Abstract
Cellular functions depend on numerous protein-coding and non-coding RNAs and the RNA-binding proteins associated with them, which form ribonucleoprotein complexes (RNPs). Mutations that disrupt either the RNA or protein components of RNPs or the factors required for their assembly can be deleterious. Alternative splicing provides cells with an exquisite capacity to fine-tune their transcriptome and proteome in response to cues. Splicing depends on a complex code, numerous RNA-binding proteins and an enormously intricate network of interactions among them, increasing the opportunity for exposure to mutations and mis-regulation that cause disease. The discovery of disease-causing mutations in RNAs is yielding a wealth of new therapeutic targets, and the growing understanding of RNA biology and chemistry is providing new RNA-based tools for developing therapeutics.
Introduction
The normal function of cells depends on accurate expression of a large number of protein coding RNAs (messenger RNAs; mRNAs) and non-coding RNAs. These RNAs participate in transcription (e.g., 7SK RNA), RNA processing (e.g., small nuclear RNAs; snRNAs, small nucleolar RNAs; snoRNAs) and translation (e.g., ribosomal RNAs; rRNAs, transfer RNAs; tRNAs, 7SL/SRP RNA, microRNAs; miRNAs). There are many other RNAs that are involved in other processes (e.g., telomerase, MRP, RNase P RNAs), and those whose functions are less well characterized or entirely unknown (e.g., vault RNAs, Y RNAs, Piwi-interacting RNAs; piRNAs). RNAs exist in cells as ribonucleoprotein complexes (RNPs) that are composed of one or more RNAs and typically numerous RNA-binding proteins (Dreyfuss et al., 1993; Glisovic et al., 2008). The RNPs are the functional forms of the corresponding RNAs and their normal activity depends on both the specific composition and the precise arrangement of their protein constituents. As there are numerous RNAs and a very large number of RNA-binding proteins, the biogenesis of RNPs must be orchestrated with great fidelity. Mutations that disrupt any of the components of RNPs, either RNAs or proteins, or the factors required for their assembly can be deleterious to cells and cause disease (Lukong et al., 2008; Wang and Cooper, 2007). Examples of such cases are now too numerous to afford a meaningful discussion of all but a few within the space constraints of this article. We focus here on defects in pre-mRNA splicing as these have emerged as a common disease-causing mechanism underlying many human genetic ailments. For a more general perspective, a summary of representative examples of disease-causing defects associated with other types of RNAs or RNPs is listed in Table 1. Here, we discuss several diseases that result from mutations that affect the splicing of the transcript produced from the same gene (in cis) as well as mutations in the splicing machinery and the regulatory proteins that affect splicing of other transcripts (in trans). The discovery of new disease-causing mutations in RNAs provides a range of therapeutic targets, and the continuing elucidation of RNA biochemistry is also yielding new tools that can be exploited in the development of RNA-based therapeutics.
Table 1.
Trans-acting mutations affecting RNA-dependent functions that cause disease
| DISEASE | GENE / MUTATION | FUNCTION |
|---|---|---|
| Prader Willi syndrome | SNORD116 | ribosome biogenesis |
| Spinal muscular atrophy (SMA) | SMN2 | splicing |
| Dyskeratosis congenita (X-linked) | DKC1 | telomerase / translation |
| Dyskeratosis congenita (autosomal dominant) | TERC | telomerase |
| Dyskeratosis congenita (autosomal dominant) | TERT | telomerase |
| Diamond-Blackfan anemia | RPS19, RPS24 | ribosome biogenesis |
| Shwachman-Diamond syndrome | SBDS | ribosome biogenesis |
| Treacher-Collins syndrome | TCOF1 | ribosome biogenesis |
| Prostate cancer | SNHG5 | ribosome biogenesis |
| Myotonic dystrophy, type 1 (DM1) | DMPK (RNA gain-of-function) | protein kinase |
| Myotonic dystrophy type 2 (DM2) | ZNF9 (RNA gain-of-function) | RNA binding |
| Spinocerebellar ataxia 8 (SCA8) | ATXN8 / ATXN8OS (RNA gain-of-function) | unknown / noncoding RNA |
| Huntington’s disease-like 2 (HDL2) | JPH3 (RNA gain-of-function) | ion channel function |
| Fragile X-associated tremor ataxia syndrome (FXTAS) | FMR1 (RNA gain-of-function) | translation / mRNA localization |
| Fragile X syndrome | FMR1 | translation / mRNA localization |
| X-linked mental retardation | UPF3B | translation/ nonsense mediated decay |
| Oculopharyngeal muscular dystrophy (OPMD) | PABPN1 | 3′ end formation |
| Human pigmentary genodermatosis | DSRAD | editing |
| Retinitis pigmentosa | PRPF31 | splicing |
| Retinitis pigmentosa | PRPF8 | splicing |
| Retinitis pigmentosa | HPRP3 | splicing |
| Retinitis pigmentosa | PAP1 | splicing |
| Cartilage-hair hypoplasia (recessive) | RMRP | splicing |
| Autism | 7q22-q33 locus breakpoint | noncoding RNA |
| Beckwith-Wiedemann syndrome (BWS) | H19 | noncoding RNA |
| Charcot–Marie–Tooth (CMT) Disease | GRS | translation |
| Charcot–Marie–Tooth (CMT) Disease | YRS | translation |
| Amyotrophic lateral sclerosis (ALS) | TARDBP | splicing, transcription |
| Leukoencephalopathy with vanishing white matter | EIF2B1 | translation |
| Wolcott–Rallison syndrome | EIF2AK3 | translation (protease) |
| Mitochondrial myopathy and sideroblastic anemia (MLASA) | PUS1 | translation |
| Encephalomyopathy and hypertrophic cardiomyopathy | TSFM | translation (mitochondrial) |
| Hereditary spastic paraplegia | SPG7 | ribosome biogenesis |
| Leukoencephalopathy | DARS2 | translation (mitochondrial) |
| Susceptibility to diabetes mellitus | LARS2 | translation (mitochondrial) |
| Deafness | MTRNR1 | ribosome biogenesis (mitochondrial) |
| MELAS syndrome, deafness | MTRNR2 | ribosome biogenesis (mitochondrial) |
| Cancer | SFRS1 | splicing, translation, export |
| Cancer | RBM5 | splicing |
| Multiple disorders | mitochondrial tRNA mutations | translation (mitochondrial) |
| Cancer | miR-17-92 cluster | RNA interference |
| Cancer | miR-372 / miR-373 | RNA interference |
In humans and other complex metazoans, the vast majority of protein-coding genes contain many segments (introns) that are part of the primary transcript (premRNA) but are not included in the mRNA. The removal of introns and the joining together of the sequences included in the mRNA (exons) that contain the protein-coding open reading frame and the 3′- and 5′- untranslated regions (UTRs), is accomplished by pre-mRNA splicing. Splicing must be both rapid and precise, a demanding feat considering that introns are typically much larger than exons and are strewn with translation termination codons, which would result in truncated proteins if not accurately removed. The definition of exons and the execution of the splicing reaction are mediated by the spliceosome, a large and modular molecular machine composed of snRNAs and splicing factors, and by numerous RNA-binding proteins (see Review by M. C. Wahl, C. L. Will and R. Luhrmann on page XXX of this issue). The biosynthetic and metabolic energy cost of splicing and of the surveillance necessary to eliminate imprecisely spliced mRNAs is very high, and so is the potential harm from defects in these processes. It was therefore not self-evident at the outset why splicing arose, persisted and expanded in the number and size of introns with the evolution of more complex organisms. The real payoff of splicing, which complex organisms have evolved to capitalize on (or perhaps capitalized on for their evolution), seems to come from the capacity for alternative splicing. Alternative splicing allows individual genes to express multiple mRNAs and encode numerous proteins that differ by either large or small peptides or have different untranslated regions (UTRs). Alternative splicing provides tremendous opportunities for enrichment of the transcriptome and proteome without the need for expansion of the genome. Importantly, alternative splicing can also be regulated differently in cell type-, developmental stage-, or signal-dependent patterns. This elaborate scheme shifts the balance of control of gene expression from largely transcriptional, the production of a co-linear transcript of DNA, to a largely post-transcriptional non-linear complex and intricate processing of the pre-mRNA. Recent studies using high throughput deep sequencing indicate that the extent of alternative splicing and thus spliced isoforms is much greater in humans than was previously estimated (Pan et al., 2008; Wang et al., 2008). In some cases, a large number of splicing events are required to produce an mRNA and the number of splicing options can be vast. Examples of proteins encoded by human genes containing a large number of exons include Titin (Ttn; 316 exons), the ryanodine receptor 1 (Ryr1; 106 exons) and dystrophin (DMD; 79 exons). However, with this enormous increase in complexity comes an increased susceptibility to malfunction. Indeed, a large number of human diseases result from mutations or mis-regulation of the splicing process.
The major splicing apparatus (spliceosome) is composed of 5 small nuclear ribonucleoprotein complexes (snRNPs) each containing one or two snRNAs (U1, U2, U4/U6, and U5) and numerous (likely >100) protein factors. These protein factors include RNA-binding proteins (e.g., U2AF, SF1 and SRFs) and enzymes (helicases/RNPases, kinases and phosphatases, etc.). Thesemodulate the structure and the orderly stepwise associations, dissociations and conformational transitions of the premRNA,snRNAs and protein complexes to facilitate the splicing reaction, proofreading and substrate release (Nilsen, 2003; Staley and Guthrie, 1998; Will and Luhrmann, 2001). A minor splicing pathway, the unique components of which are about 100-fold less abundant compared to those in the major splicing pathway, operates in parallel using a similar scheme, but it recognizes different canonical splice site sequences and uses mostly different snRNAs (U11, U12, U4atac/U6atac and U5) (Patel and Steitz, 2003). Each snRNP, except U6 and U6atac, have in common a stable seven-membered ring of Sm proteins (the Sm core), which forms around a conserved sequence called the Sm site, as well as several snRNA-specific proteins. The biogenesis of snRNPs is a highly complex process involving export of the nascent pre-U snRNAs to the cytoplasm where assembly of Sm cores takes place and, following processing of the snRNAs, re-import of the snRNPs to the nucleus to function in splicing. Sm core assembly is not a spontaneous process; it is performed by the SMN (survival of motor neurons) complex, an ATP-dependent assemblyosome comprised of SMN, Gemins 2-8 and Unrip, that specifically identifies snRNAs and brings them together with Sm proteins, a recognition aided by a specific arginine modification in Sm proteins by the methylosome/PRMT5 (Neuenkirchen et al., 2008; Yong et al., 2004). Considerable insight into this pathway has come from studies of a devastating disease in which its namesake protein, SMN, is deficient.
Pre-mRNAs contain a “splicing code” made up of loosely defined consensus sequences that define the splice junctions and a bewildering number and diversity of relatively short cis-acting elements within exons as well as introns (Wang and Burge, 2008). These elements are bound by RNA-binding proteins, which have either positive (primarily SR proteins) or negative (primarily hnRNP proteins) effects on spliceosome assembly in their vicinity to maintain appropriate constitutive splicing or to regulate alternative splicing. Cells contain a large assortment of these RNA-binding proteins and their relative stoichiometries play important roles in determining splicing outcomes for many genes (Smith and Valcarcel, 2000). The repertoire of these is unique to each cell type and thus the regulation of expression and activity of these proteins is critical for normal alternative splicing. Thus, in addition to diseases caused by mutations in exons, including missense, nonsense, and frame-shift mutations in the open reading frame, which give rise to defective proteins, and mutations in UTRs that affect translation efficiency or mRNA stability, a large number of diseases are now known to result from intronic or exonic mutations that disrupt normal splicing patterns.
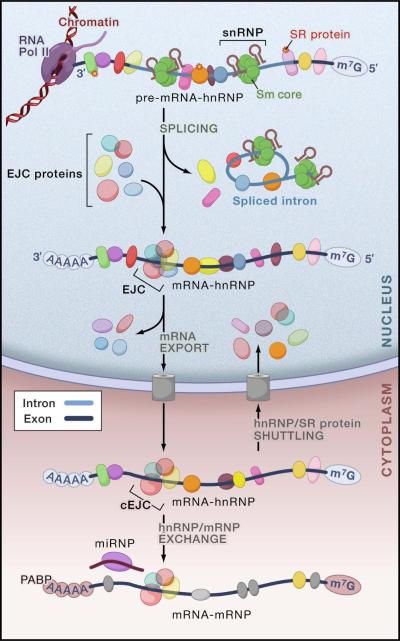
Co-transcriptionally, pre-mRNAs become associated with numerous RNA-binding proteins (hnRNP proteins) that influence their structure and interactions and play key roles in their splicing and polyadenylation. Upon splicing, additional proteins are acquired at the splice junctions (the exon junction complex; EJC) and numerous proteins are removed, and the metamorphosis of the mRNA-protein complexes (mRNPs) continues as they transit to the cytoplasm (Figure 1) (Dreyfuss et al., 2002; Maquat, 2004; Tange et al., 2004). Many of these proteins shuttle between the nucleus and the cytoplasm (Pinol-Roma and Dreyfuss, 1992), including hnRNP proteins and SR (serinearginine-rich domain-containing) proteins. The functions they perform in the nucleus are not their last, as they also have roles in mRNA translation, stability and localization in the cytoplasm. RNA editing provides yet another mode to increase the complexity of the transcriptome (Schaub and Keller, 2002), and a further layer of regulation has now been revealed with the discovery of microRNAs (miRNAs), an abundant class of non-coding RNAs that regulate mRNA translation and stability (Valencia-Sanchez et al., 2006). The large number of proteins and regulatory RNAs in post-transcriptional RNA processing and the enormously intricate network of interactions among them provide cells with exquisite capacity to fine tune their transcriptome and rapidly adjust their proteome in response to stimuli, but it also increases exposure to mutations and extends their vulnerability to mis-regulation that causes numerous diseases, particularly neuromuscular and neurodegenerative diseases and cancer.
