Abstract
Autophagy is a cellular degradation pathway for the clearance of damaged or superfluous proteins and organelles. The recycling of these intracellular constituents also serves as an alternative energy source during periods of metabolic stress to maintain homeostasis and viability. In tumour cells with defects in apoptosis, autophagy allows prolonged survival. Paradoxically, autophagy defects are associated with increased tumorigenesis, but the mechanism behind this has not been determined. Recent evidence suggests that autophagy provides a protective function to limit tumour necrosis and inflammation, and to mitigate genome damage in tumour cells in response to metabolic stress.
Autophagy is a cellular catabolic degradation response to starvation or stress whereby cellular proteins, organelles and cytoplasm are engulfed, digested and recycled to sustain cellular metabolism1,2. Constitutive, basal autophagy also has an important homeostatic function, maintaining protein and organelle quality control, acting in parallel with the ubiquitin proteasome degradation pathway to prevent the accumulation of polyubiquitinated and aggregated proteins3–8. Autophagy is also a pathway that is used for the elimination of pathogens9 and for the engulfment of apoptotic cells10. However, the effect of these events on cancer is not known. Although most evidence supports a role for autophagy in sustaining cell survival, paradoxically, cell death resulting from progressive cellular consumption has been attributed to unrestrained autophagy11–13.
Complicating this situation further, cytotoxic events often induce autophagy, but whether this is a death mechanism or a futile effort at cellular preservation is often unclear2. Another enigma has been the role of autophagy in tumour suppression; allelic loss of the essential autophagy gene beclin1 (BECN1, also known as ATG6) is found with high frequency in human breast, ovarian and prostate cancers14,15, and autophagy-defective Becnl-heterozygous16,17 and autophagy-related 4C (Atg4C)-deficient18 mice are prone to tumours. Paradoxically, most evidence supports a role for autophagy in maintaining tumour cell survival in response to metabolic stress in vitro, and in hypoxic tumour regions in vivo19–23. Resolution of these paradoxes surrounding autophagy function has proved challenging.
The mechanisms that regulate the mutually opposed survival-supporting and death-promoting roles for autophagy are still far from resolution. The most plausible explanation is that catabolism through autophagy is predominantly survival-supporting, but that an imbalance in cell metabolism, where autophagic cellular consumption exceeds the cellular capacity for synthesis, promotes cell death. Although experimental evidence to support this is currently lacking, insight into the role of autophagy in tumour suppression is beginning to evolve.
How loss of the pro-survival function of autophagy promotes tumorigenesis is partly explained by the stimulation of necrotic cell death and an inflammatory response in tumours with defects in autophagy and apoptosis19. Preventing survival under starvation through autophagy, and diverting apoptosis-defective tumour cells to a necrotic cell fate, generates chronically necrotic tumours. This can corrupt a normal wound-healing response to support tumour growth, representing a possible means by which autophagy defects provide a non-cell-autonomous mechanism for stimulating tumorigenesis24–26. In contrast to apoptosis, necrosis and cell lysis causes nuclear HMGB1 to be released from cells, and this and other events stimulate the innate immune response, the recruitment of inflammatory cells, cytokine production and nuclear factor-κB (NFκB) activation, which in some cases is linked to increased tumorigenesis27–29. Indeed, blocking autophagy with constitutive activation of Akt in apoptosis-defective cells results in necrosis in response to metabolic stress in vitro, and in tumours in vivo this necrosis is coincident with NF-κB activation and promotion of tumorigenesis19. How different cell-death processes interface with the immune system and tumour micro-environment to modulate tumour growth is far from clear and is an important area for future investigation.
How defective autophagy and compromised survival to stress can promote tumour progression despite reduced cellular fitness is suggested by the increased rate of cellular damage accumulation. In tumour cells in which cell-cycle checkpoints are inactivated, autophagy limits the accumulation of genome damage and suppresses the mutation rate20,23. This supports the role for autophagy in protecting the genome in a cell-autonomous mechanism of tumour suppression. It is currently unclear how autophagy limits genome damage, but this could involve maintaining energy homeostasis or preventing the damaging effects of oxidative stress from defective organelle and unfolded protein accumulation. As we begin to define the role of tumour cell metabolism in response to stress, the rational ability to modulate the autophagy pathway in cancer therapy is emerging.
Apoptosis and metabolic stress
A common cellular response to metabolic stress is cell death by apoptosis, and in tumour cells in particular this apoptosis is crucial to suppressing tumorigenesis30 (FIG. 1a). The tumour-suppressive role for apoptosis is well known, originating with the realization that many tumours have apoptosis suppressed by various mechanisms, including overexpression of the apoptosis inhibitor BCL2, and that preventing apoptotic cell death allows tumour cells to survive the stress of oncogene activation, uncontrolled proliferation (FIG. 2a) and chemotherapy31. Indeed, BCL2 antagonists have entered the clinic as part of the armamentarium to functionally restore the apoptotic pathway to resistant tumours32.
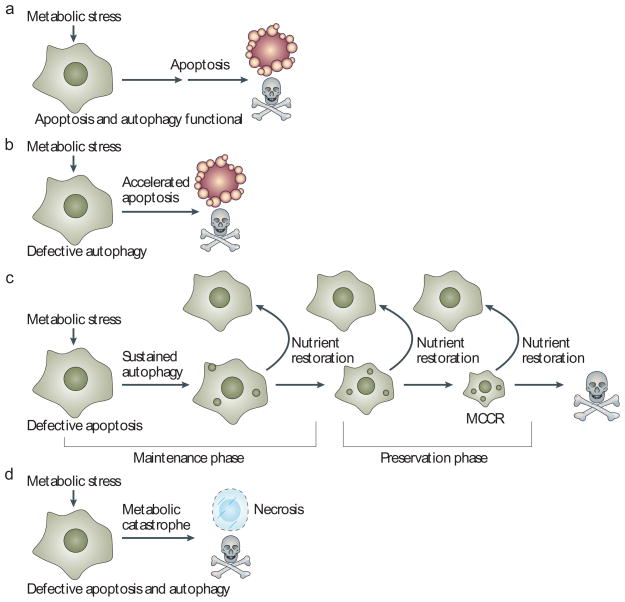
Figure 1. Altered capacity for apoptosis and autophagy can dictate cell fate in response to metabolic stress.
