Abstract
Repair and regeneration are key processes for tissue maintenance, and their disruption may lead to disease states. Little is known about the molecular mechanisms that underline the repair and regeneration of the digestive tract. The sea cucumber Holothuria glaberrima represents an excellent model to dissect and characterize the molecular events during intestinal regeneration. To study the gene expression profile, cDNA libraries were constructed from normal, 3-day, and 7-day regenerating intestines of H. glaberrima. Clones were randomly sequenced and queried against the nonredundant protein database at the National Center for Biotechnology Information. RT-PCR analyses were made of several genes to determine their expression profile during intestinal regeneration. A total of 5,173 sequences from three cDNA libraries were obtained. About 46.2, 35.6, and 26.2% of the sequences for the normal, 3-days, and 7-days cDNA libraries, respectively, shared significant similarity with known sequences in the protein database of GenBank but only present 10% of similarity among them. Analysis of the libraries in terms of functional processes, protein domains, and most common sequences suggests that a differential expression profile is taking place during the regeneration process. Further examination of the expressed sequence tag dataset revealed that 12 putative genes are differentially expressed at significant level (R > 6). Experimental validation by RT-PCR analysis reveals that at least three genes (unknown C-4677-1, melanotransferrin, and centaurin) present a differential expression during regeneration. These findings strongly suggest that the gene expression profile varies among regeneration stages and provide evidence for the existence of differential gene expression.
Keywords: intestinem, cDNA librariesm, echinoderm
Each year, 60 million to 70 million people are affected by digestive diseases, and over 234,000 deaths, including those from gastrointestinal cancers, are registered (2, 42). Many of the digestive diseases are due to malfunctions of intestinal cell growth and repair mechanisms (28). When the repair of the mucosal epithelium is disturbed, as occurs in diseases such as inflammatory bowel disease or chronic colitis, persistent ulcer lesions can appear (40). Another example is a series of digestive system diseases caused by deficient cellular control mechanisms that give rise to diverticuli, fistulas, and cancers (9, 41). Repair mechanims are not only involved in healing from disease but are also crucial after digestive tract surgery (18). Fast and efficient regeneration of the digestive tissues would minimize surgical complications, reduce hospitalization times, and improve general patient well-being following common surgeries such as colonoscopy or those associated with birth defects or cancers. In view of the importance of cell growth and repair control associated with digestive tract problems, it is surprising how little is known about gastrointestinal regeneration and the mechanisms underlying the healing process in this system (41).
To understand the mechanisms involved in intestinal regeneration, our laboratory has spearheaded the use of a novel model system, the echinoderm Holothuria glaberrima (19, 20, 46). These animals are phylogenetically related to vertebrates, and their digestive tracts are similar to those of vertebrates in structure and tissue composition. Their extraordinary capacity for gastrointestinal regeneration provides a model system for the study of the underlying mechanisms in this process. When holothurians are exposed to adverse stimuli, they respond by ejecting most of their internal organs. This evisceration process is followed by a period of regeneration where the ejected organs are replaced, the gastrointestinal tract being the first to regenerate (19, 20). During regeneration, all intestinal tissues are replaced, including the luminal epithelia, the visceral muscle, and the enteric nervous system.
Previous studies in our laboratory identified at least six stages of intestinal regeneration in the sea cucumber H. glaberrima (19). These stages are marked by cellular events that occur during the regeneration process. Stage 1 involves the process of wound healing, which corresponds to 0–3 days postevisceration (dpe). During this stage the mesenterial wound is closed by cells of the coelomic epithelia and an immune response is mounted against pathogens that may have entered during the evisceration process. In stage 2 (3–7dpe), the free edges of the mesenteries thicken and dedifferentiation and cell division of epithelial and muscle cells occur (11). Stage 3 (7–10 dpe) is characterized by the initial appearance of the luminal epithelium and remodeling of the extracellular matrix in the submucosa (46). Stage 4 (9–14 dpe) involves the proliferation and migration of the luminal epithelium, as well as the deposition of new extracellular matrix components (11, 46). In stage 5 (15–21 dpe), the lumen is continuous, but the layers have not yet acquired the properties of the noneviscerated organ. Thus, differential cellular growth and division occur. At stage 6 or growth phase, the layer composition of the intestine is similar to that of the noneviscerated gut, and the tissue grows to reach full size.
Although the cellular events underlining the regeneration process have been extensively studied in our laboratory (10, 11, 39), little is known about the genes involved. Previous attempts to identify the genes associated with intestinal regeneration in H. glaberrima have been made in our laboratory using a gene by gene strategy, which is time consuming and provided limited information (37, 49, 51). Expressed sequence tags (ESTs) provide an alternative method for identifying regeneration-associated genes. EST analysis allows rapid gene discovery, clarification of gene function, and the identification of stage specific gene expression profiles (1, 29!). EST analysis was used to test our hypothesis that the genetic expression profile changes during the different stages of regeneration. We generated an EST dataset from three cDNA libraries: two libraries from stages 1 and 2 of regenerating intestine and one from normal noneviscerated intestine of H. glaberrima. The ESTs obtained provide a preliminary view of the gene expression pattern in regenerating vs. normal animals and a valuable insight into the genes that may be involved in the regeneration process. The gene expression profile helps identify genes differentially expressed during the initial regeneration stages and in particular serves to discover genes homologous to those associated with disease or healing in humans.
Materials and Methods
Animals
Adult sea cucumbers, H. glaberrima, were obtained from the coastal waters of Puerto Rico and maintained in seawater aquaria at 22–24°C. Evisceration was induced by injections (3–5 ml) of 0.35 M KCl into the coelomic cavity. Prior to dissection, animals were anesthetized by placement in ice cold water for 1 h and then killed by cutting through the calcareous ring. The intestines of noneviscerated and regenerating animals at 3dpe and 7dpe were removed under RNase-free conditions and stored in RNAlater (Ambion, Austin, TX) until use.
cDNA libraries
Unidirectional cDNA libraries were constructed in the bacteriophage λ-ZAPII according to the procedures indicated in the cDNA synthesis and in vitro phage packaging of Stratagene's Uni-Zap kit (Stratagene, La Jolla, CA). The 7dpe (37) and the noneviscerated (normal) (49) cDNA libraries were previously described. The 3dpe cDNA library was constructed by Amplicon Express (Pullman, WA) using 2 mg of total RNA extracted with the RNAeasy midi prep kit from Qiagen (Valencia, CA). Mass excisions of the three cDNA libraries were performed using the Escherichia coli strain XL-1 Blue and phage supernatant according to the cDNA library manual (Stratagene). All three libraries were amplified but not normalized.
Transformed bacteria were plated in medium containing ampicillin to select for growth of those with an insert. Individual colonies were randomly picked and inoculated into 96-well microtiter plates. Colony PCRs were performed using M13 primers (Sigma Genosys, St. Louis, MO). The PCR products were analyzed by electrophoresis using 1.0% agarose gels and cleaned by ExoSap (USB, Cleveland, OH) prior to sequencing. The percentage of insert amplification was determined by dividing the number of amplified clones by the number of total clones submitted to PCR.
Nucleotide sequencing
Single-pass sequencing was performed on each PCR template using the T3 primer (5′-AATTAACCCTCACTAAAGGG-3′). PCR products were sequenced in 96-well microplates using DYEnamic ET dye terminator kit diluted four times with reaction buffer (1 M Tris·HCl, pH 9.0, 1 M MgCl) (Amersham Pharmacia Biotech, Piscataway, NJ) Sequencing was performed on a MegaBACE 500 capillary sequencer (Amersham Pharmacia Biotech). Samples were injected at 3 Kv for 50 s and run at 9 Kv for 120 min.
Sequence analysis, contig assembly, and BLAST similarity
The chromatogram produced for each clone was analyzed using Chromas 2.13 (Technelysium Pty, Helensvale, Australia). Quality control of sequenced ESTs was performed with Phred (21) with a cutoff of 20 for trimming low-quality regions, and Cross-Match (16) for vector trimming.
ESTAP (EST Analysis Pipeline) (35), a specialized database for ESTs, was used for assembly and analysis of the sequences. The ESTs obtained from cDNA sequencing were grouped into three projects: two for regenerating tissues (3 days and 7 days) and one for normal intestinal tissue. Initially, two subprojects for the 7dpe cDNA library (7dpe A and 7dpe B performed at different times and conditions) were maintained to corroborate the reproducibility of the results. The 7dpe A project mass excision and sequencing had been performed 2 yr before the 7dpe B project. After reproducibility was determined, the projects were fused together for further analysis. The contig assembly for the three libraries was performed using CAP3 (25) set to the default parameters. The contigs for each library were analyzed using a Venn diagram to determine the percentage of similarity among them (21, 54, 61).
Each cDNA sequence was queried against the nonredundant protein database at the National Center for Biotechnology Information (NCBI, Bethesda, MD) using the BLASTX algorithm (4). In both cases, the default BLAST parameters were used. BioParser, an EST database manager (12) was used to classify Blast results using an E-value of 1e-3 or less. After analysis, a putative product was assigned to the results from the similarity search (BLASTX). ESTs with an E-value < 1e-7 were classified according to their probable function. The ESTs significantly similar to hypothetical proteins were placed in the “unclassified” category. Sequences were also queried against those at the NCBI for the Holothuroidea and against a custom database of all known Strongylocentrotus purpuratus (NCBI, taxonomy ID: 7668), sequences extracted from GenBank (8) with the NCBI taxonomy Browser (62). The sea urchin S. purpuratus was selected because of its phylogenetically proximity and the extent of the sequencing efforts in this organism (45).