Figure 1. The mRNA biogenesis pathway and post-transcriptional gene regulation.
Overview of the dynamic interactions of the major RNPs involved in pre-mRNA processing, mRNA biogenesis and regulation of gene expression. The nascent transcript (pre-mRNA), capped with a 7-methyl guanosine (m7G) cap, is bound by hnRNP proteins and SR (serine-arginine-rich domain-containing) proteins. Small nuclear ribonucleoprotein particles (snRNPs) bind to the splice sites at the 5′ and 3′ ends of introns. The spliced introns are excised as lariats, which are then debranched and degraded. The exon junction complex (EJC) assembles on spliced mRNA ~20 nucleotides upstream of joined exons, followed by the export of the mRNA to the cytoplasm. MicroRNAs (miRNAs) in the form of ribonucleoprotein complexes (miRNPs) bind to target mRNAs to regulate translation and mRNA stability. The poly(A)-binding protein (PABP) binds to the poly(A) tail of cytoplasmic mRNAs. The SR domains are indicated with a star. cEJC indicates the remaining stable EJC on mRNA in the cytoplasm, which is removed by translating ribosomes.
Mutations that cause splicing defects in disease
Cis-acting mutations that disrupt splicing
Three consensus sequences are found at all exon-intron boundaries: the 5′ and 3′ splice sites at the 5′ and 3′ ends of the intron, respectively, and the branch point sequence (BPS) which is typically located within 30-50 nucleotides upstream of the 3′ splice site. These sequences contain a considerable amount of the information required for precise exon joining [estimated to be about half in some cases (Lim and Burge, 2001)] and are recognized through RNA-RNA and RNA-protein interactions with components of the spliceosome. The 5′ splice site binds to the U1 snRNP early during spliceosome assembly, then to U6 snRNP; the BPS binds SF1 and then U2 snRNP, and U2AF binds near the 3′ splice site. Mutations within the sequences that disrupt these interactions are responsible for 9-10% of the genetic diseases that are caused by point mutations (Cartegni et al., 2002; Wang and Cooper, 2007). The remaining information required for splicing is contained within relatively short (~6 nucleotide) sequences located within both exons and introns that either enhance or suppress splicing (Wang and Burge, 2008). Exonic splicing enhancers (ESEs) and exonic splicing silencers (ESSs) activate or repress splicing, respectively, from within exons while intronic splicing enhancers (ISEs) and silencers (ISSs) function from within introns. In contrast to the consensus splice sites, which are relatively well defined in sequence and position, the intricate code formed by these auxiliary splicing elements is only partially understood. While a significant portion of the code has been deciphered, it is not yet possible to determine from genomic sequence whether a disease-associated mutation will disrupt splicing. This is the case in part because these elements are composed of diverse classes of sequences, the full complement of which remains to be identified. There is also an incomplete understanding of the subtleties of the code including extensive interdependency between and within elements (Roca et al., 2008). These elements function through interactions with RNA-binding proteins and while the preferred binding sites for more than 40 proteins are known, primarily for SR and hnRNP proteins (Gabut et al., 2008), cognate binding proteins have been identified for only a minority of the growing list of putative splicing elements.
Essentially all exons, whether alternatively or constitutively spliced, contain splicing elements (Wang and Burge, 2008). As a result, exons are a virtual minefield with regard to the effects of nucleotide substitutions on splicing. Up to 25% of synonymous (in terms of amino acid coding) substitutions can disrupt normal splicing as can nonsynonomous and termination codons (Pagani et al., 2005), emphasizing the importance of considering silent mutations as mediators of pathogenic effects. For some genes, up to 50% of point mutations within exons affect splicing and it has been hypothesized that more than half of known disease-causing mutations disrupting splicing (Lopez-Bigas et al., 2005). A large number of exonic mutations that result in aberrant splicing have been documented (Cartegni et al., 2002; Wang and Cooper, 2007). Particularly instructive are disease-causing exonic mutations that have been studied extensively and have become focal points for various RNA-based therapeutic approaches.
A striking example of the detrimental effect that mutations in exonic splicing signals can have is a single nucleotide substitution in the survival of motor neuron 2 (SMN2) gene and its critical role to spinal muscular atrophy (SMA) (Lefebvre et al., 1995). SMN is a ubiquitously expressed protein that plays a critical role in snRNP biogenesis and is essential for viability of all cells in divergent eukaryotes (Neuenkirchen et al., 2008; Yong et al., 2004). SMA is a common, often fatal, motor neurodegenerative disease and a leading genetic cause of infant mortality (Talbot and Davies, 2001; Wirth et al., 2006). SMA severity corresponds to the degree of functional SMN protein deficiency. In humans, there are two SMN genes, SMN1 and SMN2, both of which encode the same open reading frame. The vast majority of SMA patients have deletions of the SMN1 gene but retain SMN2, exposing the functional deficiency of a single nucleotide substitution, a C->T change at position 6 in exon 7 of SMN2. This mutation, though it does not change the amino acid coding, significantly alters the splicing pattern of the SMN2 pre-mRNA, causing frequent skipping of exon 7 that produces an inactive and unstable protein lacking the last 16 amino acids (Figure 2A). Thus, SMN2 is much less effective than SMN1 in producing SMN protein (Wirth et al., 2006). Two models have been proposed to explain exon 7 skipping in SMN2. One is that the substitution disrupts an ESE, which the splicing activator, ASF/SF2 binds (Cartegni and Krainer, 2002) and the other is that it creates an ESS to which the splicing suppressor hnRNP A1 binds (Kashima and Manley, 2003; Kashima et al., 2007). Though they appear contradictory, these models, in fact, are compatible (Cartegni et al., 2006) and underscore the precariousness of splicing signals in that a single nucleotide change can convert an ESE into an ESS. It is intriguing that critical RNA binding sites for ubiquitous, abundant but functionally opposing splicing factors differ by a single nucleotide, and it can be anticipated that there are many more cases where splicing is altered because of the ease with which the balance of positively and negatively-acting splicing factors (ASF/SF2-hnRNP A1, and many others) can be shifted. Because of its critical function in the biogenesis of snRNPs and splicing regulation, this SMN2 substitution has significant ramifications to splicing (described below).
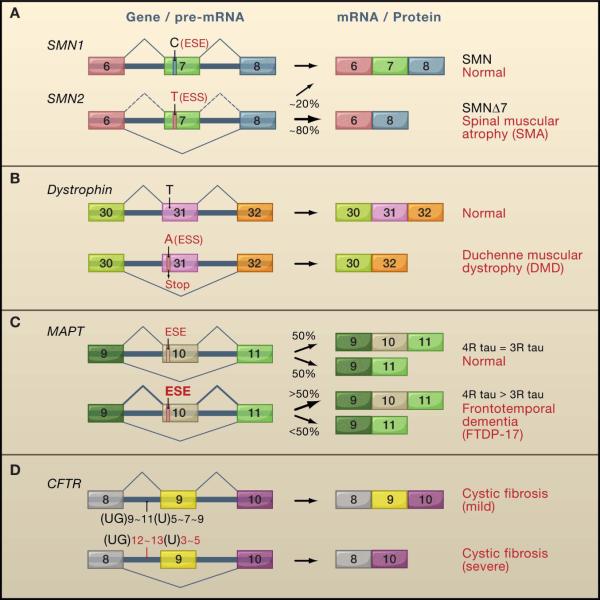
Figure 2. Mutations disrupting the cis-acting splicing code cause splicing defects and disease.
A. The motor neuron degenerative disease SMA results from deletion of the SMN1 gene. The remaining SMN2 gene has a C->T substitution in exon 7 that inactivates an exonic splicing enhancer (ESE), and creates an exonic splicing silencer (ESS), leading to exon 7 skipping and a truncated protein (SMNΔ7).
B. A T->A substitution in exon 31 of the dystrophin gene simultaneously creates a premature termination codon (STOP) and an ESS, leading to exon 31 skipping. This mutation causes a mild form of DMD because the mRNA lacking exon 31 produces a partially functional protein.
C. Mutations within and downstream of exon 10 of the MAPT gene encoding the tau protein affect splicing regulatory elements and disrupt the normal 1:1 ratio of mRNAs including or excluding exon 10. This results in a perturbed balance between tau proteins containing either four or three microtubule-binding domains (4R-tau and 3R-tau, respectively), causing the neuropathological disorder FTDP-17. The example shown is the N279K mutation which enhances an ESE function promoting exon 10 inclusion and shifting the balance toward increased 4R-tau.
D. Polymorphic (UG)m(U)n tracts within the 3′ splice site of the CFTR gene exon 9 influence the extent of exon 9 inclusion and the level of full-length functional protein, modifying the severity of cystic fibrosis (CF) caused by a mutation elsewhere in the CFTR gene.
The massive (2.4 Mb) dystrophin gene, most of which is in its 78 introns, is a splicing accident waiting to happen, and it does with an incidence of 1:3,000 of male births in Duchenne muscular dystrophy (DMD). DMD is caused by loss-of-function mutations and while >65% of DMD mutations are genomic deletions, a large number of exonic and intronic point mutations cause disease through aberrant splicing. Dystrophin is positioned at the cytoplasmic side of the skeletal muscle sarcolemma where it communicates signals between the extracellular matrix and the cellular contractile apparatus and it stabilizes the cell membrane, which must withstand repeated contraction-induced distortions. The protein contains four domains: an actin-binding domain at the N-terminus, a linker region of twenty-four spectrin-like repeats, a cysteine rich domain and a C-terminal domain (Nowak and Davies, 2004). A particularly revealing T->A substitution in exon 31 not only creates a premature termination codon (PTC), but also introduces an ESS that binds to hnRNP A1, resulting in partial exon skipping (Disset et al., 2006) (Figure 2B). Interestingly, mRNAs lacking this exon lose coding for one spectrin-like repeat but retain the correct reading frame to produce a partially functional protein, explaining why an individual harboring this mutation has a milder form of the disease than would be expected given the presence of a PTC. In this example, the specific mechanism of pathogenesis would have been missed if only the PTC mutation detected in genomic DNA had been considered, demonstrating the need to determine the effects of mutations on splicing directly by using RNA from the affected tissues. Examples such as this one also suggested the potential therapeutic benefits of inducing exon skipping to restore reading frame (see below).
As another example, mutations within and downstream from the alternatively spliced exon 10 of the (Microtubule associated protein tau) MAPT gene encoding the tau protein disrupt the 1:1 ratio required for mRNAs that include or exclude this exon. Exon 10 encodes the fourth of four microtubule-binding domains (R) and disruption of the balance between 4R-tau and 3R-tau isoforms (Figure 2C) results in hyperphosphorylation and aggregation of tau proteins into neurofibrillary tangles that are hallmarks of several neurodegenerative diseases such as Alzheimer’s disease (AD). This exon and surrounding introns are dense with splicing regulatory elements, which is indicative of the degree to which splicing of this exon is controlled. Numerous mutations within and around MAPT exon 10 disrupt exonic and intronic splicing elements and cause the inherited neuropathological disorder frontotemporal dementia with Parkinsonism linked to chromosome 17 (FTDP-17), demonstrating a direct relationship between aberrant tau expression due to alternative splicing disruption and neuropathology (Liu and Gong, 2008).
Also high on the menu of disease-causing splicing mutations is the (cystic fibrosis transmembrane conductance regulator) CFTR gene, which encodes a transmembrane chloride channel required for proper function of secretory epithelium in multiple tissues including lung, small intestine, and testes. A tryptophan deletion (•F508) causes a severe loss of CFTR protein function that is responsible for more than 50% of the cases of cystic fibrosis (CF) in the United States. Scores of mutations elsewhere in the gene produce milder forms of the disease (atypical CF), however, individuals with the same mutation were found to exhibit significant differences in disease severity. One mechanism for these differences was found to involve polymorphic (UG)m and (U)n tracts within the 3′ splice site of CFTR exon 9, which exhibits slight exon skipping even from the normal allele in unaffected individuals. Individuals with a longer (UG)m tract exhibit more exon skipping in part through increased binding of TDP-43 (Figure 2D) (Buratti et al., 2004). The severity of the effects of mutations elsewhere in the CFTR gene is modulated by the level of exon 9 inclusion and ultimately the amount of full-length functional protein that is expressed.
Splicing as a modifier of disease susceptibility and severity
Genetic variation is the predominant determinant of individual phenotypic differences, including those that are relevant to disease: disease severity (modifiers), disease susceptibility, development of the multigenic common diseases, and differences in positive and negative responses to therapeutic compounds. Recent results indicate an unexpected level of variability in the expression of different splice variants between individuals, strongly suggesting a significant role for splicing in phenotypic differences that are relevant to disease (Graveley, 2008; Wang and Cooper, 2007). An early estimate of the effects of genetic variation on splicing found that up to ~21% of alternatively spliced genes could be influenced by local polymorphisms (Nembaware et al., 2004). Combined analyses of mRNA expression (including splicing differences) using exon tiling microarrays linked with polymorphism determination using the CEU HapMap revealed that genetic variants affected differences in mRNA structure due to alternative splicing and in 5′ and 3′ end selection more than differences in whole gene mRNA levels (Kwan et al., 2008). Particularly telling is a recent analysis using deep sequencing for global quantification of alternative splice variants in cerebellar tissue from six individuals which revealed that up to 30% of the splicing events exhibited individual-specific differences reflective of striking variability in cis as well as trans-acting splicing environments (Wang et al., 2008). These studies strongly support the contention that polymorphisms located within introns and exons have a significant impact on individual variation of gene expression at the level of RNA processing and strongly contribute to individual differences in disease severity, susceptibility, and therapeutic responses. The future goal of providing personalized medicine will require a complete understanding of the effects of genetic variation on the splicing code as well as on the nuclear splicing machinery that interprets the code.
