Abstract
Pyrroline-5-carboxylate synthase (P5CS) is a bifunctional enzyme that exhibits glutamate kinase (GK) and γ-glutamyl phosphate reductase (GPR) activities. The enzyme is highly relevant in humans because it belongs to a combined route for the interconversion of glutamate, ornithine and proline. The deficiency of P5CS activity in humans is associated with a rare, inherited metabolic disease. It is well established that some bacteria and plants accumulate proline in response to osmotic stress. The alignment of P5CSs from different species and analysis of the solved structures of GK and GPR reveal high sequence and structural conservation. The information acquired from different mutant enzymes with increased osmotolerant properties, together with the position of the insertion found in the longer human isoform, permit the delimitation of the regulatory site of GK and P5CS and the proposal of a model of P5CS architecture. Additionally, the GK moiety of the human enzyme has been modeled and the known clinical mutations and polymorphisms have been mapped.
Keywords: proline, hypoprolinemia, glutamate kinase, γ-glutamyl phosphate reductase, p53, P5CS deficiency
Introduction
Pyrroline-5-carboxylate synthase (P5CS) is the key enzyme in the synthesis of proline and ornithine (Fig. 1). One of the final products, proline, and the intermediate substrate, pyrroline-5-carboxylate (P5C), have noteworthy properties related to redox balance and osmotic stress and have been the focus of numerous studies related with both human health and agriculture.1–10
Figure 1.
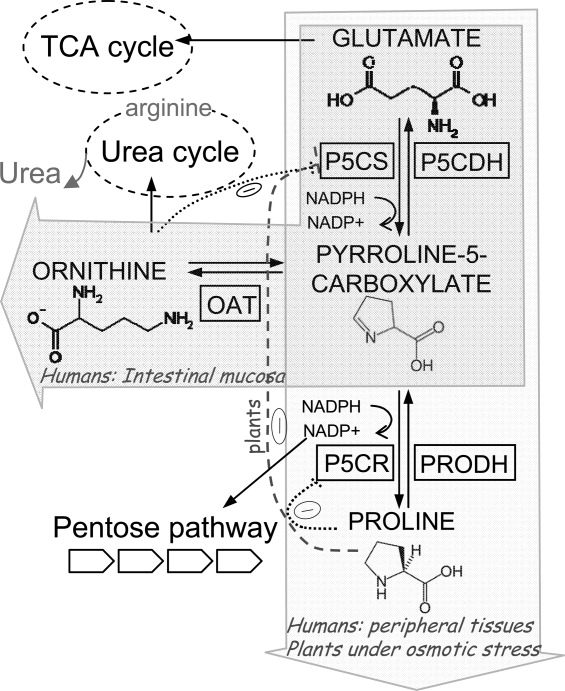
General pathway of the proline metabolism in higher eukaryotes, with the regulation of P5CS and P5CR in plants and mammals.
P5CS, evolved from two bacterial polypeptides encoded by a single operon,11 has two sequential activities: glutamate kinase (GK) and γ-glutamyl phosphate reductase (GPR). It produces P5C from glutamate, and synthesizes the labile intermediate γ-glutamyl phosphate.12 The N-terminal moiety contains the kinase domain responsible for substrate phosphorylation and houses the regulatory binding site for either proline or ornithine. The reduction reaction is performed by the C-terminus of P5CS which also harbors the cofactor binding site (Fig. 2). The kinetics of various members of the P5CS family have been characterized7,13–16 and their catalytic constants illustrate a capacity for adaptation to specific environments. Furthermore, a number of mutations have been described for the plant enzyme, which affect proline regulation.7,17 However, the identification of the proline binding site in P5CS and the mechanism responsible for its inhibition remain critical themes particularly in terms of plant technological development.
Figure 2.
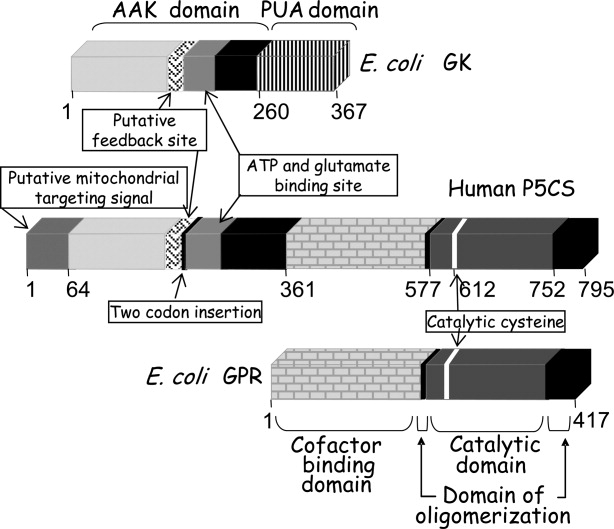
Schematic representation of human P5CS and bacterial GK and GPR, showing the location of certain structural features.