Domain and electronic annotation for contigs
Domains were searched with RPS-BLAST against the conserved domain database (35) from NCBI. BLAST, Gene Ontology (GO) annotation, and domain search output files were parsed for homologous sequences, with an E-value of 1e-7 as a cutoff for BLASTX searches against the sequence databases. The default cut-off of 0.01 was considered to yield significant similarity to conserved domains from the Conserved Domain Database (CDD). Biological process, molecular function, and cellular component were assigned to contigs based on the GO annotation of the closest annotated homolog (6). For this purpose, the GenBank annotation files from the GO database were downloaded and parsed for the gene identifier numbers of previously identified homologs.
In silico differential display of H. glaberrima contigs from regenerating and normal intestine
The statistic R test (55) was used to compare the abundance of genes in the three cDNA libraries (3dpe, 7dpe, and normal) and between normal and regenerating datasets under the null hypothesis that all libraries are equally distributed. A high R value indicates high divergence of the gene between libraries. The statistic R was obtained using the logarithmic likelihood ratio between the observed amount of fragments for a specific gene and its expected values. This statistic tends asymptotically to a X2 distribution (55). To validate the statistic R and select the significance threshold, the two methods suggested by Stekel and colleagues (55) were applied. First, a randomization procedure to determine the true differential expression and, second, application of the theory of large deviations.
We randomly generated 1,000 datasets and calculated the R statistic under the null hypothesis. The number of genes for each R value was averaged over the 1,000 runs and the three libraries. The significance was established under 99% of believability. The biological significance is given by the putative genes for which the statistic R value was greater than the threshold and consequently in the 1% of the sample distribution. In our case, the statistic significant values of R were >6.0.
Gene expression analysis
The differential gene expression of several genes (unknown c-4766-1, unknown NADH, melanotransferrin, and centaurin) was semiquantitatively determined by gene-specific relative RT-PCR using cytochrome b (168 bp) for normalization. Total RNA was extracted from the intestine tissues after homogenization using Tri-Reagent (MRC, Cincinnati, OH), following the manufacturer's specifications. Quality of the extracted RNA was evaluated by agarose electrophoresis, and quantity was determined by spectrophotometry at 260-nm optical density in a NanoDrop (ND-1000). For gene-specific relative RT-PCR, first-strand synthesis was performed with the RETROscript First Strand Synthesis Kit following the manufacturer's specifications (Ambion). In brief, 1 μg of total RNA was incubated with random decamers for 3 min at 80°C. After incubation, dNTPs, buffer, RNase inhibitor (Ambion), and reverse transcriptase were added, and the reaction was incubated at 44°C for 1 h. This was followed by a 10-min incubation at 92°C. The single-stranded cDNA obtained was amplified by PCR as described below. The primers were designed for optimal performance using the Whitehead Institute's Primer3 and Net primer. The primers were design to amplify a region of 207 bp of the unknown C-4766-1, 237 bp for unknown 4860-1, 241 bp for NADH, 335 bp for melanotransferrin, and 257 for centaurin. Primers for melanotransferrin, centaurin, and cytochrome b were synthesized at Sigma Genosys (St. Louis, MO). Primers for NADH and the unknown C-4766-1 at AlfaDNA (Montreal, Canada). Cycling conditions for the amplified products were as follow: unknown C-4766-1, unknown 4860-1, and NADH 94°C 45 s, 55°C 1 min, 72°C; melanotransferrin 94°C 45 s, 58°C 1 min, 72°C; centaurin 94°C 45 s, 56°C 1 min, 72°C; all performed for 35 cycles.
Data analysis for RT-PCR was carried out using the SPSS Base 8.0.1 statistical package (SPSS). Values are presented as the means ± SE. Differences between the means of each group were analyzed using the Student t-test.
Results
cDNA libraries and EST sequence quality analysis
Three independent cDNA libraries from normal and regenerating intestine (stage 1, 3dpe and stage 2, 7dpe) were sequenced to generate an EST dataset. A summary comparison of the three cDNA libraries is provided on Table 1. We found that the titers and the percent of phagemids with inserts were similar for the normal, 3dpe, and 7dpe intestine cDNA libraries (106–107 and 90–98%). However, the average insert size varied among libraries, with the shortest for the 3dpe cDNA library (Table 1). Single-pass sequencing was performed from 5′-end of 6951 cDNA clones randomly picked from the unidirectional, nonnormalized cDNA libraries. High-quality sequences were obtained from 5,173 clones, 1,704 from the normal cDNA library, 1,252 from the 3dpe cDNA library, and 2,217 from the 7dpe cDNA library. The read length of reliable sequence for each cDNA library ranged from 400 to 700 nt (Fig. 1) with quality averages that ranged from 37 to 50 (Table 1).
Table 1. Comparison of the 3dpe, 7dpe, and nonevicerated cDNA libraries.
| cDNA Library | pfu | % of Insert Amplification | Average Insert Size | # of Sequenced Clones | # of Clean ESTs | Read Sequence Length Average | Average Sequence Quality |
|---|---|---|---|---|---|---|---|
| 3dpe | 106 | 90 | 800 | 2,161 | 1,252 | 400 | 37 |
| 7dpe | 107 | 97 | 1,249 | 2,884 | 2,217 | 600 | 45 |
| Normal | 106 | 98 | 1,278 | 1,906 | 1,704 | 700 | 50 |
Percentage of amplification and insert size as well as the number of expressed sequence tags (ESTs) and their sequence quality are shown for the 3 cDNA libraries. pfu, Plaque-forming unit; dpe, day postevisceration.
Fig. 1.
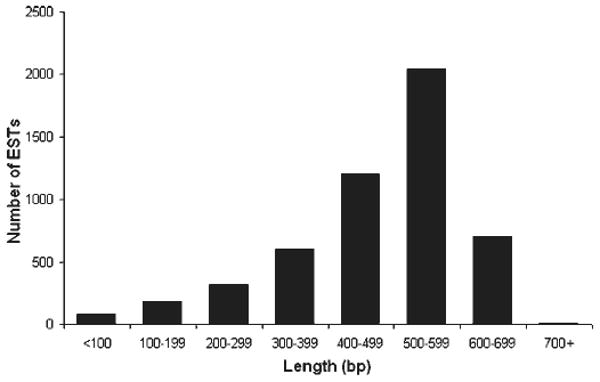
Distribution of sequence length for the dataset of Holothuria glaberrima. The average length of the expressed sequence tag (EST) sequence for the virtual project was 500 bp after quality assessment.
cDNA efficiency assessment
The efficiency and reproducibility of our protocols were corroborated by comparing two sets of ESTs for the 7dpe library performed at different times. The 7dpe (A) dataset had 773 ESTs and the 7dpe (B) had 1,444 ESTs, for a total of 2,217 sequences. The efficiencies of the two datasets were similar (72% for 7dpe A and 80% for 7dpe B). Moreover, genes that were previously reported in the 7dpe (A) such as actin, ferritin, thymosin, and QM protein were also identified in the 7dpe (B) dataset as well. In addition, genes such as collagen, serum amyloid, and ependymin, which were previously found by Northern blot and/or RT-PCR analyses to be expressed at 7dpe (49, 51, 56), were identified in both EST datasets.
EST BLAST analysis
ESTs were subjected to BLASTX searches against the nonredundant database (NCBI) for the identification of putative homologies to known proteins as of August 2006. Of the total dataset, 3,242 sequences (72.2%) had no similarity with known genes. About 90% of these sequences presented an open reading frame (Table 2). A total of 1,112 sequences (21%) shared similarity with previously described proteins. The E-value distribution of these 1,112 sequences in the nonredundant database is shown in Fig. 2A. The 3% of the ESTs that matched a protein with an E-value < 1e-100 could be considered true orthologs (23). About 65% of the ESTs had an E-value between 1e-20 and 1e-99, and 32% E-value 1e-03 to 1e-19, presenting significant and weak similarity, respectively. We also compared our ESTs dataset against the ESTs database of the sea urchin S. purpuratus (NCBI), finding that 1,905 (38%) of the sequences shared similarity with a putative protein in the sea urchin database. The E-value distribution of these 1,905 sequences in the sea urchin is shown on Fig. 2B. About 2% of the ESTs have an E-value < 1e-100, which might be considered as true orthologs, 77% had E-values between 1e-20 and 1e-99, presenting significant similarity, and 21% had weak similarity (E-value 1e-03 to 1e-19). Our dataset was also searched against the 657 ESTs and 14 genes reported in the NCBI for different species of holothurians and found that 97.4% of these ESTs are present in our dataset. These sequences correspond to 639 ESTs in our dataset; thus 87% of our sequences are novel, enriching the existing database for holothurians with ∼4,534 new sequences. From these new 4,534 sequences, 2,348 are nonredundant.
Table 2. Match to GenBank sequences of ESTs from 3dpe, 7dpe, and noneviscerated intestines cDNA libraries.
| cDNA Library | % of Identified Proteins, E-value < 10-7 | % of Hypothetical Proteins, E-value < 10-4 | % of Unidentified Sequences | % Sequences With ORF |
|---|---|---|---|---|
| 3dpe | 35.6 (9.37 ribosomal) | 12.4 | 52.0 | 93.5 |
| 7dpe | 26.2 (10.70 ribosomal) | 8.6 | 65.2 | 90.7 |
| Normal | 46.2 (4.96 ribosomal) | 5.5 | 48.3 | 89.0 |
Percentage of the identified proteins, hypothetical proteins, and unidentified sequences present in the 3 cDNA liraries. “% of Hypothetical Proteins” represents the group of ESTs in each cDNA library that presents a match with a hypothetical protein in the database. “% of Unidentified Sequences” represents the group of ESTs in each cDNA library without any similarity match in GenBank (unknowns). “% Sequences with ORF” represents the group of unknown ESTs with an open reading frame (ORF).
Fig. 2.
