Abstract
The development of an automated, high-throughput fractionation procedure to prepare and analyze natural product libraries for drug discovery screening is described. Natural products obtained from plant materials worldwide were extracted and first prefractionated on polyamide solid-phase extraction cartridges to remove polyphenols, followed by high-throughput automated fractionation, drying, weighing, and reformatting for screening and storage. The analysis of fractions with UPLC coupled with MS, PDA and ELSD detectors provides information that facilitates characterization of compounds in active fractions. Screening of a portion of fractions yielded multiple assay-specific hits in several high-throughput cellular screening assays. This procedure modernizes the traditional natural product fractionation paradigm by seamlessly integrating automation, informatics, and multimodal analytical interrogation capabilities.
Natural products are a vast resource of compounds with seemingly unlimited chemical and functional diversity, and have been a rich source for lead molecules in drug discovery programs.1–4 Sixty percent of new drugs for cancer and 75% of those for infectious diseases have originated from natural sources.5,6 Between 2001 and 2005, 23 natural product based drugs were launched in Europe, Japan, and the United States for treating various disorders such as cancer, diabetes, dyslipidemia, atopic dermatitis, Alzheimer’s disease, bacterial and fungal infections, genetic diseases such as tyrosinemia, and Gaucher’s disease.7
However, during the last two decades, research efforts in the discovery of therapeutic natural products have waned because of the complications and significant time requirements inherent in compound isolation. Primary screening of crude plant extracts or microbial fermentations, followed by bioassay-guided fractionation, purification, and structure elucidation of novel bioactive compounds can take several months.8 The required scale of isolation has been too large to be implemented effectively in an automated, high-throughput fashion. The combination of these and other factors has led to a lagging emphasis in natural product discovery. However, recent advances in high-throughput screening (HTS) technology have enabled biological assays to be conducted in 384- or 1536-well microtiter plates that require only nanograms of test samples. Simultaneously, the development of new analytical and automation technologies has revolutionized sample fractionation and processing, providing a new opportunity to reestablish natural products as a viable source of novel lead compounds in drug discovery programs.7,9,10
Natural product extracts present several problems with respect to modern drug discovery programs. First, polyphenols (vegetable tannins), which are often present in considerable quantities in ethanol extracts of plants, can cause false-positive results in both enzymatic and cellular screening procedures due to non-selective enzyme inhibition and changes in cellular redox potential. Second, the chemical diversity found in a single extract may represent several different classes of molecules that exhibit different (and sometimes opposing) biological activities. Third, biologically active compounds may be present in crude extracts at extremely low concentrations that are below the detection threshold for bioactivity screening.
Several reports have described improved fractionation methods for natural products with single- or multi-step solid phase extraction (SPE),11 multi-channel counter-current chromatography (CCC),5 or flash chromatography/preparative HPLC.8 The single or multi-step SPE methods11 are simple in that natural product samples are eluted with solvent mixtures with increasing amounts of methanol (20% – 100%), and then concentrated via SPE resins. Such methods often take at least four days for fractionation and concentration, and therefore suffer from both low resolution and low efficiency. The other two methods (CCC and HPLC)5,8 are efficient for fractionation and structure determination of natural products that have potential therapeutic activities. The major active components can be isolated effectively to allow structure elucidation. However, these methods require large amounts of raw natural product materials, and their protocols are complicated and offer relatively low throughput. Thus, they have limited utility for generating diverse natural product fractions for general screening. Therefore, high-throughput and high-productivity automated natural products fractionation systems are needed to match the current HTS capacity and drug discovery needs.
Herein, we propose a high-resolution and high-throughput fractionation strategy to address these problems12,13 and report the development of such a natural product fractionation system. The major consideration in designing this system was applying high-throughput techniques to natural product fractionation and reformatting processes, and thus providing an automated and high-throughput method that afforded high-quality samples compatible with current state-of-the-art screening. Our processing system can fractionate 2600 unique natural product samples per year, providing 62,000 fractions in 0.5–10 mg scale for creation of libraries that will serve as a long-term biological screening resource. The method is useful for the primary screening of a large number of natural product samples never (or rarely) studied previously.