Although all P5CSs share high sequence identity (Supporting Information Fig. S1), ornithine inhibits human P5CS instead of proline, and a two amino acid insertion renders one of the human forms insensitive to ornithine,3 suggesting that structural and functional differences exist between plant and animal enzymes. In this review, we will discuss some novel information about the allosteric regulation of P5CS, model the GK moiety of the human enzyme, analyze functional and structural data to understand the pathological consequences of P5CS deficiency in humans, and propose a scheme for the architectural organization of this enzyme.
P5CS
P5CS is well conserved among eukaryotes, with a protein sequence identity higher than 65% when compared to human P5CS. However, in prokaryotes (and also in lower eukaryotes, as yeast), two separate enzymes exist, GK and GPR,18 that are homologous to both the moieties of P5CS.
The three main residues of the active center of the GK moiety of human P5CS (Lys76, Asp247, Lys311) are totally conserved, as demonstrated by the alignment of representative P5CSs and GKs (Supporting Information Fig. S1). This catalytic triad is also found among other members of the amino acid kinase (AAK) family,19 and consists of two catalytic lysine residues that are connected and mutually oriented towards the substrates by a key aspartate residue. The function of these residues has been demonstrated by site-directed mutagenesis13 which has been further confirmed by structural studies20 of Escherichia coli GK. The catalytic cysteine of the GPR moiety is conserved in P5CSs across species. Remarkably, the oligomerization domain is highly conserved (Supporting Information Fig. S1).
Homologous enzymes of P5CS in bacteria and lower eukaryotes
GK
This enzyme (EC 2.7.2.11) catalyzes the following reaction: ATP + Glutamate → ADP + γ-glutamyl phosphate.21 GK contains a ∼260-residue N-terminal AAK domain and, in most of the cases, a ∼110-residue C-terminal RNA binding domain (named PUA) that is absent in P5CS.22,23 The presence of this domain, usually found in RNA-modifying enzymes, could suggest an additional role for bacterial GK as a mediator of some cellular responses given the special functions of proline.
The 3D structures of two bacterial GKs have been solved. Despite their low sequence identity (33%), both have the same basic architecture of a α3β8α4 sandwich topology and they superimpose with an RMSD of 1.5 Å over 216 residues [Fig. 3(A,B,C)]. The Campylobacter jejuni enzyme (PDB ID 2AKO) has only an AAK domain, whereas the E. coli enzyme has an additional PUA domain (PDB ID 2J5T). In both cases, the active site forms an open pit where glutamate and ATP bind in an extended conformation.
Figure 3.
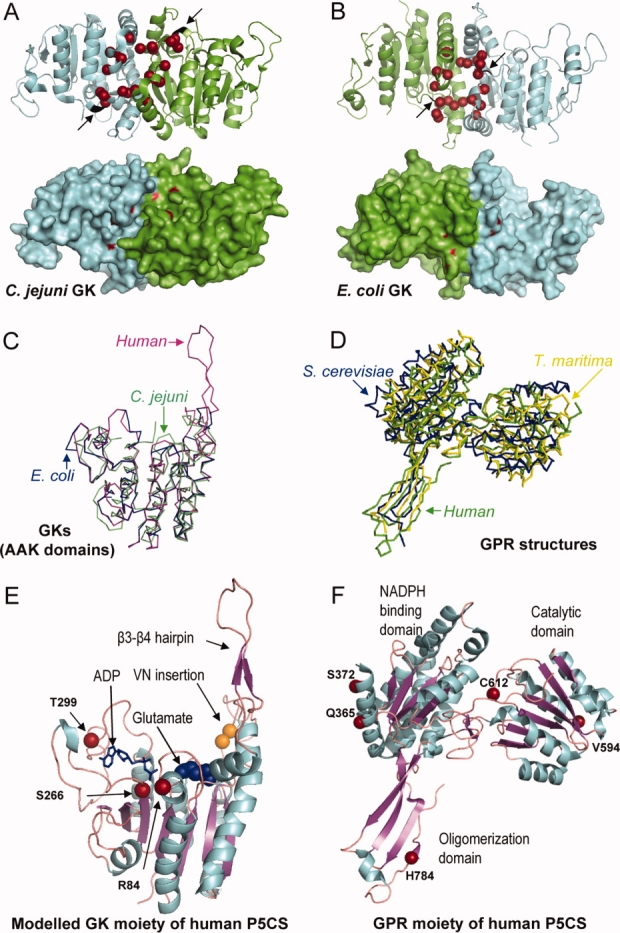
(A) A ribbon and surface representation of a GK dimer from C. jejuni and (B) from E. coli where the mutated residues found in the different GK and P5CS enzymes affecting proline regulation are drawn in red. Arrows denote the location of the VN insertion (colored in black) found in the human long P5CS isoform. C: Superposition of the AAK domain of E. coli GK colored in blue (2J5T), C. jejuni GK (2AKO) colored in green, together with the modeled human GK moiety generated by using the Swiss Model Workspace, colored in pink. D: Superposition of the GPR enzymes from T. maritima (1O20) colored in yellow, S. cerevisiae (1VLU) colored in blue and the human GPR moiety of P5CS (2H5G) colored in green. E: Ribbon representation of the modeled structure of the human GK moiety, with the ADP molecule depicted by sticks, and the glutamate by spheres, and (F) the crystal structure of the human GPR moiety with the different domains, the catalytic cysteine and some structural features indicated. The residues affecting clinical mutations and polymorphisms are shown.