Disease-causing defects in the splicing machinery
Mutations in the constituents of the splicing machinery or any of the trans-acting splicing components and their regulators have the potential to affect expression of a large number of genes, directly or through secondary effects. Important themes that have emerged over the past several years are that mutations in ubiquitous spliceosome components and splicing factors can cause tissue-specific pathogenesis, and that changes in the relative stoichiometry of general splicing factors can cause profound changes in splicing patterns and cause disease. Splicing abnormalities have been found to be particularly prevalent in neurodegenerative disease and in cancer. Though specific causality in many cases is difficult to prove, this observation brings to the forefront the enormous vulnerability of splicing, its potential pathogenicity and the potential utility of splicing patterns as signatures (biomarkers) of specific diseases and as targets for therapeutic intervention.
Mutations in spliceosome components
Although the spliceosome is composed of a large number of proteins and five snRNPs, very few mutations in core spliceosome components have been found, suggesting that such mutations are incompatible with life either at the cellular level or in early development. Interestingly, mutations in four proteins (PRPF31, PRPF8, PRPF3, RP9) are associated with disruption of basal spliceosome function and all cause autosomal dominant forms of retinitis pigmentosa (RP) (Mordes et al., 2006). All four proteins are components of the U4/U5/U6 tri-snRNP complex, which joins the assembling spliceosome to form the catalytic center. Dominant inheritance suggests that haploinsufficiency does not provide a sufficient level of spliceosome function in photoreceptor neurons, the affected cells. For example, although a general defect in splicing was not detected in lymphoblast cell lines derived from RP patients containing null alleles for PRPF31 (Rivolta et al., 2006), specific pre-mRNAs were found to be sensitive to PRPF31 mutations in transient expression assays (Mordes et al., 2007). The intriguing sensitivity of photoreceptor neurons could reflect an inability to produce sufficient levels of highly expressed mRNAs such as from the rhodopsin gene (mutations in this gene also cause dominant forms of the disease). However alternative mechanisms are possible such as photoreceptor-specific sensitivity to aggregates formed from accumulation of mutant proteins (Mordes et al., 2007).
Changes in the snRNP repertoire and splicing abnormalities in SMA
The best-characterized function of the SMN complex is its essential role in the biogenesis of snRNPs (Neuenkirchen et al., 2008; Yong et al., 2004). SMN deficiency causes a corresponding deficit in snRNP assembly capacity (Wan et al., 2005). Motor neuron developmental abnormalities could be rescued by injection of purified snRNPs in SMN-deficient zebrafish embryos (Winkler et al., 2005) and reduced snRNP assembly has been demonstrated in an SMN–deficient SMA mouse model (Gabanella et al., 2007; Zhang et al., 2008b). Recent studies in these mice also revealed profound perturbations in RNA metabolism, including changes in snRNAs and numerous splicing abnormalities (Zhang et al., 2008b). Surprisingly, instead of a uniform decrease in the steady-state levels of all the snRNAs, each snRNA is affected differently in every tissue of SMN-deficient mice, resulting in different snRNA stoichiometries and an altered snRNP repertoire in every tissue (Gabanella et al., 2007; Zhang et al., 2008b). In addition, the SMN deficiency causes widespread and cell-type specific splicing abnormalities affecting numerous mRNAs of functionally diverse genes, including shifts in the ratios of known alternatively spliced isoforms as well as many splicing defects of constitutive exons (Zhang et al., 2008b). Knockdown of spliceosomal proteins in yeast and Drosophila have shown that depletion of general and constitutive components of the splicing machinery can have differential effects on splicing of specific pre-mRNAs and on alternative splicing (Clark et al., 2002; Park et al., 2004; Pleiss et al., 2007). Although the mechanism behind the splicing defects in SMN deficiency is unknown, and SMN is not itself a splicing factor, it is reasonable to consider the possibility that changes in the snRNP repertoire affect the efficiency, rate and fidelity of spliceosome assembly on different introns. Alternatively, or in addition, SMN may have direct roles in splicing.
These observations illustrate how a deficiency in a ubiquitously-expressed housekeeping protein may cause tissue-specific outcomes and invite a new perspective on the pathophysiology of the disease spinal muscular atrophy (SMA). A conundrum in SMA has been to reconcile the known housekeeping function of SMN with the apparent selectivity of SMA pathogenesis to motor units (anterior horn α-motor neurons and the muscles they innervate). It is likely that tissue-specific factors functioning in concert with SMN determine a cell’s snRNP repertoire. Because each cell type has a unique assortment of splicing factors and SMN deficiency causes its snRNP repertoire to change in a unique way, the resulting perturbations are distinct and give rise to cell-type specific effects on splicing. These findings also reveal a key role for the SMN complex in RNA metabolism and splicing regulation, and indicate that SMA involves splicing abnormalities throughout the organism, and not only in motor units. Although recent studies suggest that the extent of alternative splicing is much greater than was previously believed, it is nevertheless clear that each tissue produces a consistent pattern of alternatively spliced mRNA isoforms (Castle et al., 2008; Pan et al., 2008; Wang et al., 2008). It is thus not apparent why other tissues in the SMA mice do not show overt pathology despite numerous splicing abnormalities. The extent to which abnormal mRNAs are translated will need to be determined to address this issue. In addition to SMN’s role in snRNP biogenesis, it likely plays a role in the biogenesis or function of other RNPs possibly including complexes containing the Sm-like proteins (Lsm proteins) with which it interacts, as well as snoRNPs, hnRNPs and mRNPs (Neuenkirchen et al., 2008; Rossoll et al., 2003; Yong et al., 2004; Zhang et al., 2003). It is therefore possible that SMN deficiency has other consequences on RNA metabolism in addition to its effects on snRNPs and splicing. Significant questions remain, including whether the change in the snRNP repertoire is the direct cause of the splicing abnormalities, if SMN or the SMN complex has other roles in splicing, and whether the demise of motor neurons is caused by mis-splicing of one or several specific mRNAs or by the cumulative effect of many splicing abnormalities.
Splicing factors in disease: mutations and changes in stoichiometry
Like the major spliceosomal snRNPs, many of the hnRNP and SR proteins that regulate splicing are present in vast excess over their high-affinity binding sites on premRNAs (Dreyfuss et al., 2002; Dreyfuss et al., 1993). Yet even moderate changes in expression of one of them, and hence their relative stoichiometry, can have significant effects on alternative splicing (Caceres and Kornblihtt, 2002; David and Manley, 2008; Martinez-Contreras et al., 2007; Smith and Valcarcel, 2000). Furthermore, many of the hnRNP and SR proteins shuttle continuously between the nucleus and the cytoplasm (Caceres et al., 1998; Pinol-Roma and Dreyfuss, 1992) and their sub-cellular distribution can shift in response to stress signals. The resulting change in stoichiometry of splicing factors in the nucleus could also modulate alternative splicing patterns. A diverse set of diseases are associated with changes in expression of RNA binding proteins involved in splicing and splicing regulation (Gabut et al., 2008; Lukong et al., 2008; Ule, 2008).
Recent studies suggest a role for several RNA-binding proteins in neurological and psychiatric diseases. TDP-43, initially identified as binding to the HIV trans-acting response element (TAR), and shown to be a transcriptional repressor of HIV, is an hnRNP protein similar in general structure to hnRNP A1 and A2. It has been found to have several different associations with disease (Buratti and Baralle, 2008). In one, TDP-43 binds to the polymorphic (UG)m repeats within intron 8 of the pre-mRNA of CFTR (the gene mutated in cystic fibrosis) such that increased binding to longer (UG)m repeats causes increased exon 9 skipping. Recent studies have shown that TDP-43, which is normally nuclear, is found in cytoplasmic inclusions in affected regions of the brains of individuals with frontotemporal dementia (FTD), Alzheimer’s disease and amyotrophic lateral sclerosis (ALS) (Ule, 2008). In these patients, TDP-43 is depleted from nuclei and the protein that accumulates in the cytoplasmic inclusions is ubiquitinated, cleaved or abnormally phosphorylated. These findings represent a significant advance and a useful histological marker that correlates well with FTD. Mutations in the TARDBP gene, which encodes TDP-43, also are associated with both sporadic and familial forms of ALS, demonstrating a role for TDP-43 in the pathogenesis of neurological diseases (Daoud et al., 2008; Kabashi et al., 2008; Rutherford et al., 2008; Sreedharan et al., 2008). It remains to be determined whether the pathogenic mechanism relates to toxicity of the cytoplasmic aggregates, loss of TDP-43′s nuclear function, or a combination of both.
Another RNA binding protein that has recently been implicated in neurological disorders is quaking (QKI). QKI is the human homologue of the gene mutated in the spontaneous mouse dysmyelination mutant, quaking viable (qkv). The protein is a member of the STAR family, which contains domains that link RNA binding with signal transduction. QKI expresses nuclear and cytoplasmic isoforms via alternative splicing and it regulates both splicing and mRNA stability; putative pre-mRNA and mRNA targets have been identified (Chenard and Richard, 2008). QKI expression in oligodendrocytes is required for normal myelination in mice and it plays a fundamental role in differentiation of oligodendrocyte precursor cells in culture for which RNA binding activity is required (Chen et al., 2007). QKI is a strong candidate gene for schizophrenia susceptibility based on genetic linkage data as well as mRNA expression studies. These findings demonstrate reduced expression of QKI mRNA as well as putative QKI target mRNAs in disease-relevant regions from post-mortem patient brain tissue samples (Lauriat et al., 2008). Interestingly, a large-scale analysis of interactions among proteins associated with inherited ataxias identified a number of RNA binding protein families including QKI as well as Nova and Fox (Lim et al., 2006). Nova is one of the best-characterized regulators of an alternative splicing network, coordinating expression of proteins modulating synaptic function (Ule et al., 2005) and Fox is an important splicing regulator in several tissues including the brain (Underwood et al., 2005). An analysis of the Fox splicing regulatory network identified thousands of putative targets with enrichment for neuromuscular functions and genes associated with diseases affecting neurons and striated muscle where Fox expression is high (Zhang et al., 2008a). In addition, mutations in the A2BP1/FOX-1 gene are associated with mental retardation, epilepsy, and autism (Bhalla et al., 2004; Sebat et al., 2007), strongly suggesting a direct role in disease. Both Nova and Fox proteins are likely to have other RNA processing roles such as regulating alternative polyadenylation site selection (Licatalosi et al., 2008). These results are highly suggestive of further roles of RNA binding proteins in causing and modifying the severity of neurological diseases.
Splicing abnormalities in cancer
Disruption of splicing has long been thought of as a hallmark of a variety of cancers, however, recent computational analyses using cancer-derived expressed sequence tags (ESTs) found that the prevalence of alternatively spliced genes was slightly lower in tumors compared to normal tissue. What differed between normal and cancerous tissues were the sets of genes rather than the number of alternatively spliced genes (Kim et al., 2008). The major difficulty has been to determine if the splicing changes detected in cancer are pathogenic, however, the cause-effect relationships between the production of cancer-associated splice variants and cancer formation and progression are becoming clearer. Cis-acting mutations cause aberrant splicing and inappropriate expression of cancer-associated genes. Several genes use a balance between hnRNP and SR proteins to regulate alternative splicing, producing appropriate ratios of pro- and anti-apoptotic isoforms (Srebrow and Kornblihtt, 2006). However, it has also become clear that the mis-regulation of alternative splicing in cancer cells is extremely complex in that the effects of a splicing regulator on the same splicing event can differ between cells. A test of the effects of knockdown of 14 hnRNP proteins on 56 alternative splicing events of apoptotic genes in three cancer/immortalized cell lines revealed a surprisingly high level of cell line-specific effects (Venables et al., 2008). The results indicate that cell-specific differences in trans-acting regulatory environments make it difficult to predict the effects of changes in individual splicing factors.
Despite these complexities, strong links have been established between altered expression of specific splicing factors, aberrant splicing of their pre-mRNA targets, and induction of signaling pathways that are relevant to transformed or malignant cellular phenotypes. A particularly coherent picture has emerged for the SR protein SF2/ASF, which is elevated in several cancers and induces transformation when overexpressed at low levels. SF2/ASF overexpression induces splicing to produce an oncogenic isoform of the ribosomal protein S6 kinase-β1 (S6K1), a regulator of translation, and knockdown experiments demonstrated that expression of this isoform is required for transformation by SF2/ASF (Karni et al., 2007). Elevated SF2/ASF also promotes splicing of the RON kinase to a constitutively active form that has been associated with invasive behavior in several cancers. The effect of SF2/ASF on RON splicing is direct via binding to an exonic splicing enhancer (ESE) within the downstream exon. The invasive phenotype of human cancer cells is reversed by knockdown of endogenous SF2/ASF or the constitutively active RON splice variant (Ghigna et al., 2005). Expression of a number of other splicing factors is also altered in cancer (Grosso et al., 2008), however, the relevance to cancer formation or progression remains to be established. It is also important to consider that most if not all proteins that regulate splicing are multifunctional. For example, recent results suggest that transformation induced by SF2/ASF could occur by a mechanism that is independent of splicing (Karni et al., 2008).
