Abstract
Background
Rare mutations in more than 20 genes have been suggested to cause dilated cardiomyopathy (DCM), but explain only a small percentage of cases, mainly in familial forms. We hypothesized that more common variants may also play a role in increasing genetic susceptibility to DCM, similar to that observed in other common complex disorders.
Methods and Results
To test this hypothesis, we performed case-control analyses on all DNA polymorphic variation identified in a resequencing study of six candidate DCM genes (CSRP3, LDB3, MYH7, SCN5A, TCAP, and TNNT2) conducted in 289 unrelated white probands with DCM of unknown cause and 188 unrelated white controls. In univariate analyses, we identified associated common variants at LDB3 site 10779, LDB3 site 57877, MYH7 sites 16384 and 17404, and TCAP sites 140 and 1735. Multivariate analyses to examine the joint effects of multiple gene variants confirmed univariate results for MYH7 and TCAP and identified a block of 9 variants in MYH7 that was strongly associated with DCM.
Conclusions
Common variants in genes known to be causative of DCM may play a role in genetic susceptibility to DCM. Our results suggest that examination of common genetic variants may be warranted in future studies of DCM and other Mendelian-like disorders.
Keywords: dilated cardiomyopathy, genetics, multivariate analyses, resequencing
Dilated cardiomyopathy (DCM) is characterized by left ventricular enlargement and systolic dysfunction and is one of the most common causes of congestive heart failure. Patients with idiopathic DCM (IDC) are those with DCM in which all detectable causes (other than genetic) have been excluded. Patients with familial dilated cardiomyopathy (FDC) are those from families with ≥ 2 members meeting criteria for IDC. Efforts to identify genetic causes of DCM have focused primarily on FDC patients, and rare, mostly private mutations in more than 20 genes have been discovered. However, these mutations account for only a fraction of FDC cases (probably less than 25%).1
In our recent paper we selected six (CSRP3, LDB3, MYH7, SCN5A, TCAP, and TNNT2) of the previously identified FDC candidate genes for resequencing based upon their estimated mutation frequency and likelihood to show disease-associated variation.2 These genes included muscle LIM protein (CSRP3), LIM binding domain 3 (LBD3), β-myosin heavy chain (MYH7), the cardiac sodium channel (SCN5A), titin-cap or telethonin (TCAP), and cardiac troponin T (TNNT2), which have a role in cardiac muscle contraction and have been previously reviewed3. We showed that rare non-synonymous (NS) mutations were present not only in those probands with FDC but also in some IDC patients, that is, those with apparently sporadic disease. This finding not only emphasizes that the underlying etiology of IDC can be genetic, but further suggests that in some cases, rare private mutations can be observed in FDC or IDC patients. The 31 NS mutations that we identified were found in 36 of the 313 DCM patients that were resequenced but in none of the 253 controls; 30 of these mutations were found in individuals of white European descent. Genetic cause, if present, of DCM in the remaining 277 patients remains unknown, but could be due to rare mutations in other known DCM genes, mutations in other undiscovered DCM genes, or alternatively, to the influence of more common variants.
The genetics of DCM is complicated due to observed locus and allelic heterogeneity. Focusing our investigation only on rare variants that are present in DCM cases but absent in controls (i.e., a standard resequencing approach) limits the identification of other variants that may be functionally important. We hypothesized that common variants play a role in increasing genetic susceptibility to DCM, similar to other complex disorders. In an effort to test this hypothesis, we evaluated all rare and common variants identified in the six genes, including intronic variants, synonymous coding variants, and those in the 5′ and 3′ UTR regions. While this method of examining common non-coding variants is a conventional approach for genetic studies of common, complex disorders (e.g., Type 2 Diabetes or Alzheimer’s disease), it has never been applied to dilated cardiomyopathy which has long been considered a Mendelian-like disorder.