The product of the reaction catalyzed by GK, γ-glutamyl phosphate, rapidly transforms to oxoproline,24 as the enzymatic activities of the E. coli enzyme also reflect. Only 10% of glutamyl hydroxamate was found—from the reaction between γ-glutamyl phosphate and hydroxylamine—14,24 in relation to ADP production, unless GPR was included in the assay. For this reason, the existence of a complex between GK and GPR has been hypothesized,24,25 comparable to P5CS where both activities are found in the same molecule.
The GK of C. jejuni forms a dimer whereas this enzyme from E. coli forms a tetramer. The E. coli tetramer, which is flat and elongated, is formed by two dimers that interact exclusively through their AAK domains.20 In principle, the oligomeric state of these enzymes could modulate the mechanisms that regulate their activities, as described for another member of the AAK family, the N-acetylglutamate kinase (NAGK). While NAGK from Pseudomonas aeruginosa is hexameric and inhibited by arginine, this enzyme from E. coli is dimeric and insensitive to arginine.26 However, a triple mutant of E. coli GK that can form only dimers displays the same kinetic and allosteric properties as the wild-type enzyme, suggesting that the mechanism of allosteric inhibition in GK is independent of the oligomeric state of the enzyme (unpublished results from our laboratory).
The GKs of C. jejuni and E. coli diverge in the arrangement of the dimer, as shown in Figure 3(A,B). In the C. jejuni enzyme, the subunits face the same side and the active sites are exposed with one facing up and the other facing down (PDB ID 2AKO). Conversely, both of the subunits in the GK dimer from E. coli are arranged in the same direction, but face opposite sides.20 Nevertheless, the residues involved in allosteric inhibition are located within a similar region [Fig. 3(A,B)]. The kinetics of the E. coli GK have been characterized,13,14 and show cooperativity among the monomers. GK activity depends hyperbolically on the concentrations of both ATP and glutamate, but dependency on glutamate becomes sigmoidal in the presence of proline, reflecting competition between both ligands.13
The deregulation of certain bacterial GKs causes the accumulation of high concentrations of proline and appears to play a paramount role in osmotolerance.27–30 Various studies of natural and engineered strains with increased osmotolerant properties have been published. Examples of these include the halophilic bacteria Halobacillus halophilus,28 a Salmonella typhimurium strain harboring a Asp107Asn mutant of E. coli GK,27,29 and a proline auxotroph strain of E. coli expressing a mutated GK and a GPR enzymes from Bacillus subtilis.30 When these same enzymes were introduced into the plant Arabidopsis thaliana, osmotic tolerance was improved and the effect was enhanced by fusing the two enzymes into a single polypeptide.31 Recently, a yeast strain expressing a deregulated GK was generated, resulting in a greater tolerance to both freezing and oxidative stresses.32,33
GPR
This enzyme (EC 1.2.1.41) catalyzes the following reaction: γ-glutamyl phosphate + NADPH → glutamate semialdehyde + phosphate + NADP+. GPR from E. coli has been purified and characterized from its natural source15,34 and gel filtration experiments with the recombinant form have suggested that the enzyme exists as a tetramer (unpublished results from our laboratory). Like other aldehyde dehydrogenases, the GPR structure has three distinct domains (Fig. 2). Domain I houses the site for the cofactor NADPH, while domain II contains the active site where a catalytic cysteine has been identified.35,36 Both have an α/β architecture with a five-stranded parallel β-sheet. Domain III, the oligomerization domain, forms a three-stranded anti-parallel β-sheet,35 with two strands originating from the amino terminus (β1 and β2) and the third (the last β strand) from the C-terminus.
Most aldehyde dehydrogenases function as either dimers or tetramers. GPR from Saccharomyces cerevisiae forms a dimer (PDB ID 1VLU), while the enzyme from Thermotoga maritima (PDB ID 1O20) forms a tetramer made up of a dimer of dimers in which both the dimeric and the tetrameric contacts are extensive.35 These two 3D structures superimpose with an RMSD of 1.8 Å over 214 residues (predicted by using 3D-Match by Softberry, Inc.) [Fig. 3(D) and Supporting Information Fig. S2].