Importantly, cancer-related changes in splicing and splicing factors are nevertheless potentially useful biomarkers for cancer diagnosis and classification. Tumor classification is critical for accurate diagnosis, prognosis, selection of the most effective therapeutic approach, and follow-up treatment. Analysis of global alternative splicing patterns by high throughput RT-PCR, splicing-sensitive microarrays and deep sequencing provides the opportunity for an analysis of unprecedented specificity to link molecular signatures to predictions of therapeutic efficacy.
Disruption of alternative splicing by an RNA gain-of-function
In an unexpected twist for a molecule thought of as a passive substrate, RNA expressed from a mutated allele can be pathogenic. This is a not so subtle reminder that RNA is an intrinsically potent molecule. This mechanism of disease is observed in a class of so-called microsatellite expansion disorders. Microsatellites are short (1 to 10 nucleotide) repeats that vary in number between individuals and cause disease when located in a gene and expand beyond a normal threshold number of repeats. The expansions can cause disease by three mechanisms which are not mutually exclusive: loss of protein function, gain of aberrant protein function due to expansion of triplet repeats within the open reading frame, or gain-of-function of the RNA containing the expansion (Orr and Zoghbi, 2007). There is clear evidence for an RNA gain-of-function in four human diseases: myotonic dystrophy types 1 (DM1) and 2 (DM2), fragile X-associated tremor ataxia syndrome (FXTAS), and spinocerebellar ataxia 8 (SCA8). An RNA gain-of-function mechanism is also likely in SCA10, SCA12, and Huntington’s disease-like 2 (HDL2) (Figure 3) (O’Rourke and Swanson, 2008).
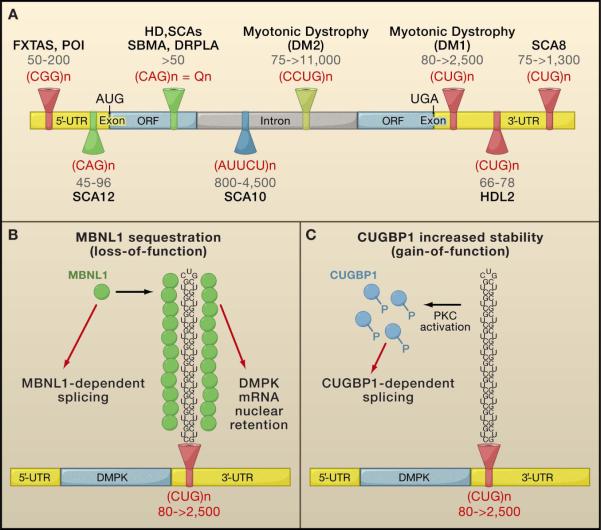
Figure 3. RNA gain-of-function.
A. Microsatellite expansions within transcribed regions with confirmed or potential RNA gain-of-function effects. The type and number of nucleotide repeats are indicated. (Fragile X-associated tremor ataxia syndrome, FXTAS; premature ovarian insufficiency, POI; Huntington’s disease, HD; spinocerebellar ataxias, SCAs; dentatorubral pallidoluysian atrophy, DRPLA; spinal and bulbar muscular atrophy, SBMA; Huntington’s disease like 2, HDL2).
B. Sequestration of MBLN1 in myotonic dystrophy (DM1).
C. Activation of protein kinase C (PKC) by the expanded CUG RNA induces hyperphosphorylation of CUGBP1, which stabilizes the protein.
The pathogenic mechanism of an RNA gain-of-function is best characterized for DM1 in which a CUG repeat located within the 3′ UTR of the DMPK mRNA expands beyond its normal range of 5-38 repeats to a pathogenic range of 50 to >2,500 repeats. This mutation results in the second most common cause of muscular dystrophy as well as a variety of multisystemic features that include cardiac arrhythmias and central nervous system dysfunction. An early hypothesis, proven to be correct, was that the RNA repeats disrupt the functions of specific RNA binding proteins in trans (Timchenko et al., 1996; Wang et al., 1995). Proteins identified based on their affinity for CUG RNA repeats include CUG binding protein 1 (CUGBP1) and muscleblind-like 1 (MBNL1) (O’Rourke and Swanson, 2008). Both proteins, or their paralogues in the CELF and MBNL gene families, are regulators of several aspects of nuclear and cytoplasmic RNA processing including alternative splicing, mRNA stability, translation, mRNA localization, and editing (Barreau et al., 2006; Pascual et al., 2006). The best-characterized pathogenic effect of expanded CUG RNA is its disruption of a program of postnatal splicing transitions in striated muscle. At the heart of this mechanism is the disrupted balance of antagonistic regulation by MBNL1 and CUGBP1 (Figure 3B). The result is the inappropriate expression of embryonic rather than adult splice variants in adult tissues and consequent manifestations of disease.
A dramatic cellular hallmark of DM1 is the accumulation of expanded CUG RNA in nuclear foci. In one of two pathogenic effects of the RNA, MBNL1 colocalizes with the RNA foci and is depleted from the nucleoplasm more than two-fold resulting in aberrant regulation of MBNL1-sensitive splicing events (Lin et al., 2006). Interestingly, MBNL1 binds to the expanded CUG repeats because the RNA forms an extended stable hairpin structure that resembles its natural intronic binding site found near targeted alternative exons (Warf and Berglund, 2007; Yuan et al., 2007). Mouse models that produce Mbnl1 depletion by either an Mbnl1 gene knockout (Mbnl1•E3/•E3E3) or expression of 250 CTG repeats within a human skeletal α actin transgene (HSALR) reproduce muscle pathology and expression of embryonic splicing patterns as observed in DM1 skeletal muscle (Kanadia et al., 2003; Lin et al., 2006). In the mouse models and individuals with DM1, the embryonic splicing pattern for the muscle specific chloride channel (Clcn1) introduces a premature termination codon resulting in loss of Clcn1 function and the myotonia that is characteristic of the disease (Lueck et al., 2007). Delivery of MBNL1 via adeno-associated virus to HSALR skeletal muscle reverses the Clcn1 and other splicing changes as well as the myotonia (Kanadia et al., 2006) demonstrating the link between MBNL1 depletion and a clinical feature of the disease. A role for MBNL1 is also supported by results from Drosophila DM1 models in which human MBNL1 homologues suppress the severe phenotype induced by CUG repeat RNA in both eye and muscle (de Haro et al., 2006). Disruption of MBNL1 function could play a pathogenic role in other microsatellite disorders such as FXTAS in which male carriers of the fragile X pre-mutation in the FMR1 gene develop a late onset neurological syndrome, whereas female carriers develop premature ovarian insufficiency (POI) (Figure 3A) (Hagerman and Hagerman, 2004). Nuclear inclusions found in affected regions of FXTAS brain samples contain MBNL1 as well as hnRNPA2 (Iwahashi et al., 2006). MBNL1 also colocalizes with nuclear RNA foci found in neurons from individuals affected with HDL2 in which the mutated junctophilin-3 (JPH3) gene contains expanded CTG repeats (Rudnicki et al., 2007).
In addition to depleting RNA binding proteins, CUG repeat RNA has the provocative effect of inducing a signaling event resulting in activation of protein kinase C (PKC) (Figure 3C). One consequence of PKC activation is hyperphosphorylation and stabilization of CUGBP1 protein, which explains the observations that CUGBP1 protein is upregulated 2-4 fold in DM1 heart and skeletal muscle without an increase in mRNA levels (Nezu et al., 2007). PKC activation and CUGBP1 hyperphosphorylation and upregulation are direct and rapid effects as all three events were observed six hours after induction of a CUG repeat-containing transgene mRNA in heart tissue from an inducible DM1 mouse model (Kuyumcu-Martinez et al., 2007; Wang et al., 2007). The mechanism by which expanded CUG RNA activates PKC is unknown. The possibility that PKC activation is secondary to depletion of MBNL1 is unlikely since neither the Mbnl1 E3/ E3 nor HSALR mouse models exhibit elevated CUGBP1 (Kanadia et al., 2003; Lin et al., 2006). Recent results demonstrate that a large subset of splicing transitions that occur during the first two weeks of postnatal heart development are controlled by MBNL1 upregulation and CUGBP1 downregulation. Interestingly, CUGBP1 mRNA levels do not change and the ten-fold decrease in protein expression appears to involve protein dephosphorylation and destabilization (Kalsotra et al., 2008; Kuyumcu-Martinez et al., 2007). These results suggest the possibility that expanded CUG repeats are aberrantly stimulating a natural signaling event that maintains CUGBP1 protein stability in embryonic striated muscle.
DM2 is caused by expanded CCTG repeats in intron 1 of the ZNF9 gene and is clinically similar to DM1 but is generally less severe and lacks a congenital form (Liquori et al., 2001). AlthoughMBNL1 is sequestered on expanded CCUG repeat RNA, which forms a hairpin structure and accumulates in nuclear foci like CUG repeat RNA, CUGBP1 is not induced (Lin et al., 2006) suggesting that DM2 is primarily a disease of MBNL depletion. The lack of CUGBP1 up-regulation suggests that PKC is not activated by expanded CCUG RNA and can provide insights into the cis requirements for PKC activation by RNA such as whether CCUG versus CUG repeats or a gene-specific context is required.
RNA generated from microsatellite expansions also has the potential to produce pathogenicity through RNA interference pathways. Expanded repeats containing CUG, CAG, and to a lesser extent CCG or CGG are processed into ~21 nucleotide RNAs after folding into single imperfect hairpins that are cleaved by Dicer (Krol et al., 2007). In addition, antisense transcripts have been detected for the expanded repeats of the DMPK, SCA8, and FMR1 genes (Cho et al., 2005; Ladd et al., 2007; Nemes et al., 2000) providing a second potential source of small RNAs. Small CUG or CAG RNAs have been detected in cells from DM1, SCA1, and Huntington’s disease patients (Cho et al., 2005; Krol et al., 2007). Evidence for downstream effects of CUG siRNAs comes from knockdown of Dicer in DM1 cells that resulted in elevated levels of normal endogenous CAG repeat-containing mRNAs (Krol et al., 2007). Although these results provide a proof of principle, targets relevant to DM1 pathogenesis remain to be identified.
With the exception of SCA12, disease-causing CAG expansions are within open reading frames and result in the production of proteins containing expanded polyglutamine tracts (polyQ) (Orr and Zoghbi, 2007). Disease-causing CAG repeat expansions can result in protein loss- or gain-of-function or both, depending on the gene affected, however, whether expanded CAG RNA can contribute to pathogenicity depends on the experimental system. The phenotype observed in a transgenic mouse model for SCA1 that expresses a pathogenic human ataxin-1 mRNA containing 82 CAG repeats [ataxin-1-(CAG)82] is not observed in mice expressing equivalent levels of mRNA containing a point mutation that inactivates a protein nuclear localization signal. These results convincingly demonstrate that the ataxin-1-(CAG)82 mRNA alone is not pathogenic in mouse Purkinje cells (Klement et al., 1998). Strong support for a CAG RNA gain-of-function mechanism comes from a Drosophila model in which expression of a truncated pathogenic human ataxin-3-(CAG)78 mRNA causes neurotoxicity (Li et al., 2008). The role for RNA toxicity came to light from modifier screens that identified mbl, the Drosophila MBNL homologue. In addition, expanded CAG repeats located in the 3′ UTR of a heterologous gene (DsRED), demonstrated not to be expressed as polyQ protein or CUG antisense RNA, were also pathogenic, directly demonstrating a pathogenic role for CAG repeat RNA. Expression of polyQ protein from nonpathogenic interrupted CAA/CAG repeat RNA induced some pathogenicity indicating that both polyQ protein and CAG RNA contribute to the full phenotype observed in the fly model. These results open the door for consideration of RNA gain-of-function effects for expanded CAG repeats in human disease. Interestingly, whereas MBNL1 suppressed CUG RNA toxicity in the Drosophila DM1 model, fly and human MBNL1 homologues enhanced the pathogenic effects of CAG RNA suggesting different pathogenic mechanisms for CUG and CAG RNA toxicity (Li et al., 2008).
A different set of RNA-binding proteins is associated with the expanded CGG repeats that cause FXTAS. HnRNP A2 and Pur-α were identified in purified nuclear inclusions from brains of a FXTAS mouse model (Jin et al., 2007). Both proteins directly interact with CGG repeat RNA and expression of each protein suppressed the neurodegeneration phenotype in a Drosophila FXTAS model (Jin et al., 2007). CUGBP1 expression also suppressed the phenotype in flies and was shown to interact with repeat RNA indirectly by binding to hnRNP A2 (Sofola et al., 2007). How these proteins fall into a mechanism of pathogenesis is not yet clear. Pur-α knockout mice exhibit an ataxia phenotype, consistent with a depletion model in FXTAS, however, additional effects of the RNA are possible as demonstrated in DM1. It is also important to consider that suppression of an RNA gain-of-function phenotype in Drosophila models, particularly when using human proteins, could result not only from restoring the function of a depleted Drosophila protein but also by rendering the RNA nontoxic by a different mechanism such as preventing RNA aggregation or interactions that lead to toxicity. Understanding the specific mechanism of suppression would provide information regarding the pathogenic mechanism of disease as well as potential therapeutic approaches.
RNAs as therapeutic targets and tools
Many approaches have been explored and many more can be envisioned to modify the splicing pattern of a mutant pre-mRNA or eliminate an mRNA that bears a disease-causing mutation to achieve therapy. Increasing knowledge of RNA biology and chemistry is stimulating renewed efforts to target the RNA itself or the splicing and translational machinery as entry points for therapeutic intervention. Here, we discuss several strategies including antisense oligonucleotides, antisense snRNAs, RNA interference and small molecules (see Analysis by L. Bonetta on page XXX of this issue). Other methods including trans-splicing and ribozymes will likely emerge as additional options in the future.