a | Apoptosis is a common response to metabolic stress in which cells activate caspases and die efficiently. For immortal epithelial cells, metabolic stress triggers apoptosis within 24 to 48 hours19,20,23. Execution of apoptosis occurs in less than an hour and cell viability loss is five to six orders of magnitude19. b | Defective autophagy (through loss of BECN1 or ATG5, for example) can increase apoptotic cell death in some cells in response to metabolic stress56. In human mammary cells in 3D culture in vitro, this accelerated apoptosis manifests as increased lumen formation in mammary acini 20. Therefore, preservation of cell metabolism through autophagy might increase the threshold for apoptosis activation. c | In cells with defects in apoptosis, survival in metabolic stress is dependent on autophagy and is prolonged for weeks19,20,22,23. During the maintenance phase, activities such as cell division and motility are sustained. Prolonged starvation and progressive autophagy causes cells to gradually shrink in size but restoration of nutrients allows recovery. In the preservation phase, cell division and motility decrease, presumably as a bioenergenic conservation effort, creating the minimal cell that is capable of recovery (MCCR). Eventually, restoration of nutrients fails to allow recovery. In this way, autophagy can be viewed as an interruptible path to cell death. d | Cells with defects in apoptosis and autophagy fail to tolerate metabolic stress, undergo metabolic catastrophe and die by necrosis36.
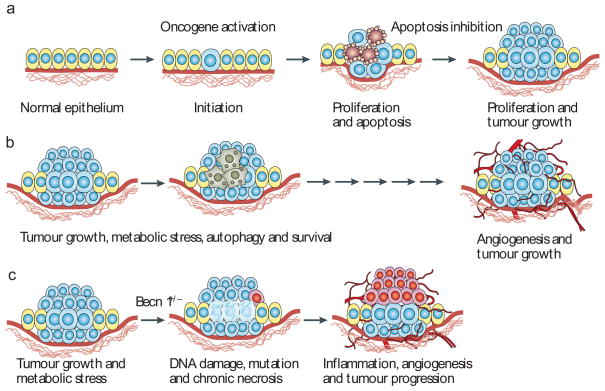
Figure 2. Role of apoptosis and autophagy in tumorigenesis.
a | Tumour-initiating mutational events such as oncogene activation promote cell proliferation, but also apoptosis, which limits tumour growth. Following the acquisition of defects in apoptosis, tumour proliferation is sustained in the absence of apoptotic cell death. b | Tumour growth is initially limited by the absence of a blood supply, which can trigger autophagy-mediated survival in the most metabolically stressed tumour regions, commonly the hypoxic center19,20,23. The eventual recruitment of a blood supply cures the tumour of hypoxia and metabolic stress, and the tumour cells formerly surviving through autophagy can emerge to contribute to tumour growth30. c | In tumours formed by cells with defects in both apoptosis and autophagy, necrotic cell death is stimulated in metabolically stressed tumour regions and this necrosis is associated with the activation of an inflammatory response, DNA damage and tumour progression19,20,23. Analogous to a wound-healing response, chronic necrosis and inflammation can stimulate angiogenesis and tumour growth24–26.
There is an entire family of multidomain (BCL2 homology regions or BH1-4) anti-apoptotic BCL2-homologous proteins that function to sequester the core pro-apoptotic regulators, multidomain BAX and BAK31. Pro-apoptotic BH3-only proteins disrupt this BAX and BAK antagonism by BCL2-like proteins and some might directly activate BAX and BAK to propagate the death signal. Once activated, BAX and BAK oligomerize and permeabilize membranes, particularly the outer mitochondrial membrane, to release pro-apoptotic factors such as cytochrome c and SMAC/DIABLO. Cytochrome c is a cofactor for the apoptosome that activates the cysteine protease caspase 9, whereas SMAC/DIABLO inhibits the caspase antagonists, the inhibitor of apoptosis proteins (IAPs). Together, this results in effector caspase activation and widespread cellular protein substrate cleavage, causing cell death31.
Exactly how metabolic stress triggers apoptosis is not completely understood, but it is associated with the induction of the pro-apoptotic BH3-only protein PUMA, requires the BH3-only protein BIM, depends on either BAX or BAK, and is inhibited by BCL2 (REFS 30,33,34). Once the apoptotic signal reaches BAX and BAK, mitochondrial membrane permeabilization, caspase activation and cell death occur in less than an hour and the process is extremely efficient and irreversible (FIG. 1a). Neighbouring cells or professional phagocytes engulf apoptotic cells, preventing the activation of an innate inflammatory response. These attributes are the reasons why apoptosis defects are selected for in tumours and why restoring the apoptotic response is desirable for cancer therapy. Tumour cells with defects in apoptosis through either deficiency in BAX and BAK or gain of BCL2 or BCL-XL (also known as BCL2L1) function are highly resistant to metabolic stress and the properties of these ‘undead cells’ are revealing insights into the mechanisms regulating metabolic stress response.
Metabolic stress has a major influence on tumours in vivo, either as a stimulator of apoptosis to limit tumour progression, or as a damaging insult in surviving apoptosis-defective tumour cells (FIGS 1c, 2a). Tumours are frequently subjected to metabolic stress, arising from the initial lack of a blood supply, vascular collapse in established tumours or therapeutic intervention. It is well known that tumour cells can be reliant on the inefficient process of glycolysis, rather than the more productive energy-generating process of oxidative phosphorylation, to support metabolism (the Warburg effect)35. As such, tumour cells are particularly vulnerable to metabolic stress, which is only exacerbated by the high energy demand of unrestrained cell growth and the potentially reduced capacity to access the nutrient recycling activity provided by autophagy36. How apoptosis-defective tumour cells adapt to metabolic stress and if or how this facilitates tumour progression has now been linked to autophagy.