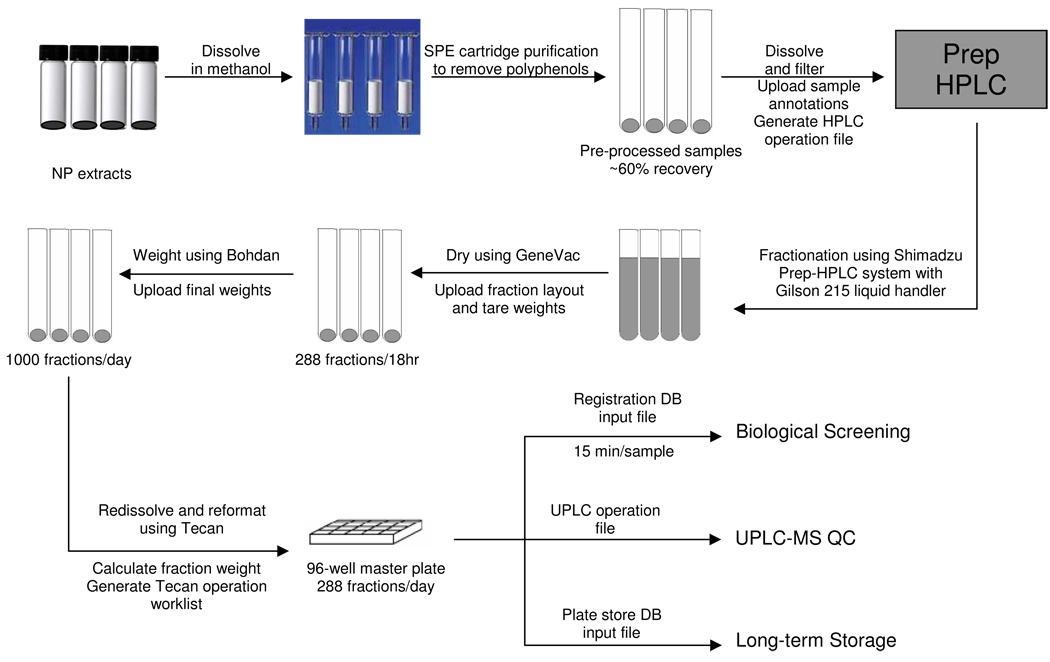
Plant materials were collected worldwide and dried materials were extracted by ethanol prior to pre-fractionation treatment (Figure 1). Extracts were dissolved and eluted from polyamide SPE cartridges to remove polyphenols. These pre-processed samples were dissolved in DMSO and fractionated by a Shimadzu preparative HPLC system with a Gilson 215 liquid handler. Fractionation was monitored by PDA and ELSD detectors. All test tubes for fraction collection were pre-weighed automatically on Bohdan weighing stations. Twenty-four plant extracts were fractionated in approximately nine hours. Fractions were collected and dried overnight in two GeneVac chambers. The final tube weight was measured automatically on a Bohdan weighing station in four hours. Dried fractions were reformatted into plates suitable for screening and storage, respectively. The screening plate was subsequently reformatted into 384-well microtiter plates for UPLC/MS analysis.
Figure 1.
Flow chart for the automated, high-throughput natural product fractionation system.
All data transactions involved in the fractionation process were managed by a web-based program, fractionation workflow application (FWA). Sample annotations such as the unique NCNPR ID (see Experimental Section), sample weight in milligrams, plant part, family, genus, and species are uploaded to the application (Table S1, Supporting Information). This information is used to generate an HPLC operations file. Final weights are appended to each fraction record following fractionation and drying. The workflow application then provided: (a) a Tecan worklist that directs fraction reformatting into the 96- and 384-well plates; (b) a visual map to guide the proper placement of reagents and consumables; (c) the input files required for compound and plate store registration, and (d) the UPLC QC operations file. In summary, the workflow application paralleled the physical flow of material through the fractionation process by keeping track of all sample data and driving the analytical and liquid handling instruments.
Polyphenols are a class of plant natural products that can bind proteins in aqueous solutions. These components may cause false-positive results during enzymatic screening. High levels of plant polyphenols are also deleterious to cell-based assays through perturbation of both cellular oxidation potential and extracellular pH. Thus, it is usually desirable to remove polyphenols from plant extracts when preparing screening libraries. In the work reported here, polyphenols were removed using a polyamide SPE cartridge before fractionation.14 Polyphenols were retained on the cartridge, while nonpolyphenolic compounds were eluted. Since compounds containing two or three phenolic hydroxy groups can also be eluted, most flavonoids could be recovered.