Antisense oligonucleotides
Antisense oligonucleotides (AOs) have been used extensively to redirect splicing of a mutation-bearing pre-mRNA to prevent it from generating a disease-causing mRNA and forcing it to splice into a disease-rescuing mRNA (Sazani and Kole, 2003). Generally, AOs have been designed to hybridize and block one or more sequences in the target pre-mRNA that are critical for the particular splicing event that one wishes to inhibit and cause the splicing machinery to select an alternative pattern whose outcome is more favorable. To be effective, the AO’s target RNA sequence needs to be accessible in the native RNP, which not all sequences are, and it must be specific. However, it is difficult to predict if these requirements will be met, as well as whether the desired splicing switch will be achieved. Thus, many different AOs need to be tested and this may require tiling of many AOs of various lengths across a considerable stretch of the target pre-mRNA. Examples of disease-causing genes targeted by AOs in vitro and in cell systems include point mutations in the β-globin gene in β-thalassemia, the CFTR gene in cystic fibrosis, and lamin A in the premature aging disease Hutchinson-Gilford progeria syndrome (HGPS). Mutations in these genes activate aberrant cryptic splice sites and prevent correct pre-mRNA splicing (Dominski and Kole, 1993; Friedman et al., 1999; Scaffidi and Misteli, 2005). An AO targeting mutations in the gene encoding the muscle-specific chloride channel (ClC-1) reverses the defect of ClC-1 alternative splicing in mouse models of myotonic dystrophy (Wheeler et al., 2007). AOs have also been extensively studied as potential treatment strategies for SMA and DMD. AOs that mask the ISS or the 3′ splice site of exon 8 of the SMN2 gene have been used to switch the splice site and promote exon 7 inclusion and thus the production of a full length functional SMN protein (Figure 4A) (Singh, 2007). An interesting mechanism-based approach has used bifunctional oligonucleotides that link an antisense sequence to exon 7 with an ESE and thereby recruit splicing-enhancing SR proteins or directly couple antisense sequence to an SR peptide to promote exon 7 inclusion (Figure 4A) (Cartegni and Krainer, 2003; Skordis et al., 2003). Similar AO approaches have also been used for DMD (Alter et al., 2006). There are also several notable examples of the use of AOs to downregulate specific mRNAs by mechanisms that do not necessarily involve modulation of splicing. An AO targeted to a superoxide dismutase 1 (SOD1) mutation that causes the neurodegenerative disease amyotrophic lateral sclerosis (ALS) administered to mice with the disease reduces mutant SOD1 mRNA and protein levels and slows disease progression (Smith et al., 2006).
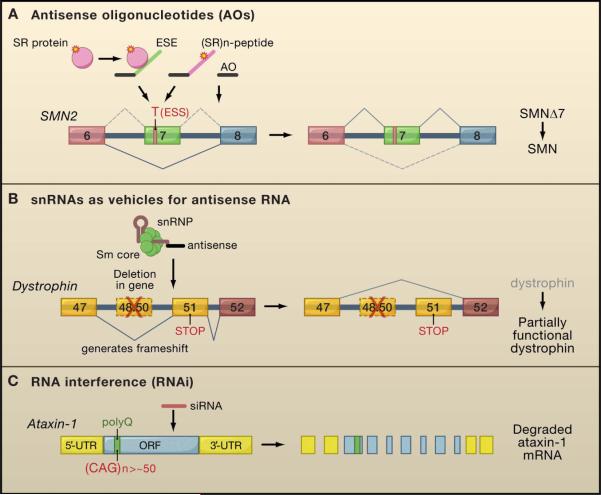
Figure 4. Therapeutic approaches that use or target RNAs.
A. Antisense oligonucleotides (AOs) or bifunctional AOs that have sequences complementary to exon 7 conjugated to splicing-enhancing effectors---e.g., serine-arginine (SR) peptide or an ESE that recruits SR proteins---are used to promote exon 7 inclusion of the SMN2 gene, decreasing the production of a truncated SMN protein (SMNΔ7) and increasing that of a full-length functional protein (SMN).
B. Modified U1 or U7 snRNA-based vectors can carry complementary sequences to the targeting RNAs to achieve the desired splicing pattern. As shown here, genomic deletions (e.g., from exon 48 to 50) in dystrophin genes of DMD patients lead to a premature termination codon (STOP) in exon 51 caused by a frameshift. Antisense U7 snRNA targeting the 3′ splice site of exon 51 prevents the inclusion of this exon and restores the reading frame, and as a result, produces a partially functional protein.
C. RNAi approaches are used to eliminate pathogenic mRNAs. siRNAs delivered to a mouse model of spinocerebellar ataxia type I (SCA1) elicit degradation and efficient knockdown of the disease-causing ataxin-1 mRNA that contains the expanded CAG repeats encoding polyglutamine (polyQ).
Although proof-of-concept experiments for efficacy of AOs have been demonstrated for many model systems and diseases, only a handful have reached the level of practical therapies in humans (see Analysis by L. Bonetta on page XXX of this issue). One successful clinical application of AOs is the development of the first FDA-approved antisense drug that targets cytomegalovirus (CMV) mRNAs to treat retinitis caused by CMV (Holmlund, 2003). The major bottlenecks in implementing AOs as therapeutics relate to tissue-specific delivery of sufficient amounts of the AOs to achieve a re-administration regimen that will sustain therapeutic levels and stability long-term. AOs with various backbone chemistries ---morpholino, peptide nucleic acid (PNA), locked nucleic acid (LNA), 2′-O-methyl, thiophosphate, and numerous other oligonucleotides in place of the naturally occurring phosphodiester ribo- or deoxy-nucleotides--- have been developed to improve affinity, boost stability in the circulation and in target cells, and enhance cell penetration and nuclear accumulation (Karkare and Bhatnagar, 2006). Improved tissue-specific delivery has also been achieved by conjugating AOs to peptides that are selectively taken up by the target tissues (Henke et al., 2008). AOs remain a promising avenue for the therapy of many genetic diseases. However, tests for specificity and undesirable off-target effects must be rigorously and carefully performed.
snRNAs as vehicles for stable antisense RNAs
RNAs can also be used as a tool to target other RNAs, including pre-mRNAs by antisense hybridization. U1 snRNA- and U7 snRNA-based vectors consisting of the native snRNAs modified to have an antisense sequence to a target pre-mRNA in place of the native 5′-end of the RNA have been developed. These modified snRNAs are transcribed from the corresponding DNA constructs in the target cells and have unique advantages as antisense vehicles. Like endogenous snRNAs, they acquire Sm cores that contribute to their stability, nuclear localization and efficient hybridization to complementary target RNAs. Anti-SMN U7 snRNAs containing sequences complementary to the 3′ splice site of exon 8 or to exon 7 linked with an ESE induced an almost complete and durable correction of SMN2 splicing in cultured cells (Madocsai et al., 2005) and prolonged survival of mice with severe SMA (Meyer et al., 2008). Unlike synthetic AOs they are RNA, not chemically modified entities, but their delivery into cells presents unique obstacles that AOs are not subject to. Viral vectors including recombinant adeno-associated virus (AAV) and lentiviruses are used as delivery vehicles to transduce antisense RNAs to the target cells (Danos, 2008). Systemic administration of AAV U7 or U1 antisense RNA into a DMD mouse model leads to the skipping of the exon containing a mutation causing frameshift and PTC, and to a sustained production of partially functional dystrophin protein and an improved phenotype (Figure 4B) (Denti et al., 2006; Goyenvalle et al., 2004). These results provide the foundation for clinical phase studies that are currently underway for the treatment of DMD (Muntoni and Wells, 2007). Another U7 antisense RNA application is inhibition of HIV-1 production. Lentiviral U7 induces a partial skipping of internal exons encoding the HIV-1 regulatory proteins Tat and Rev (Asparuhova et al., 2007). However, serious issues remain including tissue-specific targeting and immune response to the viral vectors, as is the case for conventional gene replacement therapy.
RNA interference (RNAi)
The capacity to selectively eliminate an mRNA of a disease-causing allele or to prevent translation of a deleterious protein by RNAi presents a wide range of targets for therapeutic modulation. RNAi relies on the base pairing interaction of 21-23 nucleotide RNAs, a size sufficient to uniquely target an mRNA or even a specific splice variant, and provides a versatile and potent tool (Grimm and Kay, 2007). RNAi-based strategies are applicable to all diseases in which decreasing expression of an RNA, whether from a mutant allele or an aberrantly expressed mRNA in cancer cells, would have therapeutic effects. Great progress has been made toward translating the expertise of RNAi from an extensively used experimental tool to an effective and safe treatment. The main challenges again are optimal delivery to the appropriate tissues and cells, avoiding the cellular anti-viral response to double-stranded RNA, and achieving the optimal balance of high potency without off-target effects. The three most common therapeutic approaches to RNAi-based delivery are: (i) synthetic double-stranded small interfering RNAs (siRNAs) applied directly to tissues using a permeabilizing vehicle in which exogenous siRNAs become incorporated into the endogenous RNAi machinery (RISC complex); (ii) short hairpin RNAs (shRNAs) typically delivered by expression from viral vectors, are expressed in the nucleus, exported to the cytoplasm by Exportin 5 and processed by Dicer into siRNAs; (iii) artificial miRNAs, in which the targeting hairpins are expressed in a pri-miRNA context using viral vectors, processed in the nucleus by the Drosha-DGCR8 complex, exported to the cytoplasm and then processed into functional siRNAs by Dicer (Rossi, 2008). ShRNAs are typically expressed at high levels to produce highly effective gene silencing, but they can also elicit toxicity due to saturation of the endogenous RNAi machinery (Rossi, 2008). Investigations using AAV-delivered artificial miRNAs to knockdown the expanded ataxin-1 mRNA in a SCA1 mouse model indicates that this approach produced efficient knockdown of this mRNA in cerebellar Purkinje cells without toxicity (Figure 4C) (Boudreau et al., 2009). These results provide a next-generation platform for virus-mediated delivery and expression. RNAi approaches are showing promise in several other mouse models of disease including Huntington’s disease and ALS, and in HIV-infected “humanized” mice. Direct siRNA delivery is being used in preclinical and clinical trials to treat several diseases such as macular degeneration, respiratory syncicial virus (RSV) infection, and liver cancer (Aigner, 2007; Pappas et al., 2008).
With a similar technology to that used to deliver siRNAs, it is possible to target miRNAs using complementary nucleotide sequences (anti-miRs). Anti-miRs can be used to block endogenous or exogenous (viral) miRNAs to neutralize their effects. Delivery into most tissues is difficult but delivery to the liver works well. For example, the highly expressed and liver-specific miR-122 was found to have at least two features relevant to disease: it is required for hepatitis C virus replication in cultured hepatocytes and is involved in regulating cholesterol metabolism. Delivery of LNA-modified oligonucleotides to block miR-122 demonstrated both the loss of miR-122 and a functional consequence of lowered plasma cholesterol in mice and monkeys (Elmen et al., 2008). The potential benefits of this approach for treating hypercholesterolemia as well as acute and chronic hepatitis C infections are quite promising.
There are cautionary tales as well, however. In pre-clinical testing in mice, naked siRNAs targeting VEGF or the VEGF receptor and delivered directly to the large chamber of the eye prevented the over-proliferation of blood vessels in the retina that cause some forms of macular degeneration (Reich et al., 2003; Shen et al., 2006). This approach has progressed to phase I clinical trials in humans (Whelan, 2005). However, tests in mice designed to determine how “naked” siRNAs are taken up into cells discovered that siRNAs do not actually enter cells but rather reduce vascularization through binding to the extracellular Toll-like receptor 3 and stimulating the NF-κB pathway (Kleinman et al., 2008). The results highlight the potential for adverse effects from systemic delivery and the importance of delivery systems that facilitate entry into the intended cells.
Small molecules
Alternative splicing is an attractive target for pharmacological modulation with small molecules. As splicing of most introns is strongly dependent on serine-arginine-rich (SR) proteins and hnRNP proteins, small molecules that affect their activities or their relative amounts in the nucleus can profoundly modify splicing. SR proteins bind to ESEs via their RNA-binding domains and promote exon definition as well as prevent the action of adjacent silencers by recruiting the basal splicing machinery through their SR domains. This activity of SR proteins depends on the state of serine phosphorylation in the SR domain, which is determined by the SR kinases SRPK1, SRPK2 and the Clks1-4, and by the phosphatase PP1 that dephosphorylates them. Small molecules are ideal reagents for modulating the activities of such enzymes and have been used to target these enzymes to modulate splicing patterns. Using high throughput screening, small molecule inhibitors of SR proteins (Soret et al., 2005), SRPKs (Fukuhara et al., 2006) and Clks (Muraki et al., 2004) were identified and shown to modulate alternative splicing. These compounds so far have been analyzed for only a few splicing events, for example, HIV-1 splicing which is strongly dependent on SR proteins, but these studies suggest that this approach holds considerable promise. This justifies undertaking large-scale screens on these and other enzymes that regulate the activity of the splicing machinery in search of more potent and specific modulators of alternative splicing as a therapeutic approach for treating many diseases. Other potential approaches for regulating alternative splicing with small molecules can be envisioned based on modulation of the relative stoichiometries of splicing factors. Thus, small molecules that alter the expression or modulate the nuclear localization of splicing factors would likely provide a means to modify alternative splicing of specific disease-relevant pre-mRNAs.