Methods
The study sample consisted of 188 unrelated controls of white European descent from the Coriell database and 289 unrelated white, non-Hispanic IDC probands who have been previously described by Hershberger et al2. Written, informed consent was obtained from all subjects and the OSHU Institutional Review Board approved the study. Bi-directional sequencing of all introns, exons, 2 kb upstream and 2 kb downstream regions was performed in CSRP3, LDB3, MYH7, SCN5A, TCAP, and TNNT2 in all individuals by SeattleSNPs under contract to the NHLBI resequencing service2. The number and sites of variation within each gene are described in Table 1. The genes ranged in size from 2 to 40 exons. One truncation was found in MYH7, and, in TNNT2, a frameshift mutation and two sites with three alleles each were identified.
Table 1.
Characteristics of resequenced genes.
| Gene | Chromosome Location | Sequence length (bp) | No. of exons | Total no. of sites in the discovery panel* | Polymorphic sites in cases and controls† | Non-Synonymous sites | Possibly damaging†† | Synonymous sites | 5′ UTR | 3′ UTR | Truncation | Frame shift | Tri-allelic sites |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CSRP3 | 11p15.1 | 20,568 | 6 | 32 | 9 | 3 | 2 | 5 | 1 | 6 | |||
| LDB3 | 10q22.3–q23.2 | 66,835 | 13 | 86 | 19 | 13 | 6 | 14 | 0 | 20 | |||
| MYH7 | 14q12 | 23,332 | 40 | 131 | 33 | 15 | 9 | 29 | 3 | 2 | 1 | ||
| SCN5A | 3p21 | 102,759 | 28 | 160 | 30 | 27 | 14 | 25 | 1 | 30 | |||
| TCAP | 17q12 | 3,082 | 2 | 38 | 4 | 6 | 2 | 2 | 0 | 9 | |||
| TNNT2 | 1q32 | 19,483 | 16 | 77 | 24 | 7 | 5 | 5 | 0 | 2 | 1 | 2 |
Sites in this table that did not pass QC were not analyzed;
Identified by SeattleSNPs resequencing service in 24 HapMap samples previously analyzed and not part of our sample;
Polymorphic was defined as minor allele frequency (MAF) > 0.01;
Number of nonsynonymous sites predicted to be intolerant or possibly damaging using SIFT or Polyphen.
Only variants with genotype call rates ≥ 80% and individuals with call rates ≥ 80% (after removal of low-quality variants) were included in our analyses. In a subset of 3,352 variant genotypes called in both the resequencing and available HapMap databases, we observed 99.5% genotype concordance. Rare variants were defined as those with MAF ≤ 3% in the pooled sample of cases and controls post-quality control (QC). We tested for deviation from Hardy Weinberg equilibrium (HWE) in controls using the permutation version of the exact test with 10,000 replicates in PROC ALLELE, SAS/GENETICS, version 9.1.34. Inter-variant linkage disequilibrium was also calculated in controls using PROC ALLELE, SAS/GENETICS, version 9.1.3 under the assumption of HWE4.
We performed univariate case-control analyses using exact tests on all diallelic loci to determine whether particular genotypes were statistically over or under-represented in cases compared to controls. In order to obtain estimates of effect sizes, single-variant association analyses were performed with exact logistic regression in PROC LOGISTIC, SAS/STAT, version 9.2. A network Monte Carlo algorithm with 10,000 replicates was used to obtain exact odds ratio (OR) estimates, 95% confidence intervals (CIs), and p-values for the effect of an additional minor allele under an additive, single-variant model for the log odds of DCM, which is equivalent to an exact Cochran-Armitage trend test5. A median unbiased estimate of the OR was obtained in cases where the maximum likelihood estimate did not exist6. We conducted these association analyses both with and without 30 non-Hispanic white individuals with NS mutations previously reported in (Hershberger, RE.2008) that met QC criteria as defined above.