Autophagy promotes survival
Evidence suggests that a constitutive, low level of basal autophagy in normal tissues provides an important homeostatic, housekeeping function. Targeted deletion of Atg5 or Atg7 in the brain causes the accumulation of polyubiquitylated protein aggregates and neuronal degeneration, supporting a role for autophagy in protein quality control3,4. Furthermore, ATG5 is required for maintaining T-cell survival and proliferation37. Autophagy induction in response to stress and starvation also has a crucial role in normal cells. For example, Atg5-deficient mice fail to live through the neonatal survival period, during which tissues show signs of amino-acid depletion and metabolic insufficiency38. These findings support a prosurvival role for autophagy in both normal tissues and in response to metabolic stress.
In cancer cells, metabolic stress robustly induces autophagy, which is sustained when apoptosis is blocked19,20,22,23. Importantly, autophagy is required for tumour cells to survive metabolic stress. Genetic inactivation of autophagy, either indirectly by constitutive activation of the phosphatidylinositol 3-kinase (PI3K) pathway or directly by allelic loss of Becnl or deficiency in Atg5, or by RNA interference (RNAi), prevents survival in response to metabolic deprivation even when apoptosis is inactivated19,20,22,23. Presumably, in the absence of an external nutrient source the catabolic capacity of autophagy can sustain viability, but the role of autophagy in cell damage control and mitigation in response to stress may be equally important. Amino-acid starvation, glucose and oxygen deprivation, growth-factor withdrawal and cytotoxic cellular damage are among the many stimuli that potently induce autophagy2. In the example of nutrient starvation, autophagy serves as a back-up energy reserve, whereas the autophagic response to cellular damage probably facilitates adaptation through the removal of damaged proteins and organelles.
Autophagy regulation
Autophagy is controlled mainly, but not exclusively, by the kinase mammalian target of rapamycin (mTOR; also known as FRAP1), which is a downstream component of the PI3K pathway2,39. mTOR functions in part to suppress autophagy in response to nutrient and growth-factor availability. Conditions of starvation cause the de-repression of autophagy, which initiates isolation membrane or phagophore formation and the subsequent genesis of autophagosomes. Autophagosomes are double-membrane vesicles that sequester cytoplasm and organelles. The autophagy-regulated or Atg proteins are required for the activation of autophagy, the formation of autophagosomes, the sequestration of intracellular constituents, and the targeting and fusion of autophagosomes to lysosomes where the contents are degraded and recycled39–41. For example, the serine/threonine protein kinase ATG1 (also known as ULK1) is a candidate for activation by mTOR de-repression that stimulates autophagy. ATG5 is required for autophagy and becomes covalently conjugated to the ubiquitin-like protein ATG12 by the ubiqutin-activating enzyme ATG7, but the specific role of this process in autophagy is not known. BECN1 is part of the class III PI3K VPS34 complex that is also required for autophagy. ATG8 (also known as MAP1LC3) is another ubiquitin-like protein that is cleaved, lipidated and becomes a component of the autophagosome membrane, and this membrane translocation event is commonly used to monitor autophagy42,43.
The mechanics of phagophore and autophagosome formation and the recognition and capture of autophagosome cargo are presently unclear. Autophagy may be a nonspecific, bulk degradation process in some situations, and in others it may be specific for targeting mitochondria44–46, catalase47, peroxisomes48–50, endoplasmic reticulum51, and aggregation-prone proteins and protein aggregates6–8,52–54 for autophagy-mediated degradation. In the case of polyubiquitylated proteins, the multifunctional adaptor protein p62/SQSTM1 might facilitate the specific autophagosome-targeting process53,55. If or how other aggregation-prone proteins, protein aggregates and organelles are recognized and targeted by the autophagy machinery is yet to be determined.
Autophagy in cell survival or death
In cancer cells, autophagy can, in some situations, increase apoptotic20,56 (FIG. 1b) and caspase-independent57 cell death. However, autophagy has a more prominent role in sustaining cell viability in cancer cells with defects in apoptosis19,20,22,23 (FIG. 1c). Apoptosis-defective tumour cells have the remarkable ability to tolerate long-term metabolic stress, either by cytokine deprivation in dependent lymphoid cells22 or by oxygen (hypoxia) and glucose deprivation in epithelial cells19,20,23,30,33,34. Although the absence of pro-apoptotic BAX and BAK or the gain of anti-apoptotic BCL2 or BCL-XL function adequately explains why cells fail to die when deprived, it is insufficient to explain how cell viability is maintained for weeks under the harsh metabolic stress conditions of nutrient deprivation in vitro and in vivo. It is now clear that surviving metabolic stress in vitro and in tumours in vivo is dependent on autophagy.
The availability of cells with defects in apoptosis, with or without the capacity for autophagy (Becn1+/+ or Becn1 +/− and Atg5+/+, Atg5 +/− or Atg5−/−) has allowed a prolonged evaluation of the cellular response to metabolic stress, the survival from which is autophagy-dependent. Deprived epithelial and lymphoid cells using the autophagy survival function remain viable for weeks, during which time they undergo a dramatic decrease in cell size through progressive cellular consumption19,20,22,23. Although BCL2 localized to the endoplasmic reticulum can interact with BECN1 and inhibit autophagy58, in the functional context in which the normally predominantly mitochondrial BCL2 blocks apoptosis and confers tumorigenic growth, autophagy is efficiently induced in response to metabolic stress19,20,23,30. It remains possible that the regulation of BECN1 by BCL2 is more subtle or context-dependent.
It is clear from the analysis of time-lapse microscopy of apoptosis-defective immortal epithelial cells undergoing autophagy-mediated survival as a result of starvation that this is a complex, poorly characterized and prolonged process19,36. Initially, autophagy supports the continuation of cell proliferation under metabolic stress, which is consistent with a role for autophagy in the maintenance of homeostasis to support normal cell function during intermittent interruptions in nutrient availability (FIG. 1c). This ‘maintenance phase’ might also be crucial for sustaining ATP levels and cardiac function during the neonatal starvation period in mice38.