The historic success of small molecule pharmacological reagents, including the fact that they circumvent the major issues of delivery into most tissues encountered by nucleic acids (oligonucleotides, siRNAs and DNA constructs), makes small molecules highly attractive as a therapeutic modality. Robust, quantitative and specific cell-based alternative splicing assays will be required to realize this potential. New screens for this purpose have been recently described and deployed, so far on a modest scale, to identify modulators of alternative splicing (O’Brien et al., 2008; Stoilov et al., 2008).
Aberrant splicing frequently generates mRNAs containing PTCs. Approximately 30% of mutations causing inherited disease have PTCs and their presence leads to degradation by nonsense mediated decay and the loss-of-function of the mutant allele (Frischmeyer and Dietz, 1999). In most cases, nonsense mediated decay is protective by preventing expression of truncated proteins that could have dominant negative activity or form toxic aggregates. However, it is estimated that 5% to 15% of individuals with one of ~2300 inherited diseases carry a PTC mutation that would benefit from readthrough of the stop codon to produce a protein that is at least partially functional. For example, approximately 7% of DMD and 10% of CF cases are due to a PTC within an otherwise in-frame mRNA. Small molecules that allow readthrough of premature termination codons have been identified and their potential utility for specific diseases are being tested. In particular aminoglycoside antibiotics bind to the decoding site of the small ribosomal subunit, and induce translation readthrough of termination codons (Hainrichson et al., 2008; Zingman et al., 2007). Clinical trials using aminoglycosides for treating several inherited diseases, including the use of gentamicin to treat DMD and CF, have provided encouraging results. However, the high levels of drug required can produce significant side effects as well as increasing the levels of PTC-containing mRNAs due to inhibition of nonsense mediated decay (Zingman et al., 2007). The design of aminoglycosides with improved activity and specificity holds considerable promise (Hainrichson et al., 2008). A structurally unrelated compound, PTC124, was identified by its suppression of a PTC-containing luciferase reporter in a high throughput screen and has been shown to increase production of the dystrophin protein from PTC containing mRNAs in cultured cells and in a DMD mouse model (Welch et al., 2007). It is orally bioavailable, and is currently in clinical trials for DMD (Kerem et al., 2008). Although nonsense suppression appears to be a potentially viable therapeutic approach, the possibility that a drug affects translation termination in other mRNAs in addition to the intended target needs to be carefully evaluated. The potential for side effects is particularly important in diseases in which life-long administration of the drug will be necessary.
Concluding remarks
Disrupted functions of RNAs and RNPs are the cause of numerous maladies, and they provide a wealth of new opportunities for therapies as well as tools for treatment of human diseases. We have argued here that the intricate process of alternative splicing, in particular, while providing tremendous advantages in affording an almost explosive capacity for generating transcriptome and proteome diversity, is also fraught with the risk of malfunction. In many cases, RNAs and RNPs may provide a more readily accessible target for therapy than the defective or deficient proteins they encode. Reversal of defective protein or RNA toxicity or restoration of normal activity levels could potentially be achieved more specifically and efficaciously by eliminating or redirecting the splicing of the pre-mRNA. Much, in fact most, of what has been learned came from basic research on the biology and chemistry of RNAs and the proteins they interact with in the cellular environment. It is in the public’s interest that such fundamental research should be vigorously supported.
Acknowledgements
T.A.C is supported by the NIH and the Muscular Dystrophy Association. G.D. is supported by the Association Française Contre les Myopathies (AFM). G.D. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aigner A. Applications of RNA interference: current state and prospects for siRNA-based strategies in vivo. Appl Microbiol Biotechnol. 2007;76:9–21. doi: 10.1007/s00253-007-0984-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alter J, Lou F, Rabinowitz A, Yin H, Rosenfeld J, Wilton SD, Partridge TA, Lu QL. Systemic delivery of morpholino oligonucleotide restores dystrophin expression bodywide and improves dystrophic pathology. Nat Med. 2006;12:175–177. doi: 10.1038/nm1345. [DOI] [PubMed] [Google Scholar]
- Asparuhova MB, Marti G, Liu S, Serhan F, Trono D, Schumperli D. Inhibition of HIV-1 multiplication by a modified U7 snRNA inducing Tat and Rev exon skipping. J Gene Med. 2007;9:323–334. doi: 10.1002/jgm.1027. [DOI] [PubMed] [Google Scholar]
- Barreau C, Paillard L, Mereau A, Osborne HB. Mammalian CELF/Bruno-like RNA-binding proteins: molecular characteristics and biological functions. Biochimie. 2006;88:515–525. doi: 10.1016/j.biochi.2005.10.011. [DOI] [PubMed] [Google Scholar]
- Bhalla K, Phillips HA, Crawford J, McKenzie OL, Mulley JC, Eyre H, Gardner AE, Kremmidiotis G, Callen DF. The de novo chromosome 16 translocations of two patients with abnormal phenotypes (mental retardation and epilepsy) disrupt the A2BP1 gene. J Hum Genet. 2004;49:308–311. doi: 10.1007/s10038-004-0145-4. [DOI] [PubMed] [Google Scholar]
- Boudreau RL, Martins I, Davidson BL. Artificial MicroRNAs as siRNA Shuttles: Improved Safety as Compared to shRNAs In vitro and In vivo. Mol Ther. 2009;17:169–175. doi: 10.1038/mt.2008.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratti E, Baralle FE. Multiple roles of TDP-43 in gene expression, splicing regulation, and human disease. Front Biosci. 2008;13:867–878. doi: 10.2741/2727. [DOI] [PubMed] [Google Scholar]
- Buratti E, Brindisi A, Pagani F, Baralle FE. Nuclear factor TDP-43 binds to the polymorphic TG repeats in CFTR intron 8 and causes skipping of exon 9: a functional link with disease penetrance. Am J Hum Genet. 2004;74:1322–1325. doi: 10.1086/420978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres JF, Kornblihtt AR. Alternative splicing: multiple control mechanisms and involvement in human disease. Trends Genet. 2002;18:186–193. doi: 10.1016/s0168-9525(01)02626-9. [DOI] [PubMed] [Google Scholar]
- Caceres JF, Screaton GR, Krainer AR. A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes Dev. 1998;12:55–66. doi: 10.1101/gad.12.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- Cartegni L, Hastings ML, Calarco JA, de Stanchina E, Krainer AR. Determinants of exon 7 splicing in the spinal muscular atrophy genes, SMN1 and SMN2. Am J Hum Genet. 2006;78:63–77. doi: 10.1086/498853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartegni L, Krainer AR. Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat Genet. 2002;30:377–384. doi: 10.1038/ng854. [DOI] [PubMed] [Google Scholar]
- Cartegni L, Krainer AR. Correction of disease-associated exon skipping by synthetic exon-specific activators. Nat Struct Biol. 2003;10:120–125. doi: 10.1038/nsb887. [DOI] [PubMed] [Google Scholar]
- Castle JC, Zhang C, Shah JK, Kulkarni AV, Kalsotra A, Cooper TA, Johnson JM. Expression of 24,426 human alternative splicing events and predicted cis regulation in 48 tissues and cell lines. Nat Genet. 2008;40:1416–1425. doi: 10.1038/ng.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Tian D, Ku L, Osterhout DJ, Feng Y. The selective RNA-binding protein quaking I (QKI) is necessary and sufficient for promoting oligodendroglia differentiation. J Biol Chem. 2007;282:23553–23560. doi: 10.1074/jbc.M702045200. [DOI] [PubMed] [Google Scholar]
- Chenard CA, Richard S. New implications for the QUAKING RNA binding protein in human disease. J Neurosci Res. 2008;86:233–242. doi: 10.1002/jnr.21485. [DOI] [PubMed] [Google Scholar]
- Cho DH, Thienes CP, Mahoney SE, Analau E, Filippova GN, Tapscott SJ. Antisense transcription and heterochromatin at the DM1 CTG repeats are constrained by CTCF. Mol Cell. 2005;20:483–489. doi: 10.1016/j.molcel.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Clark TA, Sugnet CW, Ares M., Jr. Genomewide analysis of mRNA processing in yeast using splicing-specific microarrays. Science. 2002;296:907–910. doi: 10.1126/science.1069415. [DOI] [PubMed] [Google Scholar]
- Danos O. AAV vectors for RNA-based modulation of gene expression. Gene Ther. 2008;15:864–869. doi: 10.1038/gt.2008.69. [DOI] [PubMed] [Google Scholar]
- Daoud H, Valdmanis PN, Kabashi E, Dion P, Dupre N, Camu W, Meininger V, Rouleau GA. Contribution of TARDBP mutations to sporadic amyotrophic lateral sclerosis. J Med Genet. 2008 doi: 10.1136/jmg.2008.062463. [DOI] [PubMed] [Google Scholar]
- David CJ, Manley JL. The search for alternative splicing regulators: new approaches offer a path to a splicing code. Genes Dev. 2008;22:279–285. doi: 10.1101/gad.1643108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haro M, Al-Ramahi I, De Gouyon B, Ukani L, Rosa A, Faustino NA, Ashizawa T, Cooper TA, Botas J. MBNL1 and CUGBP1 modify expanded CUG-induced toxicity in a Drosophila model of myotonic dystrophy type 1. Hum Mol Genet. 2006;15:2138–2145. doi: 10.1093/hmg/ddl137. [DOI] [PubMed] [Google Scholar]
- Denti MA, Rosa A, D’Antona G, Sthandier O, De Angelis FG, Nicoletti C, Allocca M, Pansarasa O, Parente V, Musaro A, et al. Body-wide gene therapy of Duchenne muscular dystrophy in the mdx mouse model. Proc Natl Acad Sci U S A. 2006;103:3758–3763. doi: 10.1073/pnas.0508917103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disset A, Bourgeois CF, Benmalek N, Claustres M, Stevenin J, Tuffery-Giraud S. An exon skipping-associated nonsense mutation in the dystrophin gene uncovers a complex interplay between multiple antagonistic splicing elements. Hum Mol Genet. 2006;15:999–1013. doi: 10.1093/hmg/ddl015. [DOI] [PubMed] [Google Scholar]
- Dominski Z, Kole R. Restoration of correct splicing in thalassemic premRNA by antisense oligonucleotides. Proc Natl Acad Sci U S A. 1993;90:8673–8677. doi: 10.1073/pnas.90.18.8673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G, Kim VN, Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- Dreyfuss G, Matunis MJ, Pinol-Roma S, Burd CG. hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjarn M, Hansen HF, Berger U, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- Friedman KJ, Kole J, Cohn JA, Knowles MR, Silverman LM, Kole R. Correction of aberrant splicing of the cystic fibrosis transmembrane conductance regulator (CFTR) gene by antisense oligonucleotides. J Biol Chem. 1999;274:36193–36199. doi: 10.1074/jbc.274.51.36193. [DOI] [PubMed] [Google Scholar]
- Frischmeyer PA, Dietz HC. Nonsense-mediated mRNA decay in health and disease. Hum Mol Genet. 1999;8:1893–1900. doi: 10.1093/hmg/8.10.1893. [DOI] [PubMed] [Google Scholar]
- Fukuhara T, Hosoya T, Shimizu S, Sumi K, Oshiro T, Yoshinaka Y, Suzuki M, Yamamoto N, Herzenberg LA, Herzenberg LA, Hagiwara M. Utilization of host SR protein kinases and RNA-splicing machinery during viral replication. Proc Natl Acad Sci U S A. 2006;103:11329–11333. doi: 10.1073/pnas.0604616103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabanella F, Butchbach ME, Saieva L, Carissimi C, Burghes AH, Pellizzoni L. Ribonucleoprotein assembly defects correlate with spinal muscular atrophy severity and preferentially affect a subset of spliceosomal snRNPs. PLoS ONE. 2007;2:e921. doi: 10.1371/journal.pone.0000921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabut M, Chaudhry S, Blencowe BJ. SnapShot: The splicing regulatory machinery. Cell. 2008;133:192, e191. doi: 10.1016/j.cell.2008.03.010. [DOI] [PubMed] [Google Scholar]
- Ghigna C, Giordano S, Shen H, Benvenuto F, Castiglioni F, Comoglio PM, Green MR, Riva S, Biamonti G. Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene. Mol Cell. 2005;20:881–890. doi: 10.1016/j.molcel.2005.10.026. [DOI] [PubMed] [Google Scholar]
- Glisovic T, Bachorik JL, Yong J, Dreyfuss G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008;582:1977–1986. doi: 10.1016/j.febslet.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyenvalle A, Vulin A, Fougerousse F, Leturcq F, Kaplan JC, Garcia L, Danos O. Rescue of dystrophic muscle through U7 snRNA-mediated exon skipping. Science. 2004;306:1796–1799. doi: 10.1126/science.1104297. [DOI] [PubMed] [Google Scholar]