The optimal loading of polyamide columns for polyphenol removal was tested. A FeCl3 solution was applied to identify polyphenols in aqueous solution. An aliquot of ethanol extract was diluted with water and 1–2 drops of 9% FeCl3 solution were added. For hydrolyzable tannins (having a carbohydrate core linked with galloyl and/or hexahydroxydiphenoyl moieties), the solution showed a bluish black color, whereas the solution turned brownish black or greenish black with condensed tannins (consisting of multi-flavonoid moieties linked through carbon-carbon interflavanyl bonds). The present results indicated that 700 mg of polyamide were enough to remove all polyphenols from a 100 mg extract sample. The recovery rates after removing polyphenols were in the range of 49.3–84.4%. The average recovery was about 60%. These data also showed that there were large amounts of polyphenols in the ethanol extracts of plants. Thus, the polyphenol removal step is required to prevent interference in biological screen runs.
The reversed-phase preparative HPLC system used methanol and water as the mobile phase, with a gradient from 2% to 100% methanol. The methanol concentration was kept at 100% for 6 min in order to elute most of the low-polarity compounds. No mobile-phase additives were used since single natural product ethanol extracts may contain various kinds of compounds with variable pKa values such as alkaloids and organic acids. With such diverse compounds, no single additive would be applicable to all components. The chromatographic gradient was designed to provide a method that allows the fractionation of most of plant natural product components. Extracts with predominantly polar (Figure S1A, Supporting Information), non-polar (Figure S1B, Supporting Information), or mixed polarity (Figure S1C, Supporting Information) compounds were fractionated well under such chromatographic conditions.
Fractions were collected for each extract from 1.2–12.4 min every 30 sec, yielding 24 fractions with a volume of 11.5 mL each. A photodiode array (PDA) and an evaporative light scattering detector (ELSD) were applied to monitor the fractionation process. Typical chromatograms are shown in Figure S2 (Supporting Information). In general, the ELSD signal intensity correlated well with fraction mass, which is consistent with previous findings. 9,15–17
Fractions were dried using a carefully controlled GeneVac evaporator in pre-weighed tubes. The dried sample tubes were weighed on a Bohdan weighing station to tare each fraction. Currently, most drug discovery programs are capable of screening large numbers of compounds against multiple targets with nanogram quantities of materials used for each assay. Thus, 0.5 mg samples provide enough material for several hundred screens. More than 85% of the fractions in the natural product library that has been generated contained greater than 0.5 mg of sample. High-throughput assays using pin-tool liquid transfers consume only nanograms of material per screen; therefore, the present fractionation process will generate enough material for long-term biological screening efforts.
The acquisition of detailed analytical information is essential for characterization of natural products, for analysis of screening data, and for assistance in the structure elucidation of compounds in active fractions. To enable the analysis of the large numbers of natural product fractions contained in the library, UPLC was applied to perform a rapid separation, while PDA, ELSD, and mass spectrometry were used for detection so that the maximum amount of information could be obtained in a single run. For ESIMS detection, both positive- and negative-ionization modes were applied simultaneously so that compounds with different properties could be ionized and analyzed to facilitate the identification of their molecular weights and fragmentation patterns. Three tailor-made UPLC gradients following an “accelerated retention window” principle18 were used for fractions with different polarities for efficiency and to ensure a good separation for the major components in fractions. The FWA program generated automatically the inlet method for individual fractions based on the fraction IDs 1 to 24.
To validate the use of this fractionation procedure in high-throughput screening, cytotoxicity of 140 randomly selected fractions from 14 natural product extracts were tested in a panel of five cell-based assays [Trypanosoma brucei (T. brucei), Plasmodium falciparum (P. falciparum), RAJI, Hep G2, and BJ (Figure S3, Supporting Information)]. Fractions were screened in a full 10-point dose response (1:3 dilution series), with an average maximum concentration of 80 µg/mL in 384-well plates. Twelve fractions exhibited high potencies (EC50 <10 µg/mL) against T. brucei. In the RAJI model cancer cell line, four fractions displayed high potencies, of which three were also potent against T. brucei. Three fractions displayed potencies below 2 µg/mL against P. falciparum, of which two were inactive in all other cell lines. Importantly, fractions in the library displayed different activity profiles against each cell line, thereby simplifying the process of identifying extracts with interesting biological activities.