We modeled the joint contributions of multiple variants in each gene by applying the Combined Multivariate and Collapsing approach (CMC)7 to the current resequenced data. In our application of the CMC method, we collapsed rare variants (MAF≤0.03) into two binary indicators of the presence or absence of any minor alleles of a given type (either “non-synonymous, frameshift, and truncation” or “other”) in a gene. If genotypes were missing at some loci in a group, these indicators took on a value of 1 if a minor allele was found at any nonmissing locus and were missing if no minor allele was found at any nonmissing loci. Common variants were scored for the number of minor alleles. We then applied Hotelling’s T2 test, a multivariate extension of the common Student’s t-test, to compare the mean minor allele counts and binary indicators between cases and controls simultaneously across all common and rare sites in a given gene. The CMC method was applied to the entire dataset, including the 30 individuals with previously identified NS mutations. Hotelling’s T2 test was performed using PROC GLM, SAS/STAT, version 9.1.3.
Results
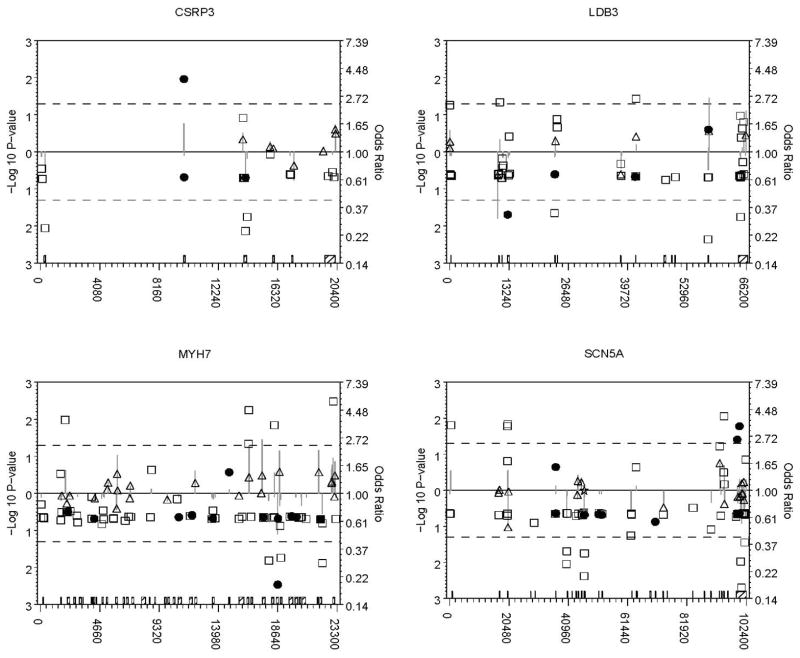
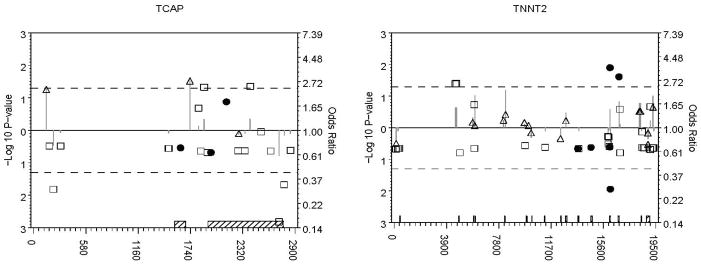
In total, 331 diallelic/polymorphic loci of the 467 loci in 466 of the 477 subjects that met QC criteria were analyzed. Figure 1 shows single-marker association results for all loci that passed QC. Loci with p-values < 0.05 in the primary or secondary univariate analysis are highlighted in Table 2. Full resequencing information for these variants is available in Supplementary Table 1. We identified a common variant at LDB3 site 10779 for which the minor allele was under-represented in cases and common variants at LDB3 site 57877, MYH7 sites 16384 and 17404, and TCAP sites 140 and 1735, for which the minor allele was over-represented in cases compared to controls. Effect sizes were similar after excluding 30 individuals with NS rare variants previously reported in Hershberger (2008), suggesting that the effects of the common variants found here are independent and not simply due to linkage disequilibrium with previously identified rare variants (Table 2).