Beyond two days of starvation cell division ceases, cells shrink markedly (FIG. 1c), motility is suppressed and cells aggregate into small clusters19,20,23,36. These aggregated clusters of starved cells often show evidence of cells consuming other cells, or heterophagy, particularly when cells are in a confined space such as spheroid growth in Matrigel20. Whether these cells are dead or alive at the time of consumption or if the persistence of these corpses reflects degradation failure due to defective autophagy remains to be determined. Furthermore, if or how this contributes to viability of the cell population is not determined. This ‘preservation phase’ is probably vital for controlling cellular consumption and mitigating protein and organelle damage while suppressing metabolism and energy use to prolong cell viability and enable recovery (FIG. 1c).
Autophagy enables stress recovery
Remarkably, autophagy affords cells a resilient capacity for regeneration, whereby restoration of nutrients results in an increase in cell size to that before starvation (FIG. 1c) and resumption of cell proliferation19,22,36. This recovery process is rapid and efficient, and dramatically impaired by defects in autophagy (Becn1+/− or Atg5−/−). Thus, autophagy-deficient cells not only fail to tolerate metabolic stress but are also defective in the recovery process23. Mitigation of the damaging effects of stress, including damaged protein, DNA and organelle accumulation during this phase is probably crucial for enabling recovery.
Following prolonged starvation and progressive autophagy, what defines the minimal cell that is capable of recovery and what events eventually lead to cellular demise are currently unknown. This capacity for durable, long-term survival of metabolic stress through autophagy might be vital for the survival of tumour cells that remain viable following treatment, for metastatic tumour cells (FIG. 3c) and possibly for stem cells. In cells using autophagy to survive metabolic stress there is a gradual erosion of cell viability with time (FIG. 1c), but whether this is due to autophagic cell death or eventual cellular attrition through atrophy is not clear. Finally, the interaction of cells undergoing progressive autophagy, either dead or alive, with the immune system is unknown.
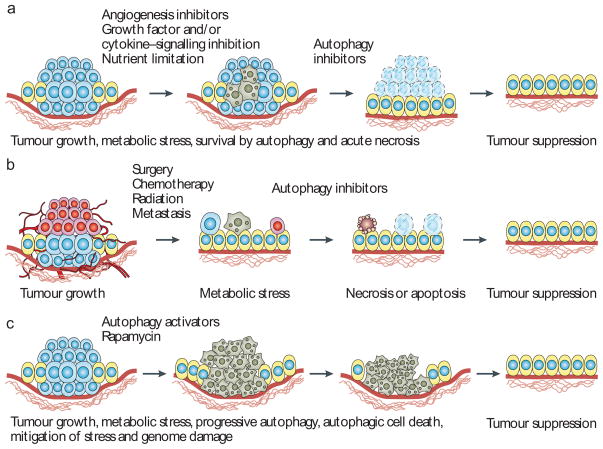
Figure 3. Application of autophagy modulation to cancer therapy.
a | In apoptosis-defective tumours that are reliant on autophagy to survive metabolic stress, autophagy inhibitors can be used to induce acute necrotic cell death that may be facilitated by proteasome inhibition, enabling tumour eradication. b | In the adjuvant setting, and after elimination of a large proportion of the tumour by radiation and chemotherapy, the remaining cells can reside in a disrupted and stressed environment, susceptible to inhibition of the autophagy survival mechanism. Tumour cells in the process of metastasis can be similarly vulnerable. c | Autophagy stimulators may be therapeutically useful to either promote autophagic cell death or to prevent the damaging effects of autophagy deficiency and mismanagement of metabolic stress leading to DNA damage and tumour progression. By limiting protein, organelle and ultimately DNA damage, autophagy stimulators may suppress tumour progression. In human breast, ovarian and prostate cancers, where allelic loss of BECN1 occurs with high frequency, correction of the autophagy deficiency with autophagy stimulators may delay tumour progression by reducing the rate at which tumour-promoting mutations accumulate.
Autophagy and tumour dormancy
One of the most daunting clinical problems is the frequent re-emergence of tumours following treatment, often after prolonged dormancy59. How residual tumour cells cope with metabolic stress and remain viable yet dormant needs to be determined, as elimination of these tumour cells might be essential to achieving durable treatment responses. The survival of tumour cells through autophagy may be a key mechanism to enable long-term tumour-cell survival and eventual re-growth and relapse. Thus, autophagy may allow residual or metastasizing tumour cells to tolerate metabolic deprivation with the flexibility to recover once growth conditions are favourable. This dramatic capacity for recovery afforded by autophagy vaguely resembles the process of sporulation in microorganisms, and its suppression may be essential to achieve efficient cancer eradication.
Metabolic stress and necrosis
Inactivation of autophagy, either by allelic loss of Becn1, deficiency in Atg5, RNAi knockdown of expression of essential autophagy regulators, or constitutive activation of the PI3K pathway and mTOR-mediated inhibition of autophagy, prevents cells surviving metabolic stress. In apoptosis-defective cells this results in cell death by necrosis19 (FIG. 1d). What specifically triggers necrosis is unknown, but insufficient ATP production to maintain plasma-membrane integrity resulting in metabolic catastrophe and cell lysis is highly probable36. This is important because inhibition of autophagy is a means for sensitizing tumour cells to metabolic stress that is effective even in tumour cells with defects in apoptosis that would otherwise be difficult to eliminate36,60. Although diverting apoptosis-defective tumour cells to a necrotic cell fate may not be a benign cell death61 (BOX 1), stimulation of acute necrotic cell death, if efficient enough, may be therapeutically useful (FIG. 3a). Indeed, the mechanism of cell death induction by alkylating agents involves poly(ADP-ribose) polymerase (PARP) activation and ATP consumption resulting in acute necrotic tumour cell death, which probably accounts for their success in the clinic62. Combining autophagy inhibitors with metabolic stress conditions might be a similarly effective means of promoting acute necrotic tumour cell death.
Box 1. Necrotic cell death stimulates inflammation.