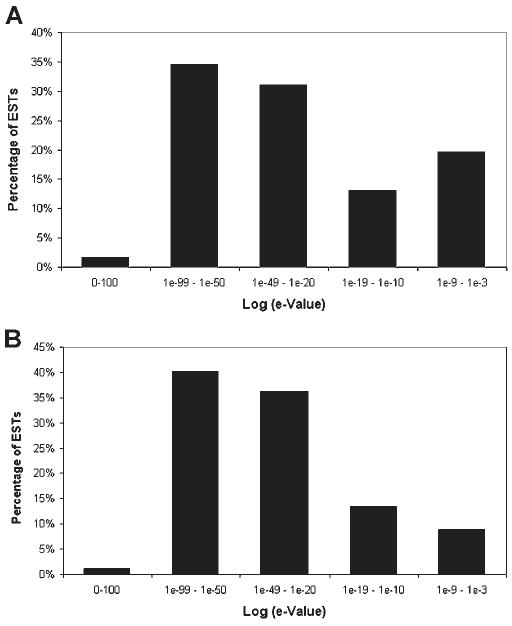
Distribution of hits per E-value of the ESTs from sea cucumber H. glaberrima. A: distribution of E-value from the first identified hit in the protein nonredundant database that was assigned a putative identity to the EST. A total of 1,112 sequences were found to have a match at the database. B: distribution of E-value from the first identified hit in the National Center for Biotechnology Information (NCBI) Strongylocentrotus purpuratus database that was assigned a putative identity to the EST. A total of 1,905 sequences present a match to the sea urchin database. For both, distributions homologies with E-value 1e-3 or less were considered. The majority of the ESTs in both analysis present an E-value between 1e-20 and 1e-99.
We analyzed the extent of overlap among the three cDNA libraries generated from regenerating and normal intestine to determine the percentage of shared transcripts between them. The libraries were ∼10% similar, which suggested that stage-specific gene expression is represented in each cDNA library. To further characterize genetic profile of the libraries, the 10 contigs with the highest EST frequency for each library were determined (Table 3). As expected, some contigs such as cytochrome c oxidase, ATP synthase, and NADH dehydrogenase were shared among the libraries. Also, even though ribosomal ESTs were present among the libraries, some ribosomal sequences were only identified in one of the libraries (L6 for normal, L36 for 3dpe, and L4 for 7pde), suggesting a differential expression profile among them.
Table 3. Summary of most abundant contigs among normal, 3dpe, and 7dpe cDNA libraries of Holothuria glaberrima.
| No. of ESTs, % | Identity |
|---|---|
| Normal | |
| 138 (8.1) | cytochrome c oxidase subunit 1 |
| 96 (5.6) | unknown (4277-3) |
| 58 (3.4) | cytochrome c oxidase subunit 3 |
| 33 (1.9) | ferritin |
| 33 (1.9) | cytochrome c oxidase subunit 2 |
| 21 (1.2) | cytochrome b |
| 14 (0.8) | thymosin beta-4 (T beta 4) |
| 13 (0.8) | unknown (125-4) |
| 13 (0.8) | ATP synthase a chain |
| 11 (0.6) | NADH dehydrogenase subunit 4 |
| 3dpe | |
| 104 (8.3) | unknown (4277-3) |
| 18 (1.4) | unknown (125-4) |
| 14 (1.1) | unknown (4911-1) |
| 14 (1.1) | cytochrome b |
| 13 (1.0) | unknown (4791-1) |
| 13 (1.0) | cytochrome c oxidase subunit III |
| 12 (1.0) | ferritin GF1 |
| 12 (1.0) | ATP synthase F0 subunit 6 |
| 12 (1.0) | unknown (4766-1) |
| 11 (0.9) | ribosomal protein L7-like |
| 7dpe | |
| 122 (5.5) | unknown (4277-3) |
| 44 (2.0) | cytochrome c oxidase subunit i |
| 31 (1.4) | cytochrome b |
| 19 (0.9) | unknown (5020-1) |
| 17 (0.8) | ATP synthase F0 subunit 6 |
| 16 (0.7) | cytochrome c oxidase subunit II |
| 14 (0.6) | cytochrome c oxidase subunit III |
| 13 (0.6) | unknown (125-4) |
| 11 (0.5) | actin |
| 10 (0.5) | NADH dehydrogenase subunit 4 |
The 10 most frequent ESTs are shown for each library. Percentages are based on the total ESTs for each cDNA library. Percentage of the identified proteins, hypothetical proteins, and unidentified sequences present in the 3 cDNA ibraries.
The ESTs were assembled into contigs to identify sequences representing the same transcript. Contig assembly was performed for each library, and 154, 134, and 262 contigs were found in the normal, 3dpe, and 7dpe cDNA libraries, respectively (Table 2). To determine the diversity of the libraries, redundancy was estimated based on the fraction of sequences that assembled into contigs. The estimated redundancy of the normal, 3dpe, and 7dpe cDNA libraries was 55, 58, and 53%, respectively. Some contigs were highly represented in one library but not the others. For example, sequences that have a match with actin were one of the largest contigs in the 7dpe cDNA library, while sequences with similarity to B-thymosin formed one of the largest contigs in the normal (noneviscerated) cDNA library. Interestingly, contigs without similarity to sequences in the databank were also among the largest in all three libraries. Some of these (4911-1, 4791-1, and 4766-1) were found to be mostly represented in the 3dpe library.
Common domains in the H. glaberrima dataset
Potential domains for the H. glaberrima contigs were identified performing an RPS-BLAST search against the conserved database (CDD, NCBI). About 42.9, 54.5, and 42.8% of the contigs of 3dpe, 7dpe, and normal libraries had a known protein domain. Domains such as Cox and ribosomal were identified in all three libraries (Table 4). Cellular functions were assigned to the identified domains and analyzed among the contigs. The most common domains over all three libraries were in the “oxidoreductase activity and protein biogenesis” category, which may have resulted from redundant annotations of cytochromes and ribosomal sequences. The “generation of precursor metabolites and energy” was the second most abundant category followed by “mitochondrial electron transport” category. Among the protein domains identified in the 3dpe library are nucleoplasmin, SAA, GroEL, A2M, and EGF-Lam. Also, proteins domain such as Band 7-prohibitin, vWFA, SapB, PP2c, and NDK were found only in the 7dpe library. Interestingly, one of the unknown contigs (#4766-1) highly represented in the 3dpe library presented a EF-hand domain, however, further characterization of this contig is needed.
Table 4. Total EST clones displaying similarity (BLASTX, E-value < 1E–07) to proteins in the nonredundant protein NCBI database, grouped into functional categories according to expressed gene anatomy database, partial representative list.
| Clone | Protein Match | Species Match | E-value | Accession No. |
|---|---|---|---|---|
| Chromosome structure/Signal transduction | ||||
| NP8F3 | histone protein Hist2 h3c1 | Gallus gallus | 9e-36 | XP_425464 |
| NP9C12 | H3.3 histone | Paracentrotus lividus | 1e-35 | CAAS3692 |
| 7dpB2D4 | development-specific protein-217 | Tripneustes gratilla | 4e-31 | A30033 |
| 7dpB11F7 | MAP kinase | Mus musculus | 4e-22 | NP_067437 |
| 7dpA15B6 | G protein | Xenopus laevis | 5e-92 | AAH41541 |
| NP7H1 | calreticulin | Strongylocentrotus purpuratus | 9e-54 | NP_999643 |
| Stress response/Cell defense | ||||
| NP18F1 | cathepsin C | Gallus gallus | 7e-36 | XP_417207 |
| 3dp13f3 | alpha-2-macroglobulin | Apis mellifera | 7e-18 | XP_392454 |
| 3dp8A6 | serum amyloid A protein | Holothuria glaberrima | 2e-41 | AAG24633 |
| 7dpB9A1 | fibrinogen-related protein | Parastichopus parvimensis | 6e-28 | A35084 |
| 7dpB13A10 | lactotransferrin | Strongylocentrotus purpuratus | 2e-47 | XP_786179 |
| NP15H1 | lactadherin | Strongylocentrotus purpuratus | 3e-21 | XP_793834 |
| NP14A6 | Fc fragment IgG | Strongylocentrotus purpuratus | 2e-16 | XP_780608 |
| NP8E3 | Tep2 | Drosophila yakuba | 7e-13 | AAR09708 |
| NP13F3 | similar to interferon | Strongylocentrotus purpuratus | 4e-10 | XP_783602 |
| NP16C11 | similar to fibronectin | Strongylocentrotus purpuratus | 6e-15 | XP_797088 |
| NP16E2 | GalNac-specific lectin | Asterina pectinifera | 7e-12 | BAB78598 |
| 3dp13D10 | superoxide dismutase | Gryllotalpa orientalis | 6e-57 | AAV73809 |
| Protein synthesis, processing, storage, transport, and degradation | ||||
| NP2E7 | proteasome | Strongylocentrotus purpuratus | 6e-61 | XP_780413 |
| 3dp8G2 | poly(A)-binding protein 1 | Gallus gallus | 9e-28 | XP_428547 |
| 3dp21A2 | heterogeneous nuclear ribonuleoprotein K | Homo sapiens | 5e-23 | CAI16019 |
| 3dP20C1 | translation elongation factor 1-alpha | Malassezia pachydermatis | 4e-26 | AAY46260 |
| 3dp24H9 | protein transport (SEC61b) | Strongylocentrotus purpuratus | 6e-17 | XP_786999 |
| 7dpA3C7 | transcription factor BIGMAX | Drosophila melanogaster | 5e-19 | AAG40148 |
| NP2D2 | vacuolar sorting protein | Bos taurus | 2e-52 | XP_615172 |
| 7dpB6A3 | QM | Pinctada fucata | 2e-94 | AAN85578 |
| 7dpA15 h3 | SSR gamma | Homo sapiens | 7e-39 | AAP97205 |
| 7dpB9D10 | solute carrier protein | Paralichthys olivaceus | 3e-45 | BAE48204 |
| 7dpB8G2 | matrix metalloproteinase | Notophthalmus viridescens | 3e-27 | AAX14804 |
| 7dpB5G7 | ferritin | Crassostrea gigas | 2e-55 | AAP83793 |
| 7dpB6E3 | alpha 1 (V) collagen | Gallus gallus | 7e-24 | AAF28099 |
| 3dp23B3 | actin | Hydroides elegans | 3e-28 | BAE19915 |
| NP2F2 | myosin light chain | Gallus gallus | 2e-11 | P02605 |
| NP2C3 | tubulin beta2 | Danio rerio | 2e-85 | AAQ97859 |
| NP5C5 | thymosin beta-4 precursor | Rattus norvegicus | 7e-11 | I52084 |
| 7dpA4F9 | tektin | Strongylocentrotus purpuratus | 2e-59 | NP_999787 |
| 7dpA2E2 | dynein | Canis familiaris | 9e-44 | XP_537691 |
| Cell proliferation/Apoptosis | ||||
| 7dpA3B9 | translationally control tumor protein | Branchiostoma belcheri | 7e-23 | AAK84394 |
| 7dpB10F8 | survivin | Xenopus laevis | 2e-22 | |
| 7dpA8F11 | programmed cell death 4 | Xenopus tropicalis | 5e-11 | AAH76698 |
| 7dpA15D10 | novel death regulatory protein (GRIM-19) | Rattus norvegicus | 7e-35 | XP_214305 |
| Nervous system | ||||
| 7dpA15B8 | slit | Gallus gallus | 2e-16 | XP_416992 |
| 7dpB6E10 | ependymin | Holothuria glaberrima | 4e-55 | AAR89380 |
| 7dpB10H6 | laminin | Tripneustes gratilla | 4e-65 | U02371 |
| 7dpA1D9 | synaptotagmin | Danio rerio | 6e-14 | NP_999861 |
| Development | ||||
| 7dpA8F5 | vitellogenin | Pseudocentrotus depressus | 4e-44 | AAK57983 |
| 3dp7D7 | Par-3 | Strongylocentrotus purpuratus | 6e-13 | XP_785679 |
| 7dA9D3 | Wnt-14 | Gallus gallus | 1e-37 | |
| 7dB13E9 | stromal derived factor | Strongylocentrotus purpuratus | 2e-39 | XP_784191 |
| 3dp17C1 | calponin | Branchiostoma belcheri | 3e-23 | BAC16745 |
NCBI, National Center for Biotechnology Information.