The relative number of hits found in this suite of assays is related to cell-doubling times. T. brucei has a very rapid growth rate, with a typical doubling time of approximately 8 h. As it is an extracellular parasite, test compounds do not have to penetrate multiple plasma membranes, as in the case of P. falciparum. Thus, cytostatic as well as cytotoxic compounds can be more easily identified in T. brucei than in the more slowly growing human cancer cell lines, which have doubling times of more than 24 h.
All operations and data for the prefractionation, fractionation, and sample processing of natural product samples were managed with FWA, which allows database driven tracking of receipt of extracts, registration of samples, and provides standard data manipulations for driving and tracking fractionation, fraction quality control, fraction reformatting, fraction storage and fraction retrieval. Since Pipeline Pilot is widely used in academia and now free to academic labs, this allows the facile sharing of all data manipulation methods.
Experimental Section
General Experimental Procedures
The plant extracts used in this study were obtained from the natural products repository of the National Center for Natural Products Research (NCNPR) at the University of Mississippi. Plant materials were collected from various origins worldwide and their voucher specimens are available at NCNPR or collaborating institutions. Plant extracts were obtained according to reported methods. 19
Prefractionation with Polyamide SPE Cartridges
Polyphenols were removed using a 700 mg polyamide-filled cartridge (Sigma-Aldrich, St. Louis, MO) and a 48-place positive-pressure SPE manifold (SPEware Corporation, Baldwin Park, CA). The ethanol extracts (~100 mg) were dissolved, and brought onto a polyamide SPE cartridge. The column was then rinsed with five column volumes of methanol. The effluent was collected and dried under a stream of nitrogen using a Zymark TurboVap LV Concentration Workstation (Caliper Life Sciences, Hopkinton, MA), which can dry 50 samples simultaneously in 5 h.
Preparative Reversed-Phase HPLC Fractionation
After prefractionation, samples were dissolved in 2 mL dimethyl sulfoxide (DMSO) before fractionation. Each sample was separated into 24 fractions and collected in preweighed 16 × 100 mm disposable glass tubes (Fisher Scientific, Hampton, NH). One batch (12 samples) of natural product extracts could be fractionated automatically in 5 h.
Preparative HPLC separations were performed on a Gemini 5 µm C18 110A column (30 × 50 mm, 5 µm, Phenomenex, Inc., Torrance, CA). A Shimadzu LC-8A binary preparative pump with a Shimadzu SCL-10A VP system controller was connected to the Gilson 215 auto sampler and Gilson 215 fraction collector (Gilson, Inc., Middleton, WI). Detections were performed by a Shimadzu SPD-M20A diode-array detector and a Shimadzu ELSD-LT II evaporative light scattering detector (Shimadzu Corp., Kyoto, Japan). The mobile phase consisted of water (A) and methanol (B): 0 min, 98:2; 0.5 min, 98:2; 6.5 min, 0:100; 12.3 min, 0:100; 12.5 min, 98:2; 12.95 min, stop). The flow rate was 25 mL/min.
Solvent Evaporation
The collected fractions were dried using a GeneVac HT series II high performance solvent evaporation system (GeneVac Inc., Gardiner, NY). The chamber was preheated to and maintained at 35 °C. The SampleGuard Control temperature was set at 40 °C, and the CoolHeat Enable pressure was set at 40 mbar. The running time was 18 h, and 288 tubes (12 samples) could be dried simultaneously.
Automatic Weighing of Natural Product Fractions
Fraction-collection tubes were pre-weighed using a Bohdan BA-200 Balance Automator (Mettler-Toledo AutoChem, Columbia, MD) and held in a custom Gilson 207 test tube rack (Gilson Inc., Middleton, WI). Then, tubes with natural product fractions were reweighed using the Bohdan BA-200. The net weight of fraction was calculated from the difference between the 2 weights by using a FWA program developed on a Pipeline Pilot platform (version 7.5.2, Accelrys). Two Gilson 207 test tube racks of 150 glass tubes could be weighed automatically in 1 h.