Necrosis typically results from physical injury in which cell lysis and the release of intracellular contents, including HMGB1 (REF. 80), activate the innate immune system and a wound-healing response61. This recruitment of inflammatory cells provides cytokines, fostering cell growth to replace damaged tissue while removing cell debris24–26. Once the tissue damage is repaired, the wound-healing response abates. Tumours, however, often show persistent, chronic necrosis and inflammation in a corrupted version of a wound-healing response. Necrotic tumours are associated with poor prognosis and the persistent inflammatory infiltration and cytokine production are thought to promote tumour growth81. A high proportion of tumours have constitutive activation of the phosphatidylinositol-3 kinase pathway, which inhibits the induction of autophagy in response to metabolic stress, and many tumours also have defects in apoptosis. This generates a necrotic cell-death response to metabolic stress. A chronic necrotic response to persistent metabolic stress is created by rapid tumour growth and high metabolic demand that outpaces ATP production by glycolysis, angiogenesis and nutrient availability. Thus, a necrotic cell fate is a common event in tumorigenesis, and evidence suggests that this alters the tumour–microenvironment interaction, although the mechanisms involved are poorly understood.
Autophagy and tumour suppression
The role for autophagy as a survival mechanism in normal cells and in tumour cells seems to contradict the observation that loss-of-function mutations in the autophagy pathway are associated with tumour progression14–17,19,20. Furthermore, constitutive activation of the PI3K pathway is one of the most common events in human cancer63, and the downstream kinase mTOR restricts autophagy induction in response to starvation64. How loss of this autophagy-mediated survival pathway promotes tumorigenesis was initially difficult to reconcile; however, two non-mutually exclusive possibilities have emerged. One explanation is that stimulation of necrotic cell death and inflammation caused by defects in apoptosis and autophagy provides a cell with non-autonomous means of tumour promotion through induction of a chronic wound-healing response19. Another explanation is that proper management of metabolic stress through autophagy is required in tumour cells to suppress the accumulation of deleterious mutations, perhaps caused by the increased oxidative stress that can drive tumour progression20,23,36. As such, overall cellular viability is compromised in tumour cells with defects in autophagy, but this initial disadvantage is overcome by an increased mutation rate resulting from failure of stress management in a cell-autonomous mechanism of tumour promotion. This is analogous to the mechanism by which defects in DNA repair cause sensitivity to DNA damage, yet the accelerated rate of mutation that results from deficient DNA repair confers an increased incidence of tumour formation65.
Autophagy limits genome damage
Autophagy is required in stressed cells for maintaining protein and organelle quality control and energy homeostasis2. A possible reconciliation of the pro-survival and tumour suppression functions of autophagy is that some aspect of the mismanagement of metabolic stress in autophagy-deficient tumour cells leads to genome damage and tumour progression20,23,60,66. This could occur through protein, organelle and DNA damage, or insufficient ATP levels for essential cellular functions that are required to maintain genome integrity, such as mitosis and DNA replication and repair. This notion is supported by upregulation of the DNA damage response, evidence of DNA double-strand breaks, and aneuploidy in autophagy-defective immortal epithelial cells in association with increased tumorigenesis20,23 (FIG. 2c). This genome damage is manifested most obviously in cells with a defect in apoptosis, which would otherwise eliminate most of these damaged, abnormal cells, and is probably also facilitated by cell-cycle checkpoint inactivation (immortalization through RB1 and p53 loss). Increased genome damage resulting from an autophagy defect, however, does not require a defect in apoptosis23, consistent with the tumour-prone state of Becn1-heterozygous mutant mice that have the apoptotic response intact16,17. The origin of the increased DNA damage in autophagy-defective cells is not yet known. Malfunctioning organelles, accumulation of toxic protein aggregates, generation of reactive oxygen species and oxidative stress, and failure of energy homeostasis are all potential contributors to induction of genome damage when autophagy is defective.
Genetic instability and an enhanced mutation rate promote tumour-cell evolution and adaptation to drive progression and resistance to therapy (FIG. 2c). It is the rare, adapted, resistant tumour cells, which emerge typically following therapy, that are lethal. Autophagy deficient, immortal epithelial cells show an increased rate of gene amplification20,23, the most common mechanism of oncogene activation in human tumours67, rendering this scenario plausible. Cancer is a disease where it is often the case that what grows back kills you, and what grows back is a mutated and more aggressive version of the original tumour generated by mutation, selection and genome instability. Despite the reduced cellular fitness caused by deficient autophagy, the poor survival but superior adaptation through an increased mutation rate might be the key advantage that promotes tumorigenesis (FIG. 2c). Interestingly, autophagy is associated with longevity, suggesting a role in the suppression of ageing phenotypes68,69. As DNA damage accelerates both cancer and ageing, this supports a general role for autophagy in protecting cellular and genome fitness to prevent cancer and extend lifespan.
Autophagy inhibition in cancer therapy
The ability to inhibit autophagy and sensitize even apoptosis-resistant tumour cells to metabolic stress is a promising avenue for cancer therapy. As many current cancer therapies (angiogenesis and growth factor and receptor inhibitors, for example) inflict metabolic stress, to which tumours are particularly vulnerable because of high metabolic demand and reliance on glycolysis, autophagy inhibitors may be particularly useful (FIG. 3a). Even conventional surgery, radiation and chemotherapy disrupt tumour architecture and vascularization leaving any remaining tumour cells potentially susceptible to metabolic stress and autophagy-inhibitory therapy (FIG. 3b). Furthermore, the process of cancer metastasis necessarily requires that tumour cells survive in isolation from the primary tumour, without its nutrient support system. This metastasis-prone state may be particularly susceptible to autophagy inhibition as cells in isolation are expected to be more reliant on autophagy, although this remains to be investigated (FIG. 3b). Although inhibition of autophagy can promote apoptosis under nutrient-deprived conditions20,56, there is the advantage that this is also effective on tumour cells with defects in apoptosis through induction of acute necrotic cell death (FIG. 3a). Preclinical studies with chloroquine, which inhibits lysosome acidification and thereby autophagy, in conjunction with alkylating agents, showed remarkable efficacy inhibiting tumour growth in mice70. Synergy between chloroquine and the HDAC inhibitor SAHA in killing imatinib-refractory chronic myeloid leukaemia cells also indicates the protective role for autophagy, supporting the therapeutic use of autophagy inhibitors in cancer therapy71. Development of specific inhibitors that target the autophagy pathway directly, such as inhibitors of the kinases ATG1 and VPS34, which positively regulate autophagy, warrant similar investigation.