Functional category analysis
The GO database was used to identify the biological process of the putative homologous proteins found among the ESTs libraries. We examined the assigned biological process to determine differences in the distribution of these processes among the libraries. For this purpose, the biological process was split into different categories (Fig. 3). The most prominent functional category in the normal cDNA library was metabolism. In the 3dpe and 7dpe libraries, metabolism and protein metabolism/modification appeared to have a similar representation. The percentage of ESTs involved in cell proliferation/apoptosis, cell adhesion, and intracellular signaling appeared to be higher in the 7dpe (7.9, 8.6, and 3.7%, respectively) than in the 3dpe (4.0, 0.5, and 1.9%, respectively) and normal (0.2, 1.4, and 0.7%, respectively) libraries (Fig. 3). In the 3dpe library the percentages of ESTs involved in inflammatory response were higher (3.5%) when compared with the 7dpe (2.0%) and normal (2.2%) libraries.
Fig. 3.
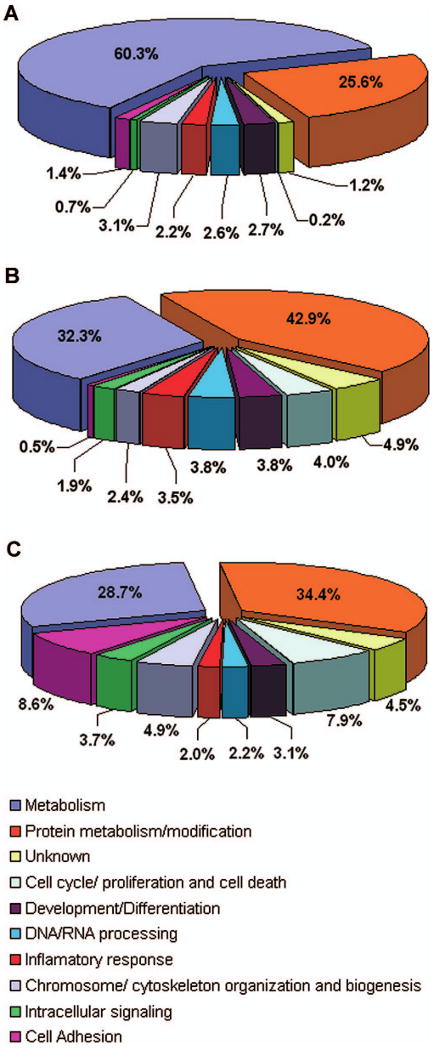
Distribution of expressed sequence tags according to the various functional categories after Gene Ontology (GO) analysis. Differences in the distribution of functional categories in the noneviscerated (normal, A), 3 days postevisceration (dpe, B), and 7dpe (C) cDNA libraries were determined for ESTs with an E-value of 1e-7.
In addition to this overview of the expression profiles, we also examined our datasets for individual sequences potentially useful in studying regeneration, healing, and developmental processes (Table 4). We searched for sequences involved in regulating cell cycle, proliferation, and apoptosis or those that could play a role during extracellular matrix remodeling and signal transduction pathways. Several sequences matched putative homolog genes involved in the processes mentioned above such as matrix metalloproteinases (MMPs), melanotransferrin, and centaurin (Table 5).
Table 5. Partial list of the protein domains identified in the H. glaberrima contigs and their cellular function.
| 3dpe | 7dpe | ||||
|---|---|---|---|---|---|
| Normal Domain | Normal Cellular Function | Domain | Cellular Function | Domain | Cellular Function |
| FRQ1 | signal transduction mechanisms/cytoskeleton/cell division and chromosome partitioning | SOUL | heme-binding proteins | TR-Fer | iron ion homeostasis |
| Euk-ferritin | metal ion binding | SAA | acute-phase response | Band-7-prohibitin | regulation of apoptosis/histone deacetylation/signal transduction |
| Epend | cell-matrix adhesion | EGF-Lam | ell adhesion, growth migration, and differentiation | vWFA | ell adhesion, migration, homing, pattern formation, and signal transduction |
| Beta-tubulin | major components of microtubules | A2M | endopeptidase inhibitor activity | ANK | transcription initiation, cell-cycle regulation, cytoskeletal, ion transport and signal transduction |
| H3 | nucleosome assembly/chromosome organization and biogenesis | GroEL | cellular protein metabolism | PP2Cc | Intracellular signalling |
| Ubiquitin | ubiquitin conjugating enzyme activity | Sec61-beta | intracellular protein transport | SOUL | heme-binding proteins |
| Cyclophilin-ABH | protein folding | Fubi | tumor suppressor protein | COL 1 | extracellular matrix structural constituent |
| WD40 | adaptor/regulatory modules in signal transduction, premRNA processing and cytoskeleton assembly | TR-Fer | iron ion homeostasis | NDK | nucleoside-diphosphate kinase activity, ATP binding |
| PCBP-like HK | mRNA stabilization, translational activation, and translational silencing | nucleoplasmin | nucleic acid binding | WD40 | adaptor/regulatory modules in signal transduction, premRNA processing and cytoskeleton assembly |
| AhpC | posttranslational modification, protein turnover, chaperones | RPP1A | translation, ribosomal structure and biogenesis | FRQ1 | Signal transduction mechanisms/Cytoskeleton/Cell division and chromosome partitioning |
| CT | regulate ligand-receptor interactions for a variety of signaling pathways | Cox2 | cytochrome c oxidase activity | TRAP-beta | cotranslational protein targeting to membrane |
| Cox2 | cytochrome c oxidase activity | Euk-Ferritin | metal ion binding | Sap B | activation of various lysosomal lipid-degrading enzymes |
| Cox4 | cytochrome c oxidase activity | RPL30 | translation, ribosomal structure and biogenesis | Cox2 | cytochrome-c oxidase activity |
| CyoB | energy production and conversion | ATP-synt-A | proton transport | NADH5 | ATP synthesis coupled electron transport |
| Rplx | translation, ribosomal structure and biogenesis | RpsN | translation, ribosomal structure and biogenesis | Actin | structural constituent of cytoskeleton |
| Oxired-q2 | mitochondrial electron transport, NADH to ubiquinone | RpsK | translation, ribosomal structure and biogenesis | RPP1A | translation, ribosomal structure and biogenesis |
| Ribosomal S8 | translation, ribosomal structure and biogenesis | Cyt-c-oxidase | mitochondrial electron transport | OSCP | ATP synthesis coupled proton transport |
| Actin | structural constituent of cytoskeleton | actin | structural constituent of cytoskeleton | RAMP4 | translation, ribosomal structure and biogenesis |
| RplO | translation, ribosomal structure and biogenesis | ribosomal-S8 | translation, ribosomal structure and biogenesis | H2A | nucleosome assembly, chromosome organization and biogenesis |
| NADH5 | ATP synthesis coupled electron transport | NADHdh | electron transport | 40S-S3-KH | translation, ribosomal structure and biogenesis |
Partial list of the most relevant and abundant proteins domains identified in the 3 cDNA libraries.
In silico differential display of the regenerating and normal H. glaberrima contigs
To determine differences in sequence frequency at each stage, we compared the representation of the ESTs in the three libraries by performing an in silico differential display analysis. From 5,173 transcripts of the three cDNA libraries, 12 of them showed a significant differential expression under the null hypothesis that all genes are expressed equally among the libraries (Table 6). The statistic R resulted in a range of values from 0 to 25.75. These values were dependent on the amount of transcripts obtained in all libraries (Fig. 4). In our case, R < 6.0 meant only one false positive for every 17 significant genes. In contrast, if the statistic R > 6.0, the amount of significant genes was 12 with no false positives. Thus, our cut-off for statistical significance was R > 6.0.