- Graveley BR. The haplo-spliceo-transcriptome: common variations in alternative splicing in the human population. Trends Genet. 2008;24:5–7. doi: 10.1016/j.tig.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm D, Kay MA. Therapeutic application of RNAi: is mRNA targeting finally ready for prime time? J Clin Invest. 2007;117:3633–3641. doi: 10.1172/JCI34129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosso AR, Martins S, Carmo-Fonseca M. The emerging role of splicing factors in cancer. EMBO Rep. 2008;9:1087–1093. doi: 10.1038/embor.2008.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagerman PJ, Hagerman RJ. The fragile-X premutation: a maturing perspective. Am J Hum Genet. 2004;74:805–816. doi: 10.1086/386296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hainrichson M, Nudelman I, Baasov T. Designer aminoglycosides: the race to develop improved antibiotics and compounds for the treatment of human genetic diseases. Org Biomol Chem. 2008;6:227–239. doi: 10.1039/b712690p. [DOI] [PubMed] [Google Scholar]
- Henke E, Perk J, Vider J, de Candia P, Chin Y, Solit DB, Ponomarev V, Cartegni L, Manova K, Rosen N, Benezra R. Peptide-conjugated antisense oligonucleotides for targeted inhibition of a transcriptional regulator in vivo. Nat Biotechnol. 2008;26:91–100. doi: 10.1038/nbt1366. [DOI] [PubMed] [Google Scholar]
- Holmlund JT. Applying antisense technology: Affinitak and other antisense oligonucleotides in clinical development. Ann N Y Acad Sci. 2003;1002:244–251. doi: 10.1196/annals.1281.027. [DOI] [PubMed] [Google Scholar]
- Iwahashi CK, Yasui DH, An HJ, Greco CM, Tassone F, Nannen K, Babineau B, Lebrilla CB, Hagerman RJ, Hagerman PJ. Protein composition of the intranuclear inclusions of FXTAS. Brain. 2006;129:256–271. doi: 10.1093/brain/awh650. [DOI] [PubMed] [Google Scholar]
- Jin P, Duan R, Qurashi A, Qin Y, Tian D, Rosser TC, Liu H, Feng Y, Warren ST. Pur alpha binds to rCGG repeats and modulates repeat-mediated neurodegeneration in a Drosophila model of fragile X tremor/ataxia syndrome. Neuron. 2007;55:556–564. doi: 10.1016/j.neuron.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabashi E, Valdmanis PN, Dion P, Spiegelman D, McConkey BJ, Vande Velde C, Bouchard JP, Lacomblez L, Pochigaeva K, Salachas F, et al. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat Genet. 2008;40:572–574. doi: 10.1038/ng.132. [DOI] [PubMed] [Google Scholar]
- Kalsotra A, Xiao X, Ward AJ, Castle JC, Johnson JM, Burge CB, Cooper TA. A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc Natl Acad Sci U S A. 2008 doi: 10.1073/pnas.0809045105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanadia RN, Johnstone KA, Mankodi A, Lungu C, Thornton CA, Esson D, Timmers AM, Hauswirth WW, Swanson MS. A muscleblind knockout model for myotonic dystrophy. Science. 2003;302:1978–1980. doi: 10.1126/science.1088583. [DOI] [PubMed] [Google Scholar]
- Kanadia RN, Shin J, Yuan Y, Beattie SG, Wheeler TM, Thornton CA, Swanson MS. Reversal of RNA missplicing and myotonia after muscleblind overexpression in a mouse poly(CUG) model for myotonic dystrophy. Proc Natl Acad Sci U S A. 2006;103:11748–11753. doi: 10.1073/pnas.0604970103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karkare S, Bhatnagar D. Promising nucleic acid analogs and mimics: characteristic features and applications of PNA, LNA, and morpholino. Appl Microbiol Biotechnol. 2006;71:575–586. doi: 10.1007/s00253-006-0434-2. [DOI] [PubMed] [Google Scholar]
- Karni R, de Stanchina E, Lowe SW, Sinha R, Mu D, Krainer AR. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat Struct Mol Biol. 2007;14:185–193. doi: 10.1038/nsmb1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karni R, Hippo Y, Lowe SW, Krainer AR. The splicing-factor oncoprotein SF2/ASF activates mTORC1. Proc Natl Acad Sci U S A. 2008;105:15323–15327. doi: 10.1073/pnas.0801376105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashima T, Manley JL. A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nat Genet. 2003;34:460–463. doi: 10.1038/ng1207. [DOI] [PubMed] [Google Scholar]
- Kashima T, Rao N, David CJ, Manley JL. hnRNP A1 functions with specificity in repression of SMN2 exon 7 splicing. Hum Mol Genet. 2007;16:3149–3159. doi: 10.1093/hmg/ddm276. [DOI] [PubMed] [Google Scholar]
- Kerem E, Hirawat S, Armoni S, Yaakov Y, Shoseyov D, Cohen M, Nissim-Rafinia M, Blau H, Rivlin J, Aviram M, et al. Effectiveness of PTC124 treatment of cystic fibrosis caused by nonsense mutations: a prospective phase II trial. Lancet. 2008;372:719–727. doi: 10.1016/S0140-6736(08)61168-X. [DOI] [PubMed] [Google Scholar]
- Kim E, Goren A, Ast G. Insights into the connection between cancer and alternative splicing. Trends Genet. 2008;24:7–10. doi: 10.1016/j.tig.2007.10.001. [DOI] [PubMed] [Google Scholar]
- Kleinman ME, Yamada K, Takeda A, Chandrasekaran V, Nozaki M, Baffi JZ, Albuquerque RJ, Yamasaki S, Itaya M, Pan Y, et al. Sequence- and target-independent angiogenesis suppression by siRNA via TLR3. Nature. 2008;452:591–597. doi: 10.1038/nature06765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klement IA, Skinner PJ, Kaytor MD, Yi H, Hersch SM, Clark HB, Zoghbi HY, Orr HT. Ataxin-1 nuclear localization and aggregation: role in polyglutamine-induced disease in SCA1 transgenic mice. Cell. 1998;95:41–53. doi: 10.1016/s0092-8674(00)81781-x. [DOI] [PubMed] [Google Scholar]
- Krol J, Fiszer A, Mykowska A, Sobczak K, de Mezer M, Krzyzosiak WJ. Ribonuclease dicer cleaves triplet repeat hairpins into shorter repeats that silence specific targets. Mol Cell. 2007;25:575–586. doi: 10.1016/j.molcel.2007.01.031. [DOI] [PubMed] [Google Scholar]
- Kuyumcu-Martinez NM, Wang GS, Cooper TA. Increased steady-state levels of CUGBP1 in myotonic dystrophy 1 are due to PKC-mediated hyperphosphorylation. Mol Cell. 2007;28:68–78. doi: 10.1016/j.molcel.2007.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan T, Benovoy D, Dias C, Gurd S, Provencher C, Beaulieu P, Hudson TJ, Sladek R, Majewski J. Genome-wide analysis of transcript isoform variation in humans. Nat Genet. 2008;40:225–231. doi: 10.1038/ng.2007.57. [DOI] [PubMed] [Google Scholar]
- Ladd PD, Smith LE, Rabaia NA, Moore JM, Georges SA, Hansen RS, Hagerman RJ, Tassone F, Tapscott SJ, Filippova GN. An antisense transcript spanning the CGG repeat region of FMR1 is upregulated in premutation carriers but silenced in full mutation individuals. Hum Mol Genet. 2007;16:3174–3187. doi: 10.1093/hmg/ddm293. [DOI] [PubMed] [Google Scholar]
- Lauriat TL, Shiue L, Haroutunian V, Verbitsky M, Ares M, Jr., Ospina L, McInnes LA. Developmental expression profile of quaking, a candidate gene for schizophrenia, and its target genes in human prefrontal cortex and hippocampus shows regional specificity. J Neurosci Res. 2008;86:785–796. doi: 10.1002/jnr.21534. [DOI] [PubMed] [Google Scholar]
- Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- Li LB, Yu Z, Teng X, Bonini NM. RNA toxicity is a component of ataxin-3 degeneration in Drosophila. Nature. 2008;453:1107–1111. doi: 10.1038/nature06909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J, Hao T, Shaw C, Patel AJ, Szabo G, Rual JF, Fisk CJ, Li N, Smolyar A, Hill DE, et al. A protein-protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell. 2006;125:801–814. doi: 10.1016/j.cell.2006.03.032. [DOI] [PubMed] [Google Scholar]
- Lim LP, Burge CB. A computational analysis of sequence features involved in recognition of short introns. Proc Natl Acad Sci U S A. 2001;98:11193–11198. doi: 10.1073/pnas.201407298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Miller JW, Mankodi A, Kanadia RN, Yuan Y, Moxley RT, Swanson MS, Thornton CA. Failure of MBNL1-dependent post-natal splicing transitions in myotonic dystrophy. Hum Mol Genet. 2006;15:2087–2097. doi: 10.1093/hmg/ddl132. [DOI] [PubMed] [Google Scholar]
- Liquori CL, Ricker K, Moseley ML, Jacobsen JF, Kress W, Naylor SL, Day JW, Ranum LP. Myotonic dystrophy type 2 caused by a CCTG expansion in intron 1 of ZNF9. Science. 2001;293:864–867. doi: 10.1126/science.1062125. [DOI] [PubMed] [Google Scholar]
- Liu F, Gong CX. Tau exon 10 alternative splicing and tauopathies. Mol Neurodegener. 2008;3:8. doi: 10.1186/1750-1326-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Bigas N, Audit B, Ouzounis C, Parra G, Guigo R. Are splicing mutations the most frequent cause of hereditary disease? FEBS Lett. 2005;579:1900–1903. doi: 10.1016/j.febslet.2005.02.047. [DOI] [PubMed] [Google Scholar]
- Lueck JD, Mankodi A, Swanson MS, Thornton CA, Dirksen RT. Muscle chloride channel dysfunction in two mouse models of myotonic dystrophy. J Gen Physiol. 2007;129:79–94. doi: 10.1085/jgp.200609635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukong KE, Chang KW, Khandjian EW, Richard S. RNA-binding proteins in human genetic disease. Trends Genet. 2008;24:416–425. doi: 10.1016/j.tig.2008.05.004. [DOI] [PubMed] [Google Scholar]
- Madocsai C, Lim SR, Geib T, Lam BJ, Hertel KJ. Correction of SMN2 Pre-mRNA splicing by antisense U7 small nuclear RNAs. Mol Ther. 2005;12:1013–1022. doi: 10.1016/j.ymthe.2005.08.022. [DOI] [PubMed] [Google Scholar]
- Maquat LE. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol. 2004;5:89–99. doi: 10.1038/nrm1310. [DOI] [PubMed] [Google Scholar]
- Martinez-Contreras R, Cloutier P, Shkreta L, Fisette JF, Revil T, Chabot B. hnRNP proteins and splicing control. Adv Exp Med Biol. 2007;623:123–147. doi: 10.1007/978-0-387-77374-2_8. [DOI] [PubMed] [Google Scholar]
- Meyer K, Marquis J, Trub J, Nlend R. Nlend, Verp S, Ruepp MD, Imboden H, Barde I, Trono D, Schumperli D. Rescue of a severe mouse model for Spinal Muscular Atrophy by U7 snRNA-mediated splicing modulation. Hum Mol Genet. 2008 doi: 10.1093/hmg/ddn382. [DOI] [PubMed] [Google Scholar]
- Mordes D, Luo X, Kar A, Kuo D, Xu L, Fushimi K, Yu G, Sternberg P, Jr., Wu JY. Pre-mRNA splicing and retinitis pigmentosa. Mol Vis. 2006;12:1259–1271. [PMC free article] [PubMed] [Google Scholar]
- Mordes D, Yuan L, Xu L, Kawada M, Molday RS, Wu JY. Identification of photoreceptor genes affected by PRPF31 mutations associated with autosomal dominant retinitis pigmentosa. Neurobiol Dis. 2007;26:291–300. doi: 10.1016/j.nbd.2006.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muntoni F, Wells D. Genetic treatments in muscular dystrophies. Curr Opin Neurol. 2007;20:590–594. doi: 10.1097/WCO.0b013e3282efc157. [DOI] [PubMed] [Google Scholar]
- Muraki M, Ohkawara B, Hosoya T, Onogi H, Koizumi J, Koizumi T, Sumi K, Yomoda J, Murray MV, Kimura H, et al. Manipulation of alternative splicing by a newly developed inhibitor of Clks. J Biol Chem. 2004;279:24246–24254. doi: 10.1074/jbc.M314298200. [DOI] [PubMed] [Google Scholar]
- Nembaware V, Wolfe KH, Bettoni F, Kelso J, Seoighe C. Allele-specific transcript isoforms in human. FEBS Lett. 2004;577:233–238. doi: 10.1016/j.febslet.2004.10.018. [DOI] [PubMed] [Google Scholar]
- Nemes JP, Benzow KA, Moseley ML, Ranum LP, Koob MD. The SCA8 transcript is an antisense RNA to a brain-specific transcript encoding a novel actin-binding protein (KLHL1) Hum Mol Genet. 2000;9:1543–1551. doi: 10.1093/hmg/9.10.1543. [DOI] [PubMed] [Google Scholar]
- Neuenkirchen N, Chari A, Fischer U. Deciphering the assembly pathway of Sm-class U snRNPs. FEBS Lett. 2008;582:1997–2003. doi: 10.1016/j.febslet.2008.03.009. [DOI] [PubMed] [Google Scholar]
- Nezu Y, Kino Y, Sasagawa N, Nishino I, Ishiura S. Expression of MBNL and CELF mRNA transcripts in muscles with myotonic dystrophy. Neuromuscul Disord. 2007;17:306–312. doi: 10.1016/j.nmd.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Nilsen TW. The spliceosome: the most complex macromolecular machine in the cell? Bioessays. 2003;25:1147–1149. doi: 10.1002/bies.10394. [DOI] [PubMed] [Google Scholar]
- Nowak KJ, Davies KE. Duchenne muscular dystrophy and dystrophin: pathogenesis and opportunities for treatment. EMBO Rep. 2004;5:872–876. doi: 10.1038/sj.embor.7400221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien K, Matlin AJ, Lowell AM, Moore MJ. The biflavonoid isoginkgetin is a general inhibitor of Pre-mRNA splicing. J Biol Chem. 2008;283:33147–33154. doi: 10.1074/jbc.M805556200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke JR, Swanson MS. Mechanisms of RNA-mediated disease. J Biol Chem. 2008 doi: 10.1074/jbc.R800025200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu Rev Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- Pagani F, Raponi M, Baralle FE. Synonymous mutations in CFTR exon 12 affect splicing and are not neutral in evolution. Proc Natl Acad Sci U S A. 2005;102:6368–6372. doi: 10.1073/pnas.0502288102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- Pappas TC, Bader AG, Andruss BF, Brown D, Ford LP. Applying small RNA molecules to the directed treatment of human diseases: realizing the potential. Expert Opin Ther Targets. 2008;12:115–127. doi: 10.1517/14728222.12.1.115. [DOI] [PubMed] [Google Scholar]