Reformatting and Plating
The plant natural product fractions were plated using a Freedom Evo Tecan system (Tecan Group Ltd., Mannedorf, Switzerland). Samples in GeneVac racks were dissolved in the appropriate plating solvent (e.g., methanol/chloroform). The dissolved samples were divided into three portions and then transferred to: (a) 96-well plates for biological activity screening; (b) 96-well plates for long-term storage (−20 °C), and (c) 384-well plates for UPLC-MS analysis. One batch of fractionated samples (288 fractions) could be reformatted in 7 h in an unattended mode.
Quality Control (QC) with Ultra Performance Liquid Chromatography – Mass Spectrometry (UPLC-MS)
The final QC of natural product fractions was performed on a Waters Acquity UPLC-MS system (Waters Corp., Milford, MA), using a Waters Acquity UP LC system and an SQ mass spectrometry detector. An Acquity UPLC BEH C18 column (2.1 × 50 mm, 1.7 µm) was used. The mobile phase consisted of water containing 0.1% formic acid (A) and acetonitrile (B). The total run time for each analysis was 3.0 min.
Ionization and detection of natural product fractions were carried out on a Waters SQ mass spectrometry using both the positive and negative electrospray ionization modes (ESI). The capillary voltage was set at 3.4 kV. The extractor voltage was 2 V. Nitrogen was used as the nebulizing gas. Source temperature was set at 130 °C. The scan range was m/z 130–1400.
Data processing was performed automatically with OpenLynx by extracting all graphic information, such as retention time, UV, and ELSD peak areas, and converted to text to allow transfer to a database for storage and analysis. Each 384-well QC plate could be analyzed in 20 h.
Storage of Fractions for Screening
The fraction collection was stored in an automated storage archive, produced by REMP (Tecan Group, Ltd., Zurich, Switzerland) with a capacity of 1,000,000 300 µL tubes and 10,000,000 50 µL tubes. The large capacity tubes were individually capped using a cap that could be removed and replaced by appropriate robotics. The small capacity tubes were individually sealed using an adhesive free foil seal, and are “single use.” This system tracked each tube by positional array and stored them in 96 tube and 384 well tube racks, respectively. The individual racks were bar coded and the identity confirmed each time a tube was retrieved. In addition, the large tubes had a 2D barcode that could be used to confirm identity. Compounds could be retrieved from the system either as preformatted plates or as individual tubes in a “cherry picking” mode. All contents were addressable in either mode. The entire system operated under dry air at −20 °C.
High-throughput Screening Assays
Trypanosoma brucei assay using Trypanosoma brucei brucei (a protozoal human parasite) and cytotoxicity assays using BJ (a normal human fibroblast line), Raji (a Burkitt’s lymphoma cell line), and HepG2 (a liver cell line) cells were performed as before. 20 Plasmodium falciparum (a causative agent of malaria, strain 3D7, American Type Culture Collections (ATCC), Manassas, VA) assay was performed as described previously. 21–23
Integrated Informatics
A custom informatics workflow web application FWA was built using the Pipeline Pilot platform (version 7.5.2, Accelrys) to manage all operations and data transactions.
Supplementary Material
Acknowledgment
This work was supported by the National Cancer Institute (P30 CA021765), the American Lebanese Syrian Associated Charities (ALSAC), St. Jude Children’s Research Hospital, and the National Center for Natural Products Research, University of Mississippi.
Footnotes
Supporting Information Available: Preparative HPLC fractionation chromatograms, weights of fractions, biological screening results, and sample registration information. These materials are available free of charge via the Internet at http://pubs.acs.org.