Figure 1. P-values and odds ratios for all analyzed variants by gene.
Odds ratios are depicted on a log odds scale (right axis) by filled circles (rare non-synonymous, frameshift, or truncation variants), open squares (other rare variants), open stars (common non-synonymous, frameshift, or truncation variants), and open triangles (other common variants) relative to the corresponding variant’s local position in the gene’s sequence (horizontal axis). Grey vertical lines represent the associated −log 10 p-values (left axis). Points farther above or below the solid line at an odds ratio of 1.0 and a −log 10 p-value of 0 (P=1.0) indicate increasing effect sizes (odds ratios) and more significant p-values (lines). Odds ratios below the horizontal solid line indicate a lower minor allele frequency in cases, and those above, a higher minor allele frequency in cases. P-values are plotted in the same direction from the solid line as their corresponding odds ratio to indicate effect direction; dashed lines are included at P = 0.05. Exon boundaries (hatched boxes) spanning all sites within each gene are shown on the horizontal axis.
Table 2.
Single variant association analyses.
| Site information | Summary statistics | Caucasian DCM cases vs. Caucasian Controls | Caucasian DCM cases excluding those with 30 NS mutations vs. Caucasian Controls | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus | Location | Call rate | Major/Minor Allele* | N cases | N controls | MAF Cases | MAF Controls | P† | OR† | 95% CI† | P† | OR† | 95% CI† |
| LDB3 10779 | intron 1 | 0.89 | G/A | 257 | 159 | 0.26 | 0.34 | 0.02 | 0.66 | 0.41–0.93 | 0.02 | 0.66 | 0.40–0.94 |
| LDB3 57877‡ | intron 12 | 0.85 | T/- | 241 | 153 | 0.30 | 0.23 | 0.04 | 1.45 | 1.02–2.15 | 0.06 | 1.42 | 0.99–2.08 |
| MYH7 16384§ | intron 28 | 0.95 | T/A | 267 | 175 | 0.41 | 0.34 | 0.05 | 1.34 | 1.00–1.80 | 0.03 | 1.39 | 1.02–1.92 |
| MYH7 17404§ | intron 29 | 0.96 | A/G | 274 | 174 | 0.40 | 0.33 | 0.03 | 1.39 | 1.02–1.93 | 0.02 | 1.40 | 1.05–1.92 |
| TCAP 140§ | --- | 0.92 | A/G | 249 | 181 | 0.06 | 0.03 | 0.05 | 2.31 | 1.01–7.32 | 0.11 | 1.98 | 0.88–5.82 |
| TCAP 1735§ | intron 1 | 0.95 | C/T | 261 | 182 | 0.06 | 0.03 | 0.03 | 2.76 | 1.08–12.85 | 0.07 | 2.23 | 0.95–7.25 |
DCM = Dilated Cardiomyopathy; There was no evidence of deviation from HWE in controls at any locus (P > 0.23).
The minor allele was determined on the basis of its frequency in the pooled sample of cases and controls post-QC;
Exact logistic regression with an additive effect on the log odds for each additional copy of the minor allele.
LDB3 site 57877 has a deletion;
MYH7 sites 16384 and 17404 (r2 = 0.99) and TCAP sites 140 and 1735 (r2=1.00) were in linkage disequilibrium in controls.
Results from the CMC multivariate analyses are presented in Table 3. In the current dataset, the percentage of subjects with complete genotype data at all variants in a gene ranged from a minimum of 35% for MYH7 to a maximum of 85% for TCAP, hence a large number of observations had to be excluded from multivariate analyses based on Hotelling’s T2 in our dataset. The most interesting results were for MYH7 (p-value = 0.03, F17, 147 = 1.80) and TCAP (p-value= 0.09, F4, 392 = 2.04). The results for TCAP can be primarily explained by the strong correlations with the discriminant score (0.72) observed at sites 140 and 1735, which are in perfect LD (r2=1.0). In TCAP, these variants were also individually associated with DCM in univariate analyses (Table 2). Examination of the canonical correlation structure for MYH7 indicates a group of 9 variants with correlations between 0.25 and 0.36 with the discriminant score that contribute most to groupwise differences in this score within this gene. All of these variants were common (MAF > 0.13) and in HWE (P > 0.17) in controls. Univariate results and the LD structure of the 9 MYH7 variants showing moderate to high LD (0.27 ≤ r2≤ 1.0) are presented in Supplemental Figure 1. The results for 5 of the 9 variants, based on exact logistic regression, trend towards significance (0.03 ≤ p-value ≤ 0.09).