Table 6. Differential expression of contigs in H. glaberrima cDNA libraries.
| Gene ID | Clone Name | Statistic R | ↑ | ↓ |
|---|---|---|---|---|
| 2986-2 | cytochrome c oxidase subunit I [Cucumaria miniata] | 25.57 | 3 | N |
| 4791-1 | unknown (4791-1) | 15.95 | 7 N | 3 |
| 2986-3 | cytochrome c oxidase subunit I [Cucumaria miniata] | 13.85 | 3 7 | N |
| 94-1 | cytochrome c oxidase subunit III [Cucumaria miniata] | 12.77 | 3 | N |
| 4277-4 | unknown (4277-4) | 10.29 | 7 N | 3 |
| 4860-1 | unknown (4860-1) | 9.93 | 7 N | 3 |
| 76-2 | ferritin GF1 [Crassostrea gigas] | 8.73 | 37 | N |
| 5087-2 | toposome [Paracentrotus lividus] | 7.77 | 3 7 | N |
| 4766-1 | unknown (4766-1) | 6.91 | N | 3 |
| 5141-2 | unknown (5141-2) | 6.67 | 3 7 | N |
| 990-7 | cytocrome c oxidase subunit II [Cucumaria miniata] | 6.46 | 3 | N |
| 4911-1 | unknown (4911-1) | 6.36 | 7 N | 3 |
Data are presented from highest to lowest statistic R value. The Gene ID corresponds to local identification, the clone name shows the best BLASTX homology with the NCBI GenBank.
Fig. 4.
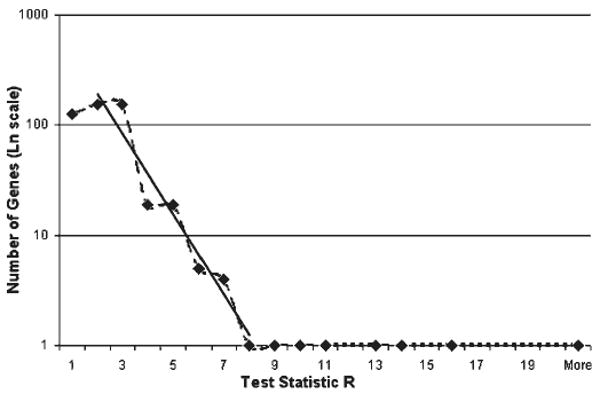
Number of genes in logarithmic scale as a function of R. The solid line represents the regression for 1 ≤ R ≤ 8 in which the slope is −0.85 with SE of 0.09. This is close to the −1 with 5% of error. For R ≥ 8 the number of genes is above this exponential curve and is greater than predicted by the large deviations calculation and thus, with true biological effect.
Gene expression analysis
Gene-specific relative RT-PCR was performed to validate the expression of some of the ESTs that were found in our analyses (Fig. 5). The sequence of the primers used is presented in Table 7. Two of these were chosen from the R analyses and represented sequences that were expected to be overexpressed in animals at 3 days postevisceration. Two other sequences corresponded to housekeeping genes, NADH and cytochrome b that according to the R analyses were not expected to change their expression during regeneration. These genes are usually used as a reference in determining expression changes. Finally, two other genes that appeared only in the regenerating libraries were analyzed. These genes had high similarities to melanotransferrin and centaurin. Melanotransferrin formed a contig of 8 ESTs that were found in the 3dpe and 7dpe libraries, but not in the normal intestinal library. Unpublished experiments in our laboratory had shown the gene to be overexpressed in coelomocytes. Centaurin, was a unique sequence found in the 7dpe library, thus we were interested in studying the possibility that genes found only within the regenerating libraries were overexpressed during the regeneration process.
Fig. 5.
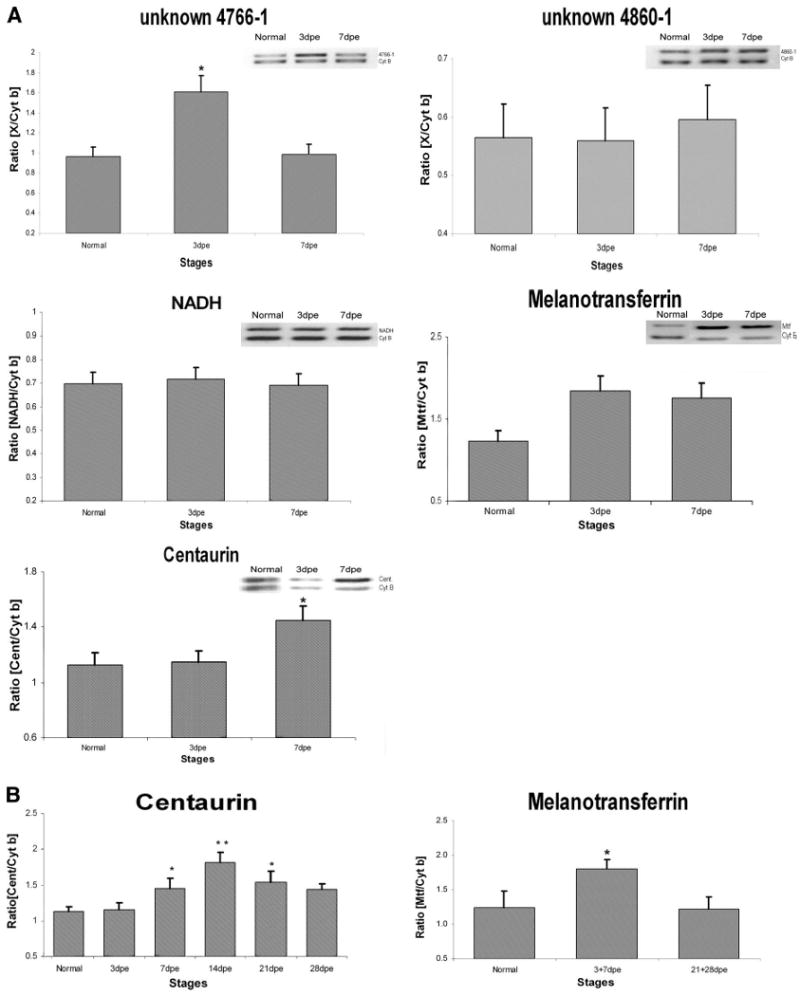
Gene expression profile analysis of unknown 4766-1, unknown 4680-1, NADH, centaurin, and melanotransferrin mRNA during intestinal regeneration. Analysis was performed for all samples by conventional RT-PCR and normalized against cytochrome b (Cyt b, 161 bp). A: compared with normal animals, unknown 4766-1 presents a significant overexpression at 3dpe (*P < 0.01, 2-tailed t-test). Centaurin expression presents a significant increase in 7dpe (*P < 0.01) compared with 3dpe and normal animals. Melanotransferrin presents an increase tendency in 3dpe and 7dpe but is not statistically significant. Unknown 4680-1 and NADH expression didn't show a significant change. B: centaurin presents a significant overexpression in stages 7dpe–21dpe, where the peak of expression appears at 14dpe (**P < 0.001, 2-tailed t-test) compared with normal animals. Overexpression of melanotransferrin was detected in 3dpe+7dpe (*P < 0.05, 2-tailed t-test) compared with later stages of regeneration (21 +28dpe) and normal animals. Values are presented as means ± SE of 5 animals per group.
Table 7. Forward and reverse primers designed for RT-PCR.
| Sequence | Primer Set |
|---|---|
| Unknown C-4766-1 | forward-5′-ATGTCGATGGTGATGAGAAGATT-3′ |
| reverse-5′-AGGTGACATGATAGGTGCAGTTC-3′ | |
| Unknown 4860-1 | forward-5′-GGTATTGTTGTGCCATTCCT-3′ |
| reverse-5′-CCTCGGTATCCTCCTCCTG-3′ | |
| Centaurin | forward –5′-GGGGCAGTACAGACAAGAT-3′ |
| reverse –5′-ATGAAGTGGTGTTCGCATA-3′ | |
| Melanotransferrin | forward –5′-TCGGAACAAGAACAGCAAAAG-3′ |
| reverse –5′-TCACCCACCCAGAAGTC-3′ | |
| NADH | forward –5′-CGGCTACTTCTGCGTTCTTC-3′ |
| reverse –5′-ATAGGCGCTGTCTCACTGGT-3′ | |
| Cytochrome | forward –5′-ACGAGGACACCCGCTATTCC –3′ |
| reverse –5′-GTCTGCTGTGTAGTGCATGGC –3′ |
Our results show that only one of the analyzed genes was overexpressed at the 3dpe stage compared with 7dpe and normal intestines. This was the unknown C-4766-1, providing some validation to our R analyses that predicted this gene to be overexpressed at 3dpe (Fig. 5A). However, a second unknown showed no significant changes in expression, which might be due to a limitation in the sensitivity of the PCR analyses, to large variability in the expression within individuals, or to the possibility that not all genes identified by R are true positives.
Two other genes showed changes in their expression. Centaurin was overexpressed in the 7dpe animals compared with 3dpe and normal intestines, and melanotransferrin showed a trend for an increase in both 3 and 7dpe compared with normal intestines (Fig. 5A). This initial observation was followed with a more complete stage analysis to determine the temporal expression profile of these genes. We found that the expression of both genes was significantly overexpressed during intestinal regeneration but their expression profile differed. Centaurin was overexpressed at 7dpe to 21dpe with a peak in expression at 14dpe (P < 0.001). Melanotransferrin was overexpressed during the first week (3–7dpe, P < 0.5) of regeneration compared with the later stages (21–28dpe) (Fig. 5B). No changes were observed in either NADH or in cytochrome b, where the latter was used for normalization purposes.
