and lectins. Glycobiology. 2005;15:31–41. doi: 10.1093/glycob/cwh143. [DOI] [PubMed] [Google Scholar]
- 10.Cravioto A, Tello A, Villafan H, Delvedovo S, Neeser JR. Inhibition of localized adhesion of enteropathogenic Escherichia coli to Hep-2 cells by immunoglobulin and oligosaccharide fractions of human colostrum and breast milk. J. Infect. Dis. 1991;163:1247–1255. doi: 10.1093/infdis/163.6.1247. [DOI] [PubMed] [Google Scholar]
- 11.Ashkenazi S, Newburg DS, Cleary TG. The effect of human milk on the adherence of enterohemorrhagic E. coli to rabbit intestinal cells. Adv. Exp. Med. Biol. 1991;310:173–177. doi: 10.1007/978-1-4615-3838-7_21. [DOI] [PubMed] [Google Scholar]
- 12.Ruiz-Palacios GM, Cervantes LE, Ramos P, Chavez-Munguia B, Newburg DS. Campylobacter jejuni binds intestinal H(O) antigen (Fuc α-1,2 Gal β-1,4 GlcNAc), and fucosyloligosaccharides of human milk inhibit its binding and infection. J. Biol. Chem. 2003;278:14112–14120. doi: 10.1074/jbc.M207744200. [DOI] [PubMed] [Google Scholar]
- 13.Hong P, Ninonuevo MR, Lee B, Lebrilla CB, Bode L. Human milk oligosaccharides reduce HIV-1-gp120 binding to dentritic cell-specific ICAM3-grabbing non-integrin. Br. J. Nutr. 2009;101:482–486. doi: 10.1017/s0007114508025804. [DOI] [PubMed] [Google Scholar]
- 14.Kunz C, Rudloff S. Biological functions of oliogsaccharides in human milk. Acta Paediatr. 1993;82:903–912. doi: 10.1111/j.1651-2227.1993.tb12597.x. [DOI] [PubMed] [Google Scholar]
- 15.Roberfroid M. Prebiotics: The concept revisited. J. Nutr. 2007:830S–837S. doi: 10.1093/jn/137.3.830S. [DOI] [PubMed] [Google Scholar]
- 16.Favier CF, de Vos WM, Akkermans ADL. Development of bacterial and bifidobacterial communities in feces of newborn babies. Anaerobe. 2003;9:219–229. doi: 10.1016/j.anaerobe.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 17.Ward RE, Ninonuevo M, Mills DA, Lebrilla CB, German JB. In vitro fermentation of breast milk oligosaccharides by Bifidobacterium infantis and Lactobacillus gasseri. Appl. Environ. Microbiol. 2006;72:4497–4499. doi: 10.1128/AEM.02515-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ninonuevo MR, Perkins PD, Francis J, Lamotte LA, LoCascio RG, Freeman SL, Mills DA, German JB, Grimm R, Lebrilla CB. Daily variations in oligosaccharides of human milk determined by microfluidic chips and mass spectrometry. J. Agric. Food Chem. 2008;56:618–626. doi: 10.1021/jf071972u. [DOI] [PubMed] [Google Scholar]
- 19.LoCascio RG, Ninonuevo M, Kronewitter S, Freeman SL, German JB, Lebrilla CB, Mills DA. A versatile and scalable strategy for glycoprofiling bifidobacterial consumption of human milk oligosaccharides. Microb. Biotechnol. 2008;2:333–342. doi: 10.1111/j.1751-7915.2008.00072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.LoCascio RG, Ninonuevo MR, Freeman SL, Sela DA, Grimm R, Lebrilla CB, Mills DA, German JB. Glycoprofiling of bifidobacterial consumption of human milk oligosaccharides demonstrates strain specific, preferential consumption of small chain glycans secreted in early human lactation. J. Agric. Food Chem. 2007;55:8914–8919. doi: 10.1021/jf0710480. [DOI] [PubMed] [Google Scholar]
- 21.Adlerberth I. Factors influencing the establishment of the intestinal microbiota in infancy. Nestle Nutr. Workshop Ser. Pediatr. Program. 2008;62:13–33. doi: 10.1159/000146245. [DOI] [PubMed] [Google Scholar]
- 22.Zhang G, Mills DA, Block DE. Development of chemically defined media supporting high-cell-density growth of Lactococci, Enterococci, and Streptococci. Appl. Environ. Microbiol. 2009;75:1080–1087. doi: 10.1128/AEM.01416-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bjursell MK, Martens EC, Gordon JI. Functional genomic and metabolic studies of the adaptations of a prominent adult human gut symbiont, Bacteroides thetaiotaomicron, to the suckling period. J. Biol. Chem. 2006;281:36269–36279. doi: 10.1074/jbc.M606509200. [DOI] [PubMed] [Google Scholar]
- 24.Salyers AA, Vercellotti JR, West SEH, Wilkins TD. Fermentation of mucin and plant polysaccharides by strains of Bacteroides from human colon. Appl.Environ. Microbiol. 1977;33:319–322. doi: 10.1128/aem.33.2.319-322.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Corfield AP, Wagner SA, Clamp JR, Kriaris MS, Hoskins LC. Mucin degradation in the human colon. Production of sialidase, sialate O-acetylesterase, N-acetylneuraminate lyase, arylesterase, and glycosulfatase activities by strains of fecal bacteria. Infect. Immun. 1992;60:3971–3978. doi: 10.1128/iai.60.10.3971-3978.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu J, Mahowald MA, Ley RE, Lozupone CA, Hamady M, Martens EC, Henrissat B, Coutinho PM, Minx P, Latreille P, Cordum H, Van Brunt A, Kim K, Fulton RS, Fulton LA, Clifton SW, Wilson RK, Knight RD, Gordon JI. Evolution of symbiotic bacteria in the distal human intestine. Plos Biol. 2007;5:1574–1586. doi: 10.1371/journal.pbio.0050156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucl. Acids Res. 2009;37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Minna MR, Miguel G, Marko K, Ulla H, Seppo JS, Erika I. Similar bifidogenic effects of prebiotic-supplemented partially hydrolyzed infant formula and breastfeeding on infant gut microbiota. FEMS Immun. Med. Microbiol. 2005;43:59–65. doi: 10.1016/j.femsim.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 29.Harmsen HJM, Wildeboer-Veloo ACM, Raangs GC, Wagendorp AA, Klijn N, Bindels JG, Welling GW. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J. Pediatr. Gastroenterol. Nutr. 2000;30:61–67. doi: 10.1097/00005176-200001000-00019. [DOI] [PubMed] [Google Scholar]
- 30.Favier CF, Vaughan EE, De Vos WM, Akkermans ADL. Molecular monitoring of succession of bacterial communities in human neonates. Appl. Environ. Microbiol. 2002;68:219–226. doi: 10.1128/AEM.68.1.219-226.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Penders J, Vink C, Driessen C, London N, Thijs C, Stobberingh E. Quantification of Bifidobacterium spp., Escherichia coli and Clostridium difficile in faecal samples of breast-fed and formula-fed infants by real-time PCR. FEMS Microbiol. Lett. 2005;243:141–147. doi: 10.1016/j.femsle.2004.11.052. [DOI] [PubMed] [Google Scholar]
- 32.Coppa GV, Zampini L, Galeazzi T, Gabrielli O. Prebiotics in human milk: a review. Dig. Liver Dis. 2006;38:S291–S294. doi: 10.1016/S1590-8658(07)60013-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.