References and Notes
- 1.Koehn FE. Prog. Drug Res. 2008;65:177–210. doi: 10.1007/978-3-7643-8117-2_5. [DOI] [PubMed] [Google Scholar]
- 2.Vuorelaa P, Leinonenb M, Saikkuc P, Tammelaa P, Rauhad JP, Wennberge T, Vuorela H. Curr. Med. Chem. 2004;11:1375–1389. doi: 10.2174/0929867043365116. [DOI] [PubMed] [Google Scholar]
- 3.Schmid II, Sattler II, Grabley S, Thiericke R. J. Biomol. Screen. 1999;4:15–25. doi: 10.1177/108705719900400104. [DOI] [PubMed] [Google Scholar]
- 4.Newman DJ. J. Med. Chem. 2008;51:2589–2599. doi: 10.1021/jm0704090. [DOI] [PubMed] [Google Scholar]
- 5.Wu S, Yang L, Gao Y, Liu X, Liu F. J. Chromatogr. A. 2008;1180:99–107. doi: 10.1016/j.chroma.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 6.Newman DJ, Cragg GM, Snader KM. J. Nat. Prod. 2003;66:1022–1037. doi: 10.1021/np030096l. [DOI] [PubMed] [Google Scholar]
- 7.Lam KS. Trends Microbiol. 2007;15:279–289. doi: 10.1016/j.tim.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 8.Eldridge GR, Vervoort HC, Lee CM, Cremin PA, Williams CT, Hart SM, Goering MG, O'Neil-Johnson M, Zeng L. Anal. Chem. 2002;74:3963–3971. doi: 10.1021/ac025534s. [DOI] [PubMed] [Google Scholar]
- 9.Cremin PA, Zeng L. Anal. Chem. 2002;74:5492–5500. doi: 10.1021/ac025516a. [DOI] [PubMed] [Google Scholar]
- 10.Bugni TS, Richards B, Bhoite L, Cimbora D, Harper MK, Ireland CM. J. Nat. Prod. 2008;71:1095–1098. doi: 10.1021/np800184g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thiericke R. J. Autom. Methods Manag. Chem. 2000;22:149–157. doi: 10.1155/S1463924600000249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harvey AL. Drug Discov. Today. 2008;13:894–901. doi: 10.1016/j.drudis.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 13.Bugni TS, Harper MK, McCulloch MW, Reppart J, Ireland CM. Molecules. 2008;13:1372–1383. doi: 10.3390/molecules13061372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hostettmann K, Marston AM, Hostettmann M. Preparative Chromatography Techniques: Applications in Natural Product Isolation. 2nd. Ed. Heidelberg: Springer; 1998. pp. 10–11. [Google Scholar]
- 15.Kibbey TCG, Yavaraski TP, Hayes KF. J. Chromatogr. A. 1996;752:155–165. [Google Scholar]
- 16.Hsu BH, Orton E, Tang SY, Carlton RA. J. Chromatogr. 1999;725:103–112. doi: 10.1016/s0378-4347(98)00529-5. [DOI] [PubMed] [Google Scholar]
- 17.Fang LL, Pan JM, Yan B. Biotechnol. Bioengin. 2000;71:162–171. doi: 10.1002/1097-0290(2000)71:2<162::aid-bit1006>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 18.Yan B, Collins N, Wheatley J, Irving M, Leopold K, Chan C, Shornikov A, Fang L, Lee A, Stock M, Zhao J. J. Comb. Chem. 2004;6:255–261. doi: 10.1021/cc0340527. [DOI] [PubMed] [Google Scholar]
- 19.Manly SP, Smillie T, Hester JP, Khan I, Coudurier L. Unique Discovery Aspects of Utilizing Botanical Sources. Boca Raton, FL: CRC Press, Taylor & Francis; 2010. pp. 213–231. [Google Scholar]
- 20.Mallari JP, Shelat AA, Obrien T, Caffrey CR, Kosinski A, Connelly M, Harbut M, Greenbaum D, McKerrow JH, Guy RK. J Med Chem. 2008;51:545–552. doi: 10.1021/jm070760l. [DOI] [PubMed] [Google Scholar]
- 21.Mallari JP, Guiguemde WA, Guy RK. Bioorg. Med. Chem. Lett. 2009;19:3546–3549. doi: 10.1016/j.bmcl.2009.04.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smilkstein M, Sriwilaijaroen N, Kelly JX, Wilairat P, Riscoe M. Antimicrob. Agents Chemother. 2004;48:1803–1806. doi: 10.1128/AAC.48.5.1803-1806.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bennett TN, Paguio M, Gligorijevic B, Seudieu C, Kosar AD, Davidson E, Roepe PD. Antimicrob Agents Chemother. 2004;48:1807–1810. doi: 10.1128/AAC.48.5.1807-1810.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.