Table 3.
Multivariate association analyses using the CMC approach.
| Gene | N (% complete data) | P* |
|---|---|---|
| CSRP3 | 366 (78.5) | 0.52 |
| LDB3 | 232 (49.8) | 0.42 |
| MYH7 | 165 (35.4) | 0.03 |
| SCN5A | 175 (37.6) | 0.96 |
| TCAP | 397 (85.2) | 0.09 |
| TNNT2 | 275 (59.0) | 0.93 |
Rare variants (MAF≤0.03) were collapsed into two binary indicators of the presence or absence of any rare variants of a given type (either “non-synonymous, frameshift, and truncation” or “other”) in a gene. Common variants were scored for the number of minor alleles. Multivariate analysis was performed using Hotelling’s T2 test to compare the mean minor allele counts and binary indicators between cases and controls simultaneously across all common and rare sites in a given gene.
Discussion
We performed a comprehensive investigation of rare and common variants in six DCM candidate genes. DCM has long been considered a Mendelian-like disorder, and the standard gene identification approach has focused on the discovery of rare mutations. Our analyses demonstrated associations of common variants in LDB3, MYH7, and TCAP with DCM. Given that DCM is relatively uncommon, few replication datasets currently exist to allow validation of our results. However, future availability of consortiums or repositories of DCM sequencing data will allow more thorough evaluation outside the scope of the current dataset. Moreover, we did not apply within-gene multiple testing corrections in single-variant analyses because we were willing to risk higher Type I error in order to increase our limited power to detect univariate associations with rare variants in our moderately sized sample. It should be noted, however, that the multivariate analyses test the joint null hypothesis that the minor allele distributions are the same in both groups at all loci and thus control the Type I error at the nominal level for each gene. Taken together, our results show suggestive, although not overwhelming evidence of associations. Given the fact that we tested variants in a priori DCM candidate genes, and in light of our modest sample size, these results should not be dismissed with overly stringent significance thresholds.
The most significantly DCM-associated site 10779 is 118 bases upstream of exon 2 in LDB3, and is close to a sequence predicted to bind to MyoD. MyoD is an important skeletal muscle-specific transcription factor that, along with dystrophin, is missing in mutant Mdx:MyoD−/− mice. These mice display skeletal muscle wasting along with cardiomyopathy, although the mechanisms by which the latter occurs is unclear. MyoD is not expressed in the heart, hence any myocardial changes apparent in mdx:MyoD−/− mice could be hypothesized to be directly related to the level of skeletal muscle damage. The remaining variants in MYH7 and TCAP are predicted by the Alternative Splice Site Predictor (ASSP) software8 to potentially interfere with normal splicing functions.