- Park JW, Parisky K, Celotto AM, Reenan RA, Graveley BR. Identification of alternative splicing regulators by RNA interference in Drosophila. Proc Natl Acad Sci U S A. 2004;101:15974–15979. doi: 10.1073/pnas.0407004101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual M, Vicente M, Monferrer L, Artero R. The Muscleblind family of proteins: an emerging class of regulators of developmentally programmed alternative splicing. Differentiation. 2006;74:65–80. doi: 10.1111/j.1432-0436.2006.00060.x. [DOI] [PubMed] [Google Scholar]
- Patel AA, Steitz JA. Splicing double: insights from the second spliceosome. Nat Rev Mol Cell Biol. 2003;4:960–970. doi: 10.1038/nrm1259. [DOI] [PubMed] [Google Scholar]
- Pinol-Roma S, Dreyfuss G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature. 1992;355:730–732. doi: 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- Pleiss JA, Whitworth GB, Bergkessel M, Guthrie C. Transcript specificity in yeast pre-mRNA splicing revealed by mutations in core spliceosomal components. PLoS Biol. 2007;5:e90. doi: 10.1371/journal.pbio.0050090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich SJ, Fosnot J, Kuroki A, Tang W, Yang X, Maguire AM, Bennett J, Tolentino MJ. Small interfering RNA (siRNA) targeting VEGF effectively inhibits ocular neovascularization in a mouse model. Mol Vis. 2003;9:210–216. [PubMed] [Google Scholar]
- Rivolta C, McGee TL, Frio T. Rio, Jensen RV, Berson EL, Dryja TP. Variation in retinitis pigmentosa-11 (PRPF31 or RP11) gene expression between symptomatic and asymptomatic patients with dominant RP11 mutations. Hum Mutat. 2006;27:644–653. doi: 10.1002/humu.20325. [DOI] [PubMed] [Google Scholar]
- Roca X, Olson AJ, Rao AR, Enerly E, Kristensen VN, Borresen-Dale AL, Andresen BS, Krainer AR, Sachidanandam R. Features of 5′-splice-site efficiency derived from disease-causing mutations and comparative genomics. Genome Res. 2008;18:77–87. doi: 10.1101/gr.6859308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi JJ. Expression strategies for short hairpin RNA interference triggers. Hum Gene Ther. 2008;19:313–317. doi: 10.1089/hum.2008.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossoll W, Jablonka S, Andreassi C, Kroning AK, Karle K, Monani UR, Sendtner M. Smn, the spinal muscular atrophy-determining gene product, modulates axon growth and localization of beta-actin mRNA in growth cones of motoneurons. J Cell Biol. 2003;163:801–812. doi: 10.1083/jcb.200304128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudnicki DD, Holmes SE, Lin MW, Thornton CA, Ross CA, Margolis RL. Huntington’s disease--like 2 is associated with CUG repeat-containing RNA foci. Ann Neurol. 2007;61:272–282. doi: 10.1002/ana.21081. [DOI] [PubMed] [Google Scholar]
- Rutherford NJ, Zhang YJ, Baker M, Gass JM, Finch NA, Xu YF, Stewart H, Kelley BJ, Kuntz K, Crook RJ, et al. Novel mutations in TARDBP (TDP-43) in patients with familial amyotrophic lateral sclerosis. PLoS Genet. 2008;4:e1000193. doi: 10.1371/journal.pgen.1000193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sazani P, Kole R. Therapeutic potential of antisense oligonucleotides as modulators of alternative splicing. J Clin Invest. 2003;112:481–486. doi: 10.1172/JCI19547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaffidi P, Misteli T. Reversal of the cellular phenotype in the premature aging disease Hutchinson-Gilford progeria syndrome. Nat Med. 2005;11:440–445. doi: 10.1038/nm1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaub M, Keller W. RNA editing by adenosine deaminases generates RNA and protein diversity. Biochimie. 2002;84:791–803. doi: 10.1016/s0300-9084(02)01446-3. [DOI] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Samul R, Silva RL, Akiyama H, Liu H, Saishin Y, Hackett SF, Zinnen S, Kossen K, Fosnaugh K, et al. Suppression of ocular neovascularization with siRNA targeting VEGF receptor 1. Gene Ther. 2006;13:225–234. doi: 10.1038/sj.gt.3302641. [DOI] [PubMed] [Google Scholar]
- Singh RN. Evolving concepts on human SMN pre-mRNA splicing. RNA Biol. 2007;4:7–10. doi: 10.4161/rna.4.1.4535. [DOI] [PubMed] [Google Scholar]
- Skordis LA, Dunckley MG, Yue B, Eperon IC, Muntoni F. Bifunctional antisense oligonucleotides provide a trans-acting splicing enhancer that stimulates SMN2 gene expression in patient fibroblasts. Proc Natl Acad Sci U S A. 2003;100:4114–4119. doi: 10.1073/pnas.0633863100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CW, Valcarcel J. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem Sci. 2000;25:381–388. doi: 10.1016/s0968-0004(00)01604-2. [DOI] [PubMed] [Google Scholar]
- Smith RA, Miller TM, Yamanaka K, Monia BP, Condon TP, Hung G, Lobsiger CS, Ward CM, McAlonis-Downes M, Wei H, et al. Antisense oligonucleotide therapy for neurodegenerative disease. J Clin Invest. 2006;116:2290–2296. doi: 10.1172/JCI25424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofola OA, Jin P, Qin Y, Duan R, Liu H, de Haro M, Nelson DL, Botas J. RNA-binding proteins hnRNP A2/B1 and CUGBP1 suppress fragile X CGG premutation repeat-induced neurodegeneration in a Drosophila model of FXTAS. Neuron. 2007;55:565–571. doi: 10.1016/j.neuron.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soret J, Bakkour N, Maire S, Durand S, Zekri L, Gabut M, Fic W, Divita G, Rivalle C, Dauzonne D, et al. Selective modification of alternative splicing by indole derivatives that target serine-arginine-rich protein splicing factors. Proc Natl Acad Sci U S A. 2005;102:8764–8769. doi: 10.1073/pnas.0409829102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srebrow A, Kornblihtt AR. The connection between splicing and cancer. J Cell Sci. 2006;119:2635–2641. doi: 10.1242/jcs.03053. [DOI] [PubMed] [Google Scholar]
- Sreedharan J, Blair IP, Tripathi VB, Hu X, Vance C, Rogelj B, Ackerley S, Durnall JC, Williams KL, Buratti E, et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley JP, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- Stoilov P, Lin CH, Damoiseaux R, Nikolic J, Black DL. A high-throughput screening strategy identifies cardiotonic steroids as alternative splicing modulators. Proc Natl Acad Sci U S A. 2008;105:11218–11223. doi: 10.1073/pnas.0801661105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot K, Davies KE. Spinal muscular atrophy. Semin Neurol. 2001;21:189–197. doi: 10.1055/s-2001-15264. [DOI] [PubMed] [Google Scholar]
- Tange TO, Nott A, Moore MJ. The ever-increasing complexities of the exon junction complex. Curr Opin Cell Biol. 2004;16:279–284. doi: 10.1016/j.ceb.2004.03.012. [DOI] [PubMed] [Google Scholar]
- Timchenko LT, Miller JW, Timchenko NA, DeVore DR, Datar KV, Lin L, Roberts R, Caskey CT, Swanson MS. Identification of a (CUG)n triplet repeat RNA-binding protein and its expression in myotonic dystrophy. Nucleic Acids Res. 1996;24:4407–4414. doi: 10.1093/nar/24.22.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J. Ribonucleoprotein complexes in neurologic diseases. Curr Opin Neurobiol. 2008;18:516–523. doi: 10.1016/j.conb.2008.09.018. [DOI] [PubMed] [Google Scholar]
- Ule J, Jensen K, Mele A, Darnell RB. CLIP: a method for identifying protein-RNA interaction sites in living cells. Methods. 2005;37:376–386. doi: 10.1016/j.ymeth.2005.07.018. [DOI] [PubMed] [Google Scholar]
- Underwood JG, Boutz PL, Dougherty JD, Stoilov P, Black DL. Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol Cell Biol. 2005;25:10005–10016. doi: 10.1128/MCB.25.22.10005-10016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- Venables JP, Koh CS, Froehlich U, Lapointe E, Couture S, Inkel L, Bramard A, Paquet ER, Watier V, Durand M, et al. Multiple and specific mRNA processing targets for the major human hnRNP proteins. Mol Cell Biol. 2008;28:6033–6043. doi: 10.1128/MCB.00726-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L, Battle DJ, Yong J, Gubitz AK, Kolb SJ, Wang J, Dreyfuss G. The survival of motor neurons protein determines the capacity for snRNP assembly: biochemical deficiency in spinal muscular atrophy. Mol Cell Biol. 2005;25:5543–5551. doi: 10.1128/MCB.25.13.5543-5551.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GS, Cooper TA. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat Rev Genet. 2007;8:749–761. doi: 10.1038/nrg2164. [DOI] [PubMed] [Google Scholar]
- Wang GS, Kearney DL, De Biasi M, Taffet G, Cooper TA. Elevation of RNA-binding protein CUGBP1 is an early event in an inducible heart-specific mouse model of myotonic dystrophy. J Clin Invest. 2007;117:2802–2811. doi: 10.1172/JCI32308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Pegoraro E, Menegazzo E, Gennarelli M, Hoop RC, Angelini C, Hoffman EP. Myotonic dystrophy: evidence for a possible dominant-negative RNA mutation. Hum Mol Genet. 1995;4:599–606. doi: 10.1093/hmg/4.4.599. [DOI] [PubMed] [Google Scholar]
- Wang Z, Burge CB. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. Rna. 2008;14:802–813. doi: 10.1261/rna.876308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warf MB, Berglund JA. MBNL binds similar RNA structures in the CUG repeats of myotonic dystrophy and its pre-mRNA substrate cardiac troponin T. Rna. 2007;13:2238–2251. doi: 10.1261/rna.610607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch EM, Barton ER, Zhuo J, Tomizawa Y, Friesen WJ, Trifillis P, Paushkin S, Patel M, Trotta CR, Hwang S, et al. PTC124 targets genetic disorders caused by nonsense mutations. Nature. 2007;447:87–91. doi: 10.1038/nature05756. [DOI] [PubMed] [Google Scholar]
- Wheeler TM, Lueck JD, Swanson MS, Dirksen RT, Thornton CA. Correction of ClC-1 splicing eliminates chloride channelopathy and myotonia in mouse models of myotonic dystrophy. J Clin Invest. 2007;117:3952–3957. doi: 10.1172/JCI33355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan J. First clinical data on RNAi. Drug Discov Today. 2005;10:1014–1015. doi: 10.1016/S1359-6446(05)03547-6. [DOI] [PubMed] [Google Scholar]
- Will CL, Luhrmann R. Spliceosomal UsnRNP biogenesis, structure and function. Curr Opin Cell Biol. 2001;13:290–301. doi: 10.1016/s0955-0674(00)00211-8. [DOI] [PubMed] [Google Scholar]
- Winkler C, Eggert C, Gradl D, Meister G, Giegerich M, Wedlich D, Laggerbauer B, Fischer U. Reduced U snRNP assembly causes motor axon degeneration in an animal model for spinal muscular atrophy. Genes Dev. 2005;19:2320–2330. doi: 10.1101/gad.342005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth B, Brichta L, Hahnen E. Spinal muscular atrophy: from gene to therapy. Semin Pediatr Neurol. 2006;13:121–131. doi: 10.1016/j.spen.2006.06.008. [DOI] [PubMed] [Google Scholar]
- Yong J, Wan L, Dreyfuss G. Why do cells need an assembly machine for RNA-protein complexes? Trends Cell Biol. 2004;14:226–232. doi: 10.1016/j.tcb.2004.03.010. [DOI] [PubMed] [Google Scholar]
- Yuan Y, Compton SA, Sobczak K, Stenberg MG, Thornton CA, Griffith JD, Swanson MS. Muscleblind-like 1 interacts with RNA hairpins in splicing target and pathogenic RNAs. Nucleic Acids Res. 2007;35:5474–5486. doi: 10.1093/nar/gkm601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Zhang Z, Castle J, Sun S, Johnson J, Krainer AR, Zhang MQ. Defining the regulatory network of the tissue-specific splicing factors Fox-1 and Fox-2. Genes Dev. 2008a;22:2550–2563. doi: 10.1101/gad.1703108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HL, Pan F, Hong D, Shenoy SM, Singer RH, Bassell GJ. Active transport of the survival motor neuron protein and the role of exon-7 in cytoplasmic localization. J Neurosci. 2003;23:6627–6637. doi: 10.1523/JNEUROSCI.23-16-06627.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Lotti F, Dittmar K, Younis I, Wan L, Kasim M, Dreyfuss G. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell. 2008b;133:585–600. doi: 10.1016/j.cell.2008.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zingman LV, Park S, Olson TM, Alekseev AE, Terzic A. Aminoglycoside-induced translational read-through in disease: overcoming nonsense mutations by pharmacogenetic therapy. Clin Pharmacol Ther. 2007;81:99–103. doi: 10.1038/sj.clpt.6100012. [DOI] [PubMed] [Google Scholar]