Discussion
EST BLAST analysis
A total of 5,173 EST sequences were analyzed to determine gene expression in normal intestine and in 3dpe and 7dpe regenerating intestine. Since the three sea cucumber libraries were not normalized, we expected a significant number of cDNAs to correspond to abundant transcripts present in intestinal tissue. However, contig analysis indicated that redundancy was relatively low among the libraries (in the range of 39–55%) compared with that reported for normalized cDNA libraries (54%) (44) and for nonnormalized libraries (74%) (32). The redundancy levels found in our libraries are similar to those seen in EST analyses from other organisms such as the fungi Mycosphaerella graminicola (56%) and Gibberella zeae (26%) (30, 58). These results suggested that the expression profile obtained for each library was representative of the intestinal regeneration stage.
Of the analyzed ESTs, a high percentage showed no significant similarity to entries in the public protein databases. This was expected in view of the small number of genetic studies in holothurians. Most of the ESTs previously reported for holothurians in the NCBI database were detected in our libraries. Thus a significant achievement from our studies is to increase the existing sequence information for this group with 4,522 new sequences. While the database for H. glaberrima is far from complete, our results enrich the current dataset from 639 to 5,161 sequences, providing a comprehensive sequence dataset for this group of organisms. These ESTs were grouped in 2,348 contigs and singletons. However, because the sequences were 5′-reads the possibility exists that clones from the same gene might be more or less complete at the 5′-end and fail to assemble into a contig. On the other hand, this number can be underestimated do to the fact that genes with high sequence similarities, such as actins, might be grouped together into one contig.
Additional comparisons of our EST dataset to the one of the sea urchin S. purpuratus, whose genome is almost complete at NCBI, showed an almost doubling of the number of sequences with shared similarities (from 1,112 to 1,905). This is to be expected in view of the evolutionary proximity between holothurians and sea urchins (5). However, the shared sequences are only about one-third of the known holothurian ESTs. This number will possibly increase as the clones are fully sequenced and a larger portion of the putative genes is available for interspecies comparisons. Nonetheless, some of our ESTs are part of gene sequences that are either holothurian specific or are associated with the process of intestinal regeneration that occurs in the sea cucumber but not in sea urchins (20, 27).
EST representation of stage-related events
This study provided a preliminary overview of the molecular genetics of regenerating and normal intestine for the sea cucumber H. glaberrima. Although, we realize that comparisons of cDNA libraries can be prone to error due to the large number of variables associated with their preparation, certain findings suggest that the ESTs found in these libraries are representative of genes expressed in the intestinal tissue from which the cDNA libraries were made. First, similar EST composition and specific putative gene homologs were found in two independent rounds of mass-excision and sequencing of the 7dpe cDNA library.
Second, the high number of ESTs associated with housekeeping processes such as metabolism and ribosomal proteins was to be expected, due to the energy production and protein synthesis requirements in both normal and regenerating organs (45). These results are in accordance with Agüero and coworkers (3), who found an even larger percentage (17.4%) of ribosomal protein in Trypanosoma cruzi total ESTs, and Chaudhuri and Lieberman (13), who showed a rise in liver ribosome and protein synthesis after partial hepatectomy. The high representation of some of these ESTs in normal compared with the regenerating tissues is also to be expected. Regenerating tissues need to express additional genes associated with this process, thus decreasing the relative expression of the housekeeping genes. The expression profile of ribosomal proteins differed among the three cDNA libraries; however, the relative percentage of ribosomal sequences increased with the regenerating stage. This increment in ribosomal proteins during regeneration may be due to the increased need for proteins synthesis during the regeneration process (13).
Third, the analysis by functional categories suggests that changes in gene expression in the regenerating libraries followed expected trends. For example, the percentage of ESTs associated with inflammatory response in the 3dpe library was almost double that of the other libraries. At this stage, wound healing is occurring and the immune response must be activated against pathogens that might have entered during evisceration (19, 52). Similarly, three functional categories increased in the 7dpe dataset: cell proliferation/apoptosis, cell adhesion, and intracellular signaling. The first was in accordance with our published observations that cell division is low at 3dpe but increases at 9dpe (19) and with our unpublished observations that apoptosis occurs mainly at the 7dpe stage. It has been reported that apoptosis may play an important role during regeneration in other organisms (26). The increase in sequences associated with cell adhesion agreed with our observations that extensive cellular migration and extracellular matrix remodeling occurs between 7 and 14dpe (10, 19, 46). Thus, during the 7dpe stage, genes associated with migration, cell adhesion, and extracellular matrix interactions can be expected to be activated.
Finally, previous work in our laboratory has identified specific genes that appeared to be associated with the regeneration process. Some of these genes, such as ependymin (56), collagen (49), and serum amyloid (49, 51), were also identified in our regenerating EST dataset. Moreover, one of the most prominent contigs in the 7dpe cDNA library corresponded to actin. Our laboratory previously identified actin clones in regenerating intestine by differential display (49), demonstrating upregulation of various actin isoforms and suggesting an important role during regeneration. The identification in this study of genes previously reported by our laboratory supported our contention that our EST dataset represented the ongoing genetic changes taking place at the time the cDNA libraries were made.
To further dissect the sequences represented in our libraries and determine a differential gene profile among them, we identified potential domains in the H. glaberrima contigs using RPS-BLAST searches. Because the libraries were not normalized, we found that Cox and ribosomal domains were among the most abundantly identified domains. However, several domains were identified in only one of the libraries but not the others, suggesting that the expression of these domains may be stage specific. Among the most relevant domains present in the 3dpe library were SAA, A2M, and EGF-lam. The SAA domain, which can induce IL-12 and 23 (24), has been reported previously by our laboratory (51). The domains A2M and EGF-lam have been associated with neuronal growth and neurodegenerative diseases, respectively (54, 63). Domains such as 7-prohibitin, PP2c, and NDK were identified only in the 7dpe library. Proteins presenting these domains have been found to be associated with apoptosis and tumorigenesis (17, 57, 64), suggesting that the expression of the corresponding genes might be highly regulated during regeneration.
Specific ESTs associated with functional categories
H. glaberrima sequences showed high similarity to genes whose function are associated with wound healing, inflammatory response, or other regeneration-related events. Among the identified sequences that appear to be involved in these processes are MMPs, melanotransferrin, and centaurin. In previous experiments, we found that during the first week of regeneration the MMP activity was increased (46). Interestingly, in the sea cucumber 7dpe dataset we identified various ESTs with high similarity to MMPs. These findings support our work and provide the tools for further exploration of the ECM remodeling processes during regeneration. These results provide the first clue to the identification of molecules that might be important in regeneration and strengthen the use of H. glaberrima as a model to study repair and regeneration of the gastrointestinal tract.
In silico differential display of the regenerating and normal H. glaberrima contigs
To identify putative genes associated with the regeneration process, an in silico differential display was performed. This method provided a more rigorous analysis of the genetic profiles of the libraries. We found that many contigs identified as the most common in the cDNA libraries were also identified by the R analysis to be differentially expressed. The R statistic showed 12 sequences to be differentially expressed at significant levels (R > 6). Some of these were overexpressed in the normal intestine, including cytochrome c oxidase subunits and ferritin. Since these proteins are known to be highly expressed in the mucosal layer of the digestive tract (14), their differential expression is probably due to regular occurrence in the luminal epithelium of normal animals, a tissue layer that would be absent in the regenerating animals.
Some of the most interesting contigs are those found to be differentially expressed by R analysis and that showed no similarity to sequences in the NCBI database. One of these, contigs #5141-2 was conformed by ESTs only found in the normal intestine library. This putative gene may be expressed by a cell type present in normal noneviscerated intestine but absent in regenerating tissues. Alternatively, it may belong to a putative gene that is underexpressed during the regeneration process. In view of our interest in finding novel genes associated with regeneration, the best candidates are those unknown contigs that were found mostly in the 3dpe or in the 7dpe libraries. Contig #5020-1 was among the most common found in the 7dpe library with a large R value (> 5.4), making it a possible putative gene involved in regeneration at 7dpe stage. Two unknowns (#4791-1 and #4860-1) were found mainly in the 3dpe library. However, the predicted overexpression of 4860-1 could not be verified by PCR analyses. The lack of significance in the expression of the unknown 4860-1 during regeneration may be due to the high variability among the animals or to limits in the sensitivity of the conventional RT-PCR. Finally, two other unknowns (#4277-4 and #4766-1) were also present in the 7dpe and normal libraries but were overrepresented in the 3dpe library. One of them (#4766-1) is of particularly interesting, first because it shows an EF-hand calcium binding domain, which have been recognized as a key player in all aspects of cell function, from mitosis to apoptotic death (22). Second, because of the increase in expression predicted by R for this clone was verified by the PCR analyses showing the clone to be significantly higher in the 3dpe than in the 7dpe and normal intestines.
Our PCR analysis also showed further evidence of differential expression that support our EST results. Clones whose expression was expected to remain stable during the regeneration process, such as NADH and cytochrome b, behaved as expected. In addition, differences in expression profiles were found with two sequences that were identified only in the regenerating libraries and that are of interest for future studies. Melanotransferrin ESTs were found only in the 3dpe and 7dpe libraries. This sequence is similar to a gene associated with angiogenesis (53), eosinophil differentiation (38), and tumor progression (15). Interestingly, the expression analysis shows that this sequence is overexpressed in the 3dpe and 7dpe stages of regeneration, supporting our ESTs results. Similarly, centaurin, a recently described gene associated mainly with cell signaling control (31), actin reorganization (60), Alzheimer's disease (47, 48), and downregulation of NF-κB (64), was found only in the 7dpe library. PCR results showed that centaurin is overexpressed at the 7dpe stage compared with normal and 3dpe, which is in accordance with our EST analysis. Moreover, the temporal expression of centaurin peaking at 14dpe and returning to levels similar to the normal animals at 28dpe clearly suggests that this gene might play an important role during intestinal regeneration.