One of the most interesting observations from this study comes from our attempt to analyze the joint contributions of multiple variants within genes. Compared to univariate analyses, methods that account for the joint effects of multiple variants either by collapsing genotypes across loci or use of multivariate analysis methods can improve power to detect association with disease phenotype. Such collapsing methods can be advantageous when rare, risk variants are combined across a gene to increase their frequency among cases. although for common variation gains in power may be offset by combining over- and under-represented alleles 7. They also have the advantage of controlling the Type I error rate over the entire gene at the nominal level without additional adjustments. Overall, the CMC results for MYH7 and TCAP appear to support the univariate findings for specific sites within each gene. We interpret our multivariate analysis results with caution however as our experience suggests that higher rates of missing genotypes, which can rapidly result in higher rates of incomplete observations as the number of loci in a gene increases, can in fact lead to reduction in power to detect multi-loci effects. It is thus important to realize that the gains in power from a multi-marker test requiring complete data will be offset to an unknown degree by the reduction in sample size due to incomplete observations. For example, it is difficult to determine whether the lack of association between LDB3 and DCM in multivariate analysis indicates false positives in univariate analyses or simply reflects a reduction in power caused by the loss of over half of the sample with incomplete observations. In some cases, this loss of power can be partly overcome by the existence of inter-variant correlation (i.e., stronger patterns of LD). For example, almost 65% of observations are lost when we attempt to analyze multiple MYH7 variants jointly. However, several of the analyzed variants (N=9) have strong canonical correlations which explain the overall multi-loci association with DCM, and these variants are in moderate to high LD (Supplemental Figure 1). Although the univariate results for these loci do not achieve the significance threshold of 0.05, many of these variants have univariate results which trend towards significance. The combined results from both univariate and multivariate analyses suggest that one or more of this cluster of common variants within MYH7 may be worthy of further investigation.
To our knowledge this is the first example in which common variants in genes known to be causative of DCM have also been found to be associated with DCM. The functional relevance of these common variants is yet unknown, however it is possible that they independently, or jointly with other common or rare variants, increase DCM susceptibility. These data taken together suggest that an analysis strategy for genetic investigations of dilated cardiomyopathy should aim to examine common variants in addition to standard rare variant analyses.
Supplementary Material
Acknowledgments
We thank the many families and referring physicians for their participation in the OHSU Familial Dilated Cardiomyopathy Research Program, without whom these studies would not have been possible. We thank Lili Tewes for editorial assistance.
Funding Sources
This work is supported by NIH awards R01-HL58626 and 5 M01 RR000334 and 1RC2HG005605-01. Resequencing services were provided by the University of Washington, Department of Genome Sciences, under U.S. Federal Government contract number N01-HV-48194 from the National Heart, Lung, and Blood Institute.
Footnotes
Disclosures
No conflicts to disclose
References
- 1.Morita H, Seidman J, Seidman CE. Genetic causes of human heart failure. J Clin Invest. 2005;115:518–526. doi: 10.1172/JCI200524351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hershberger RE, Parks SB, Kushner JD, Li D, Ludwigsen S, Jakobs P, Nauman D, Burgess D, Partain J, Litt M. Coding sequence mutations identified in MYH7, TNNT2, SCN5A, CSRP3, LBD3, and TCAP from 313 patients with familial or idiopathic dilated cardiomyopathy. Clinical and Translational Science Journal. 2008;1:21–26. doi: 10.1111/j.1752-8062.2008.00017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burkett EL, Hershberher RE. Clinical and genetic issues in familial dilated cardiomyopathy. J Am Coll Cadriol. 2005;45 (7):422–0. doi: 10.1016/j.jacc.2004.11.066. [DOI] [PubMed] [Google Scholar]
- 4.SAS Institute Inc. SAS/Genetics TM 9.1.3 User’s Guide. SAS Institute Inc; Cary, NC: SAS/Genetics. [Google Scholar]
- 5.Mehta CR, Patel N, Senchaudhuri P. Efficient Monte Carlo Methods for Conditional Logistic Regression. Journal of the American Statistical Association. 2000;95:99–108. [Google Scholar]
- 6.Hirji KF, Tsiatis AA, Mehta CR. Median unbiased estimation for binary data. The American Statistician. 2006;43:7–11. 1989. [Google Scholar]
- 7.Li B, Leal SM. Methods for detecting associations with rare variants for common diseases: Application to analysis of sequence data. Am J Hum Genet. 2008;83:311–321. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang M, Marin A. Characterization and prediction of alternative splice sites. Gene. 2006;366:219–227. doi: 10.1016/j.gene.2005.07.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.