Another angle for the use of autophagy inhibitors is their potential synergy with proteasome inhibitors. Protein turnover by lysosomal degradation through the autophagy pathway is functionally coupled to, and compensatory with, the ubiquitin-mediated proteasome protein degradation pathway72–74. Thus compromising both proteasome- and autophagy-mediated protein degradation may be a particularly lethal combination to tumour cells with a high metabolic rate or with increased susceptibility to production of unfolded proteins, which is worth testing therapeutically (FIG. 3a). The proteasome inhibitor bortezomib has the approval of the US Food and Drug Administration and shows efficacy in treating multiple myeloma75 and more proteasome inhibitors are in development that will enable testing of this possibility.
Autophagy and chemoprevention
Promoters of autophagy also have the potential for clinical benefit in the setting of cancer prevention. As autophagy is required for the effective management of metabolic stress, promoting autophagy through mTOR inhibition, for example, might be expected to limit tumour progression (FIG. 3c). This approach has shown promise in the treatment of degenerative disorders that are linked to aberrant protein accumulation, such as Huntington’s disease, that is reliant on autophagy for the removal of aggregated mutant proteins74,76. Whether autophagy stimulation can be used in individuals at risk to slow the progression of breast, ovarian and prostate cancers, which have a high incidence of allelic loss of BECN1, remains to be investigated. However, distinguishing the contribution of autophagy stimulation to tumour regression from the anti-proliferative effects of mTOR inhibition will be essential for determining whether this is a valid approach.
Therapeutic induction of autophagic cell death through over-stimulation of autophagy remains another means for tumour-cell elimination (FIG. 3c). Cytotoxic drug treatment often triggers autophagy, particularly in apoptosis-defective cells, and the excessive cellular damage and attempt to remediate that damage through progressive autophagy can promote autophagic cell death77. To this end, a better understanding of the conditions that distinguish between the survival-supporting and death-promoting roles for autophagy will be necessary. Furthermore, roles for autophagy, along with the elucidation of the signalling pathways that confer specificity to the autophagic response downstream of different stimuli, will be necessary to selectively activate the specific and therapeutic response desired.
Future directions
We are at the initial stages of understanding the complex interplay of autophagy and cancer, but it is clear that autophagy is deeply integrated into metabolism, stress response and cell-death pathways. These responses might vary with cell type and type of stress, and will undoubtedly reflect the nature of the mutational events that have occurred in the tumour cells, not only that of BECN1 and the PI3K pathway as described above, but also p53 status78,79. Establishing the mutational events and the order of those events with respect to mutational inactivation of the autophagy pathway in human tumours, and using mouse models of human cancers, may be informative. Molecular markers for the functional capacity for autophagy in human tumours and their possible association with prognosis need to be established. The connection between the mismanagement of metabolic stress and the induction of genome damage in autophagy-deficient cells is important, but the origins of the DNA damage need to be identified. Increased oxidative stress is a likely candidate, but whether this arises from protein or organelle malfunction or is a consequence of metabolic insufficiency remains to be investigated. Existing therapies indirectly inflict metabolic stress, but how this is influenced by the competency of tumour cells for autophagy is not known. How autophagy is controlled and regulated, and the specificity that is associated with cellular consumption, requires investigation. It will be important to molecularly and biochemically characterize the minimal cells that are capable of recovery and their relationship to tumour dormancy, and to test the hypothesis that inhibition of autophagy can eliminate dormant or residual tumour cells for therapeutic benefit. Moreover, the possibility of activating autophagy to promote autophagic cell death or limit genome damage and progression in the setting of chemoprevention is intriguing. Finally, the molecular events that characterize necrotic cell death and their consequences need to be identified. We are only beginning to understand the tumour-microenvironment interaction and its interface with the immune system, and it is clear that they are intimately intertwined with tumour cell metabolism, cell death mechanisms and the cellular response to stress.
Acknowledgments
Research in the White laboratory is supported by grants from the National Institutes of Health and the Department of Defense. We thank T. Sharkey for assistance with preparation of the manuscript, the members of the White laboratory and our collaborators for helpful discussions.
Footnotes
DATABASES
Entrez Gene: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=gene
ATG5 | ATG7 | ATG12 | BAK | BAX | BCL2L1 | BECN1 | FRAP1 | HMGB1 | MAP1LC3 | NFκB | ULK1
Contributor Information
Robin Mathew, University of Medicine and Dentistry of New Jersey, Robert Wood, Johnson Medical School, Piscataway, New Jersey 08854, USA.
Vassiliki Karantza-Wadsworth, Division of Medical Oncology, Department of Medicine, University of Medicine and Dentistry of New Jersey, Robert Wood, Johnson Medical School, Piscataway, New Jersey 08854, USA, Cancer Institute of New Jersey, New Brunswick, New Jersey 08901, USA.
Eileen White, University of Medicine and Dentistry of New Jersey, Robert Wood, Johnson Medical School, Piscataway, New Jersey 08854, USA, Cancer Institute of New Jersey, New Brunswick, New Jersey 08901, USA, Center for Advanced Biotechnology and Medicine, Rutgers University, Piscataway, New Jersey, USA; and the Department of Molecular Biology and Biochemistry, Rutgers University, Piscataway, New Jersey 08854, USA.