Development, homeostasis, and repair are complex processes that require precise orchestration of cell proliferation, migration, differentiation, and death. All of these events take place during intestinal regeneration. Our EST project not only has yielded new insights into the molecular basis of gastrointestinal regeneration but provides evidence for the existence of a differential expression profile during regeneration. Future studies will focus on the characterization of those sequences that were found among the most frequent in the libraries and that lack similarity to known sequences, in the hope that they will provide important information to the process of intestinal tract repair and regeneration. The analyses of regenerating and normal intestine cDNA libraries of H. glaberrima have served to increase the annotation of genes in these organisms and underscored the utility of H. glaberrima as a model system for the study of repair and regeneration of the digestive tract.
Acknowledgments
The authors thank Dr. Maria E. Perez for invaluable help in the statistical analysis. In addition we thank Dr. Sheila Ward for editorial comments.
Grants: This study was supported by National Institutes of Health (NIH)-Minority Biomedical Research Support (S06GM-08102). Partial support was also provided by DEGI-PBDT (doctoral dissertation fellowship to E. C. Suárez-Castillo), NSF-Integrative Biology and Neuroscience (0110692), NIH-Research Centers in Minority Institutions (RRO-3641-01), and the University of Puerto Rico. This project made use of the computational resources at the High Performance Computing Facility of the University of Puerto Rico supported by Idea Network of Biomedical Research Excellence (NIH-National Center for Research Resources) P20 RR-016470.
References
- 1.Adams MD, Kelley JM, Gocayne JD, Dubnick M, Polymeropoulos MH, Xiao H, Merril CR, Wu A, Olde B, Moreno RF, Kerlavage AR, McCombie WR, Venter JC. Complementary DNA sequencing: expressed sequence tags and human genome project. Science. 1991;252:1651–1656. doi: 10.1126/science.2047873. [DOI] [PubMed] [Google Scholar]
- 2.Adams PF, Hendershot GE, Marano MA. Current estimates from the National Health Interview Survey, 1996. National Center for Health Statistics Vital Health Stat 10. 1999;200:1–203. [PubMed] [Google Scholar]
- 3.Agüero F, Abdellah KB, Tekiel V, Sánchez DO, González A. Generation and analysis of expressed sequence tags from Trypanosoma cruzi trypomastigotes and amastigotes cDNA libraries. Mol Biochem Parasitol. 2004;136:221–225. doi: 10.1016/j.molbiopara.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 4.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amemiya CT, Miyake T, Rast JP. Echinoderms. Curr Biol. 2005;15:R944–R946. doi: 10.1016/j.cub.2005.11.026. [DOI] [PubMed] [Google Scholar]
- 6.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baena-López LA, Pastor-Pareja JC, Resino J. Wg and Egfr signalling antagonise the development of the peripodial epithelium in Drosophila wing discs. Development. 2003;130:6497–6506. doi: 10.1242/dev.00884. [DOI] [PubMed] [Google Scholar]
- 8.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2006;34:D16–D20. doi: 10.1093/nar/gkj157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braun S, auf dem Keller U, Steiling H, Werner S. Fibroblast growth factors in epithelial repair and cytoprotection. Philos Trans R Soc Lond B Biol Sci. 2004;359:753–757. doi: 10.1098/rstb.2004.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cabrera-Serrano A, García-Arrarás JE. RGD-containing peptides inhibit intestinal regeneration in the sea cucumber Holothuria glaberrima. Dev Dyn. 2004;231:171–178. doi: 10.1002/dvdy.20112. [DOI] [PubMed] [Google Scholar]
- 11.Candelaria AG, Murray G, File SK, Garcia-Arraras JE. Contribution of mesenterial muscle dedifferentiation to intestine regeneration in the sea cucumber Holothuria glaberrima. Cell Tissue Res. 2006;325:55–65. doi: 10.1007/s00441-006-0170-z. [DOI] [PubMed] [Google Scholar]
- 12.Catanho M, Mascarenhas D, Degrave W, de Miranda AB. BioParser: a tool for processing of sequence similarity analysis reports. Appl Bioinformatics. 2006;5:49–53. doi: 10.2165/00822942-200605010-00007. [DOI] [PubMed] [Google Scholar]
- 13.Chaudhuri S, Lieberman I. Control of ribosome synthesis in normal and regenerating liver. J Biol Chem. 1968;243:29–33. [PubMed] [Google Scholar]
- 14.Conrad ME, Weintraub LR, Crosby WH. The role of the intestine in iron kinetics. J Clin Invest. 1964;43:963–974. doi: 10.1172/JCI104982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunn LL, Sekyere EO, Rahmanto YS, Richardson DR. The function of melanotransferrin: a role in melanoma cell proliferation and tumorigenesis. Carcinogenesis. 2006;27:2157–2169. doi: 10.1093/carcin/bgl045. [DOI] [PubMed] [Google Scholar]
- 16.Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 17.Fischbach MA, Settleman J. Regulation of the nucleotide state of oncogenic ras proteins by nucleoside diphosphate kinase. Methods Enzymol. 2005;407:33–45. doi: 10.1016/S0076-6879(05)07004-7. [DOI] [PubMed] [Google Scholar]
- 18.Galko MJ, Krasnow MA. Cellular and genetic analysis of wound healing in Drosophila larvae. PLoS Biol. 2004;2:e239. doi: 10.1371/journal.pbio.0020239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.García-Arrarás JE, Greenberg MJ. Visceral regeneration in holothurians. Microsc Res Tech. 2001;55:438–451. doi: 10.1002/jemt.1189. [DOI] [PubMed] [Google Scholar]
- 20.García-Arrarás JE, Estrada-Rodgers L, Santiago R, Torres II, Diaz-Miranda L, Torres-Avillan I. Cellular mechanisms of intestine regeneration in the sea cucumber, Holothuria glaberrima Selenka (Holothuroidea: Echinodermata) J Exp Zool. 1998;281:288–304. doi: 10.1002/(sici)1097-010x(19980701)281:4<288::aid-jez5>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 21.Glassner A. Venn and Now. IEEE Comput Graph Appl. 2003;23:82–95. [Google Scholar]
- 22.Grabarek Z. Structural basis for diversity of the EF-hand calcium-binding proteins. J Mol Biol. 2006;359:509–525. doi: 10.1016/j.jmb.2006.03.066. [DOI] [PubMed] [Google Scholar]
- 23.Habermann B, Bebin AG, Herklotz S, Volkmer M, Eckelt K, Pehlke K, Epperlein HH, Schackert HK, Wiebe G, Tanaka EM. An Ambystoma mexicanum EST sequencing project: analysis of 17,352 expressed sequence tags from embryonic and regenerating blastema cDNA libraries. Genome Biol. 2004;5:R67.1–R67.19. doi: 10.1186/gb-2004-5-9-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He R, Shepard LW, Chen J, Pan ZK, Ye RD. Serum amyloid a is an endogenous ligand that differentially induces IL-12 and IL-23. J Immunol. 2006;177:4072–4079. doi: 10.4049/jimmunol.177.6.4072. [DOI] [PubMed] [Google Scholar]
- 25.Huang X, Madan A. CAP3: a DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hwang JS, Kobayashi C, Agata K, Ikeo K, Gojobori T. Detection of apoptosis during planarian regeneration by the expression of apoptosis-related genes and TUNEL assay. Gene. 2004;333:15–25. doi: 10.1016/j.gene.2004.02.034. [DOI] [PubMed] [Google Scholar]
- 27.Hyman LH. Echinodermata. New York: McGraw-Hill; 1955. The invertebrates; pp. 202–205. [Google Scholar]
- 28.Jones MK, Tomikawa M, Mohajer B, Tarnawski AS. Gastrointestinal mucosal regeneration: role of growth factors. Front Biosci. 1999;4:D303–D309. doi: 10.2741/a428. [DOI] [PubMed] [Google Scholar]
- 29.Ju Z, Karsi A, Kocabas A, Patterson A, Li P, Cao D, Dunham R, Liu Z. Transcriptome analysis of channel catfish (Ictalurus punctatus): genes, and expression profile from the brain. Gene. 2000;261:373–382. doi: 10.1016/s0378-1119(00)00491-1. [DOI] [PubMed] [Google Scholar]
- 30.Kanamarlapudi V. Centaurin-alpha1 and KIF13B kinesin motor protein interaction in ARF6 signalling. Biochem Soc Trans. 2005;33:1279–81. doi: 10.1042/BST0331279. [DOI] [PubMed] [Google Scholar]
- 31.Keon J, Antony J, Rudd J, Skinner W, Hargreaves J, Hammond-Kosack K. Analysis of expressed sequence tags from the wheat leaf blotch pathogen Mycosphaerella graminicola (anamorph Septoria tritici) Fungal Genet Biol. 2005;42:376–389. doi: 10.1016/j.fgb.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 32.Kim TH, Kim NS, Lim D, Lee KT, Oh JH, Park HS, Jang GW, Kim HY, Jeon M, Choi BH, Lee HY, Chung HY, Kim H. Generation and analysis of large-scale expressed sequence tags (ESTs) from a full-length enriched cDNA library of porcine backfat tissue. BMC Genomics. 2006;7:1–9. doi: 10.1186/1471-2164-7-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li F, Zhang D, Fujise K. Characterization of fortilin, a novel antiapoptotic protein. J Biol Chem. 2001;276:47542–47549. doi: 10.1074/jbc.M108954200. [DOI] [PubMed] [Google Scholar]
- 34.Mao C, Cushman JC, May GD, Weller JW. ESTAP - an automated system for the analysis of EST data. Bioinformatics. 2003;19:1720–1722. doi: 10.1093/bioinformatics/btg205. [DOI] [PubMed] [Google Scholar]
- 35.Marchler-Bauer A, Anderson JB, Cherukuri PF, DeWeese-Scott C, Geer LY, Gwadz M, He S, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Liebert CA, Liu C, Lu F, Marchler GH, Mullokandov M, Shoemaker BA, Simonyan V, Song JS, Thiessen PA, Yamashita RA, Yin JJ, Zhang D, Bryant SH. CDD: a conserved domain database for protein classification. Nucleic Acids Res. 2005;33:D192–D196. doi: 10.1093/nar/gki069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mattila J, Omelyanchuk L, Kyttala S, Turunen H, Nokkala S. Role of Jun N-terminal kinase (JNK) signaling in the wound healing and regeneration of a Drosophila melanogaster wing imaginal disc. Int J Dev Biol. 2005;49:391–399. doi: 10.1387/ijdb.052006jm. [DOI] [PubMed] [Google Scholar]
- 37.McNagny KM, Rossi F, Smith G, Graf T. The eosinophil-specific cell surface antigen, EOS47, is a chicken homologue of the oncofetal antigen melanotransferrin. Blood. 1996;87:1343–52. [PubMed] [Google Scholar]
- 38.Méndez AT, Roig-López JL, Santiago P, Santiago C, García-Arrarás JE. Identification of hox gene sequences in the sea cucumber Holothuria glaberrima selenka (Holothuroidea: Echinodermata) Mar Biotechnol (NY) 2000;2:231–240. doi: 10.1007/s101269900027. [DOI] [PubMed] [Google Scholar]
- 39.Murray G, García-Arrarás JE. Myogenesis during holothurian intestinal regeneration. Cell Tissue Res. 2004;318:515–524. doi: 10.1007/s00441-004-0978-3. [DOI] [PubMed] [Google Scholar]
- 40.Okamoto R, Watanabe M. Molecular and clinical basis for the regeneration of human gastrointestinal epithelia. J Gastroenterol. 2004;39:1–6. doi: 10.1007/s00535-003-1259-8. [DOI] [PubMed] [Google Scholar]
- 41.Okamoto R, Watanabe M. Cellular and molecular mechanisms of the epithelial repair in IBD. Dig Dis Sci. 2005;50:S34–S38. doi: 10.1007/s10620-005-2804-5. [DOI] [PubMed] [Google Scholar]
- 42.Pleis JR, Schiller JS, Benson V. Summary health statistics for U.S. adults: National Health Interview Survey, 2000. Vital Health Stat 10. 2003;215:1–132. [PubMed] [Google Scholar]
- 43.Pold M, Pold A, Ma HJ, Sjak-Shieb NN, Vescio RA, Berenson JR. Cloning of the first invertebrate MAGE paralogue: an epitope that activates T-cells in humans is highly conserved in evolution. Dev Comp Immunol. 2000;24:719–731. doi: 10.1016/s0145-305x(00)00027-6. [DOI] [PubMed] [Google Scholar]
- 44.Porcel BM, Tran AN, Tammi M, Nyarady Z, Rydaker M, Urmenyi TP, Rondinelli E, Pettersson U, Andersson B, Aslund L. Gene survey of the pathogenic protozoan Trypanosoma cruzi. Genome Res. 2000;10:1103–1107. doi: 10.1101/gr.10.8.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poustka AJ, Groth D, Hennig S, Thamm S, Cameron A, Beck A, Reinhardt R, Herwig R, Panopoulou G, Lehrach H. Generation, annotation, evolutionary analysis, and database integration of 20,000 unique sea urchin EST clusters. Genome Res. 2003;13:2736–2746. doi: 10.1101/gr.1674103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quiñones JL, Rosa R, Ruiz DL, García-Arrarás JE. Extracellular matrix remodeling and metalloproteinase involvement during intestine regeneration in the sea cucumber Holothuria glaberrima. Dev Biol. 2002;250:181–197. doi: 10.1006/dbio.2002.0778. [DOI] [PubMed] [Google Scholar]
- 47.Reiser G, Bernstein HG. Neurons and plaques of Alzheimer's disease patients highly express the neuronal membrane docking protein p42IP4/centaurin alpha. Neuroreport. 2002;13:2417–2419. doi: 10.1097/00001756-200212200-00008. [DOI] [PubMed] [Google Scholar]
- 48.Reiser G, Bernstein HG. Altered expression of protein p42IP4/centaurin-alpha 1 in Alzheimer's disease brains and possible interaction of p42IP4 with nucleolin. Neuroreport. 2004;15:147–148. doi: 10.1097/00001756-200401190-00028. [DOI] [PubMed] [Google Scholar]
- 49.Roig-López J, Santiago P, Jiménez C, García-Arrarás JE. Strategies to identify diferentially expressed genes during regeneration. In: Lisse Barker M., editor. Echinoderms 2000. The Netherlands: Swets & Zeitlinger; 2001. [Google Scholar]
- 50.Sala R, Jefferies WA, Walker B, Yang J, Tiong J, Law SK, Carlevaro MF, Di Marco E, Vacca A, Cancedda R, Cancedda FD, Ribatti D. The human melanoma associated protein melanotransferrin promotes endothelial cell migration and angiogenesis in vivo. Eur J Cell Biol. 2002;81:599–607. doi: 10.1078/0171-9335-00280. [DOI] [PubMed] [Google Scholar]
- 51.Santiago P, Roig-López JL, Santiago C, García-Arrarás JE. Serum amyloid A protein in an echinoderm: its primary structure and expression during intestinal regeneration in the sea cucumber Holothuria glaberrima. J Exp Zool. 2000;288:335–344. doi: 10.1002/1097-010X(20001215)288:4<335::AID-JEZ6>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 52.Santiago-Cardona PG, Berrios CA, Ramírez F, García-Arrarás JE. Lipopolysaccharides induce intestinal serum amyloid A expression in the sea cucumber Holothuria glaberrima. Dev Comp Immunol. 2003;27:105–110. doi: 10.1016/s0145-305x(02)00068-x. [DOI] [PubMed] [Google Scholar]
- 53.Saunders AJ, Bertram L, Mullin K, Sampson AJ, Latifzai K, Basu S, Jones J, Kinney D, MacKenzie-Ingano L, Yu S, Albert MS, Moscarillo TJ, Go RC, Bassett SS, Daly MJ, Laird NM, Wang X, Velicelebi G, Wagner SL, Becker DK, Tanzi RE, Blacker D. Genetic association of Alzheimer's disease with multiple polymorphisms in alpha-2-macroglobulin. Hum Mol Genet. 2006;12:2765–2776. doi: 10.1093/hmg/ddg310. [DOI] [PubMed] [Google Scholar]
- 54.Shin SJ. The Logical Status of Diagrams. Cambridge: Cambridge University Press; 1994. [Google Scholar]
- 55.Stekel DJ, Git Y, Falciani F. The comparison of gene expression from multiple cDNA libraries. Genome Res. 2006;10:2055–2061. doi: 10.1101/gr.gr-1325rr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Suárez-Castillo EC, Medina-Ortíz WE, Roig-López JL, García-Arraras JE. Ependymin, a gene involved in regeneration and neuroplasticity in vertebrates, is overexpressed during regeneration in the echinoderm Holothuria glaberrima. Gene. 2004;334:133–143. doi: 10.1016/j.gene.2004.03.023. [DOI] [PubMed] [Google Scholar]
- 57.Tamura S, Toriumi S, Saito J, Awano K, Kudo TA, Kobayashi T. PP2C family members play key roles in regulation of cell survival and apoptosis. Cancer Sci. 2006;97:563–567. doi: 10.1111/j.1349-7006.2006.00219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trail F, Xu JR, San Miguel P, Halgren RG, Kistler HC. Analysis of expressed sequence tags from Gibberella zeae (anamorph Fusarium graminearum) Fungal Genet Biol. 2003;38:187–197. doi: 10.1016/s1087-1845(02)00529-7. [DOI] [PubMed] [Google Scholar]
- 59.Tucker BA, Rahimtula M, Mearow KM. Laminin and growth factor receptor activation stimulates differential growth responses in subpopulations of adult DRG neurons. Eur J Neurosci. 2006;24:676–690. doi: 10.1111/j.1460-9568.2006.04963.x. [DOI] [PubMed] [Google Scholar]
- 60.Venkateswarlu K, Brandom KG, Yun H. PI-3-kinase-dependent membrane recruitment of centaurin-alpha2 is essential for its effect on ARF6-mediated actin cytoskeleton reorganisation. J Cell Sci. 2007;120:792–801. doi: 10.1242/jcs.03373. [DOI] [PubMed] [Google Scholar]
- 61.Venn J. On the diagrammatic and mechanical representation of propositions and reasonings. The London, Edinburgh, and Dublin Philosophical Magazine and Journal of Science. 1980;9:1–18. [Google Scholar]
- 62.Wheeler DL, Chappey C, Lash AE, Leipe DD, Madden TL, Schuler GD, Tatusova TA, Rapp BA. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2000;28:10–14. doi: 10.1093/nar/28.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yamamoto-Furusho JK, Barnich N, Xavier R, Hisamatsu T, Podolsky DK. Centaurin beta1 down-regulates nucleotide-binding oligomerization domains 1- and 2-dependent NF-kappaB activation. J Biol Chem. 2006;281:36060–36070. doi: 10.1074/jbc.M602383200. [DOI] [PubMed] [Google Scholar]
- 64.Zhu B, Fukada K, Zhu H, Kyprianou N. Prohibitin and cofilin are intracellular effectors of transforming growth factor beta signaling in human prostate cancer cells. Cancer Res. 2006;66:8640–8647. doi: 10.1158/0008-5472.CAN-06-1443. [DOI] [PubMed] [Google Scholar]