References
- 1.Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 2.Mizushima N. Autophagy: process and function. Genes Dev. doi: 10.1101/gad.1599207. (in the press) [DOI] [PubMed] [Google Scholar]
- 3.Hara T, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 4.Komatsu M, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 5.Komatsu M, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ravikumar B, Duden R, Rubinsztein DC. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum Mol Genet. 2002;11:1107–1117. doi: 10.1093/hmg/11.9.1107. [DOI] [PubMed] [Google Scholar]
- 7.Shibata M, et al. Regulation of intracellular accumulation of mutant Huntingtin by Beclin 1. J Biol Chem. 2006;281:14474–14485. doi: 10.1074/jbc.M600364200. [DOI] [PubMed] [Google Scholar]
- 8.Williams A, et al. Aggregate-prone proteins are cleared from the cytosol by autophagy: therapeutic implications. Curr Top Dev Biol. 2006;76:89–101. doi: 10.1016/S0070-2153(06)76003-3. [DOI] [PubMed] [Google Scholar]
- 9.Colombo MI. Autophagy: a pathogen driven process. IUBMB Life. 2007;59:238–242. doi: 10.1080/15216540701230503. [DOI] [PubMed] [Google Scholar]
- 10.Qu X, et al. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell. 2007;128:931–946. doi: 10.1016/j.cell.2006.12.044. [DOI] [PubMed] [Google Scholar]
- 11.Baehrecke EH. Autophagy: dual roles in life and death? Nature Rev Mol Cell Biol. 2005;6:505–510. doi: 10.1038/nrm1666. [DOI] [PubMed] [Google Scholar]
- 12.Debnath J, Baehrecke EH, Kroemer G. Does autophagy contribute to cell death? Autophagy. 2005;1:66–74. doi: 10.4161/auto.1.2.1738. [DOI] [PubMed] [Google Scholar]
- 13.Reef S, et al. A short mitochondrial form of p 19ARF induces autophagy and caspase-independent cell death. Mol Cell. 2006;22:463–475. doi: 10.1016/j.molcel.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 14.Aita VM, et al. Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics. 1999;59:59–65. doi: 10.1006/geno.1999.5851. [DOI] [PubMed] [Google Scholar]
- 15.Liang XH, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 16.Qu X, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci USA. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marino G, et al. Tissue-specific autophagy alterations and increased tumorigenesis in mice deficient in Atg4C/autophagin-3. J Biol Chem. 2007;282:18573–18583. doi: 10.1074/jbc.M701194200. [DOI] [PubMed] [Google Scholar]
- 19.Degenhardt K, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karantza-Wadsworth V, et al. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev. 2007;21:1621–1635. doi: 10.1101/gad.1565707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karantza-Wadsworth V, White E. Role of autophagy in breast cancer. Autophagy. 2007;3:610–613. doi: 10.4161/auto.4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lum JJ, et al. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 2005;120:237–248. doi: 10.1016/j.cell.2004.11.046. [DOI] [PubMed] [Google Scholar]
- 23.Mathew R, et al. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 2007;21:1367–1381. doi: 10.1101/gad.1545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balkwill F, Charles KA, Mantovani A. Smoldering and polarized inflammation in the initiation and promotion of malignant disease. Cancer Cell. 2005;7:211–217. doi: 10.1016/j.ccr.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 25.Vakkila J, Lotze MT. Inflammation and necrosis promote tumour growth. Nature Rev Immunol. 2004;4:641–648. doi: 10.1038/nri1415. [DOI] [PubMed] [Google Scholar]
- 26.Zeh HJ, 3rd, Lotze MT. Addicted to death: invasive cancer and the immune response to unscheduled cell death. J Immunother (1997) 2005;28:1–9. doi: 10.1097/00002371-200501000-00001. [DOI] [PubMed] [Google Scholar]
- 27.Ellerman JE, et al. Masquerader: high mobility group box-1 and cancer. Clin Cancer Res. 2007;13:2836–2848. doi: 10.1158/1078-0432.CCR-06-1953. [DOI] [PubMed] [Google Scholar]
- 28.Karin M. Nuclear factor-kB in cancer development and progression. Nature. 2006;441:431–436. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- 29.Karin M, Lawrence T, Nizet V. Innate immunity gone awry: linking microbial infections to chronic inflammation and cancer. Cell. 2006;124:823–835. doi: 10.1016/j.cell.2006.02.016. [DOI] [PubMed] [Google Scholar]
- 30.Nelson DA, et al. Hypoxia and defective apoptosis drive genomic instability and tumorigenesis. Genes Dev. 2004;18:2095–2107. doi: 10.1101/gad.1204904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adams JM, Cory S. Bcl-2-regulated apoptosis: mechanism and therapeutic potential. Curr Opin Immunol. 2007;19:488–496. doi: 10.1016/j.coi.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fesik SW. Promoting apoptosis as a strategy for cancer drug discovery. Nature Rev Cancer. 2005;5:876–885. doi: 10.1038/nrc1736. [DOI] [PubMed] [Google Scholar]
- 33.Degenhardt K, Sundararajan R, Lindsten T, Thompson C, White E. Bax and Bak independently promote cytochrome c release from mitochondria. J Biol Chem. 2002;277:14127–14134. doi: 10.1074/jbc.M109939200. [DOI] [PubMed] [Google Scholar]
- 34.Tan TT, et al. Key roles of BIM-driven apoptosis in epithelial tumors and rational chemotherapy. Cancer Cell. 2005;7:227–238. doi: 10.1016/j.ccr.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 35.Warburg O. On respiratory impairment in cancer cells. Science. 1956;124:269–270. [PubMed] [Google Scholar]
- 36.Jin S, DiPaola RS, Mathew R, White E. Metabolic catastrophe as a means to cancer cell death. J Cell Sci. 2007;120:379–383. doi: 10.1242/jcs.03349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pua HH, Dzhagalov I, Chuck M, Mizushima N, He YW. A critical role for the autophagy gene Atg5 in T cell survival and proliferation. J Exp Med. 2007;204:25–31. doi: 10.1084/jem.20061303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuma A, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 39.Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nature Rev Mol Cell Biol. 2007 Aug 22; doi: 10.1038/nrm2245. [DOI] [PubMed] [Google Scholar]
- 40.Klionsky DJ. The molecular machinery of autophagy: unanswered questions. J Cell Sci. 2005;118:7–18. doi: 10.1242/jcs.01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ohsumi Y, Mizushima N. Two ubiquitin-like conjugation systems essential for autophagy. Semin Cell Dev Biol. 2004;15:231–236. doi: 10.1016/j.semcdb.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 42.Mizushima N. Methods for monitoring autophagy. Int J Biochem Cell Biol. 2004;36:2491–2502. doi: 10.1016/j.biocel.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 43.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jin S. Autophagy, mitochondrial quality control, and oncogenesis. Autophagy. 2006;2:80–84. doi: 10.4161/auto.2.2.2460. [DOI] [PubMed] [Google Scholar]
- 45.Kim I, Rodriguez-Enriquez S, Lemasters JJ. Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys. 2007;462:245–253. doi: 10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y, et al. The role of autophagy in mitochondria maintenance: characterization of mitochondrial functions in autophagy-deficient S. cerevisiae strains. Autophagy. 2007;3:337–346. doi: 10.4161/auto.4127. [DOI] [PubMed] [Google Scholar]
- 47.Yu L, et al. Autophagic programmed cell death by selective catalase degradation. Proc Natl Acad Sci USA. 2006;103:4952–4957. doi: 10.1073/pnas.0511288103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Iwata J, et al. Excess peroxisomes are degraded by autophagic machinery in mammals. J Biol Chem. 2006;281:4035–4041. doi: 10.1074/jbc.M512283200. [DOI] [PubMed] [Google Scholar]
- 49.Monastyrska I, Klionsky DJ. Autophagy in organelle homeostasis: peroxisome turnover. Mol Aspects Med. 2006;27:483–494. doi: 10.1016/j.mam.2006.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sakai Y, Oku M, van der Klei IJ, Kiel JA. Pexophagy: autophagic degradation of peroxisomes. Biochim Biophys Acta. 2006;1763:1767–1775. doi: 10.1016/j.bbamcr.2006.08.023. [DOI] [PubMed] [Google Scholar]
- 51.Yorimitsu T, Klionsky DJ. Eating the endoplasmic reticulum: quality control by autophagy. Trends Cell Biol. 2007;17:279–285. doi: 10.1016/j.tcb.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 52.Berger Z, et al. Rapamycin alleviates toxicity of different aggregate-prone proteins. Hum Mol Genet. 2006;15:433–442. doi: 10.1093/hmg/ddi458. [DOI] [PubMed] [Google Scholar]
- 53.Bjorkoy G, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sarkar S, et al. Small molecules enhance autophagy and reduce toxicity in Huntington’s disease models. Nature Chem Biol. 2007;3:331–338. doi: 10.1038/nchembio883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pankiv S, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 56.Boya P, et al. Inhibition of macroautophagy triggers apoptosis. Mol Cell Biol. 2005;25:1025–1040. doi: 10.1128/MCB.25.3.1025-1040.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Colell A, et al. GAPDH and autophagy preserve survival after apoptotic cytochrome c release in the absence of caspase activation. Cell. 2007;129:983–997. doi: 10.1016/j.cell.2007.03.045. [DOI] [PubMed] [Google Scholar]
- 58.Pattingre S, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 59.Aguirre-Ghiso JA. The problem of cancer dormancy: understanding the basic mechanisms and identifying therapeutic opportunities. Cell Cycle. 2006;5:1740–1743. doi: 10.4161/cc.5.16.3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jin S, White E. Role of autophagy in cancer: management of metabolic stress. Autophagy. 2007;3:28–31. doi: 10.4161/auto.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zong WX, Thompson CB. Necrotic death as a cell fate. Genes Dev. 2006;20:1–15. doi: 10.1101/gad.1376506. [DOI] [PubMed] [Google Scholar]
- 62.Zong WX, Ditsworth D, Bauer DE, Wang ZQ, Thompson CB. Alkylating DNA damage stimulates a regulated form of necrotic cell death. Genes Dev. 2004;18:1272–1282. doi: 10.1101/gad.1199904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 65.Bartek J, Lukas J. DNA damage checkpoints: from initiation to recovery or adaptation. Curr Opin Cell Biol. 2007;19:238–245. doi: 10.1016/j.ceb.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 66.Mathew R, White E. Why sick cells produce tumors: the protective role of autophagy. Autophagy. 2007;3:502–505. doi: 10.4161/auto.4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Albertson DG. Gene amplification in cancer. Trends Genet. 2006;22:447–455. doi: 10.1016/j.tig.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 68.Finkel T, Serrano M, Blasco MA. The common biology of cancer and ageing. Nature. 2007;448:767–774. doi: 10.1038/nature05985. [DOI] [PubMed] [Google Scholar]
- 69.Melendez A, et al. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- 70.Amaravadi RK, et al. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest. 2007;117:326–336. doi: 10.1172/JCI28833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Carew JS, et al. Targeting autophagy augments the anticancer activity of the histone deacetylase inhibitor SAHA to overcome Bcr-Abl-mediated drug resistance. Blood. 2007;110:313–322. doi: 10.1182/blood-2006-10-050260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ding WX, et al. Linking of autophagy to ubiquitin-proteasome system is important for the regulation of endoplasmic reticulum stress and cell viability. Am J Pathol. 2007;171:513–524. doi: 10.2353/ajpath.2007.070188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pandey UB, et al. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447:859–863. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- 74.Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–786. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- 75.Roccaro AM, et al. Bortezomib as an antitumor agent. Curr Pharm Biotechnol. 2006;7:441–448. doi: 10.2174/138920106779116865. [DOI] [PubMed] [Google Scholar]
- 76.Rubinsztein DC, Gestwicki JE, Murphy LO, Klionsky DJ. Potential therapeutic applications of autophagy. Nature Rev Drug Discov. 2007;6:304–312. doi: 10.1038/nrd2272. [DOI] [PubMed] [Google Scholar]
- 77.Shimizu S, et al. Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nature Cell Biol. 2004;6:1221–1228. doi: 10.1038/ncb1192. [DOI] [PubMed] [Google Scholar]
- 78.Crighton D, et al. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126:121–134. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 79.Feng Z, Zhang H, Levine AJ, Jin S. The coordinate regulation of the p53 and mTOR pathways in cells. Proc Natl Acad Sci USA. 2005;102:8204–8209. doi: 10.1073/pnas.0502857102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lotze MT, Deisseroth A, Rubartelli A. Damage associated molecular pattern molecules. Clin Immunol. 2007;124:1–4. doi: 10.1016/j.clim.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Balkwill F. Cancer and the chemokine network. Nature Rev Cancer. 2004;4:540–550. doi: 10.1038/nrc1388. [DOI] [PubMed] [Google Scholar]