A study conducted in yeast revealed that either specific mutations or the disruption of GPR permitted the growth of a glutathione auxotroph strain. This was apparently achieved through the accumulation of γ-glutamyl phosphate which contributed to the production of small amounts of glutathione, thus linking the proline biosynthetic pathway with tolerance to oxidative stress.37
P5CS in plants
Intracellular proline accumulation is a widespread metabolic response of plants to osmotic stress,5 and P5CS is the key enzyme in this pathway. Several efforts have been made in different species to engineer a deregulated P5CS, with the aim of obtaining plants that are able to grow in the desert or salty land areas.5,10,38–41 Recently, the effects of cadmium and zinc stress on the GK activity of plants have been measured, revealing a potential regulatory role of P5CS in heavy metal stress.42
Two different P5CSs, resulting from independent evolutionary duplication events, have been found in many species.6 Interestingly, the duplicated enzymes studied to date seem to perform nonredundant functions and their genes present different expression patterns.9,43–45 For instance, the isoform1 of P5CS in A. thaliana is induced by dehydration and high salt, while isoform2 is essential for embryo and seedling development.8,46 Isoform1 is localized mainly to the chloroplast whereas isoform2 displays a cytoplasmic distribution. The knockout of the P5CS isoform1 in A. thaliana causes a reduction of stress-induced proline synthesis, hypersensitivity to salt stress and an accumulation of reactive oxygen species (ROS).8 Remarkably, this transgenic species displayed decreased glutathione-S-transferase (GST) and glutathione reductase (GR) activities, indicating that the glutathione detoxifying pathway was subsequently affected by the absence of P5CS isoform1. These observations suggest a link between glutathione and proline metabolism in plants; recent studies in tobacco cells have also demonstrated that proline significantly restores the activities of GST and GR under cadmium stress.47
While the primary precursor for proline biosynthesis in some plants is glutamate, ornithine serves this function in others. Nevertheless, under conditions of osmotic stress, all plants predominantly use glutamate as a substrate.38,48,49 In plants, proline is useful not only during conditions of drought and high salt osmotic stress, but also as a response to environmental changes such as low temperature,50 nutritional deficits,51 heavy metals,52 UV radiation,53 and bacterial pathogens.54 In some species, proline also accumulates during normal development in flowers,55 pollen,43 ovules,56 or fruits.57 Depending on the plant species, proline accumulation results from different combined strategies that may include any of the following changes: an increase of the P5CS transcription level, and/or an increase of P5CS activity, and/or the inhibition of proline degradation which is reduced in plants under water stress.9,39,40 In fact, high salt concentrations inhibit the activity of proline dehydrogenase (PRODH).58 In response to other situations, proline storage is produced by decreased pyrroline-5-carboxylate dehydrogenase (P5CDH) activity.59 Paradoxically, an external supply of proline has proved toxic for plants as it causes cell death induced by P5C accumulation, with an accompanying increase of ROS production.59
The P5CS from Vigna aconitifolia has been characterized.7 This hexameric enzyme has a molecular weight of 450 kDa, and is up-regulated by transcriptional mechanisms under stress conditions. As with E. coli GK,13 proline decreases the affinity of Vigna P5CS for glutamate and acts as a competitive inhibitor, whereas ADP shows a competitive inhibition of the GK activity of P5CS. However, sensitivity to proline in this species is one order of magnitude lower than the E. coli GK. In an attempt to identify the proline inhibition site of Vigna P5CS, a small region of six amino acids, that maps near the homologous residue (Asp107) of E. coli GK, was subjected to alanine scanning mutagenesis.7 One of the mutants, the F129A variant, exhibited a 200-fold lower sensitivity to proline than the wild-type enzyme while the other properties of the enzyme remained unchanged.
One unusual example of P5CS evolution is found in tomato, which has two genes for this molecule. One of these, tomPRO1, encodes two polypeptides resembling the bacterial operon, whereas tomPRO2 encodes a protein similar to eukaryotic P5CS.43 Studies of proline regulation have been performed by random mutagenesis of tomPRO1 and have revealed similar sensitivities to proline inhibition as bacterial GK (KiPro∼0.1 mM).17 The results demonstrate that most of the residues which confer sensitivity to proline were located in a small region of 16 amino acids. Most of the tomPRO1 mutants had lower specific activities and increased inhibition constants (20-fold to 3500-fold).
When the reports of the different mutations that alter the inhibitory features of the GKs and P5CSs are analyzed, a prominent common region of ∼40 amino acids can be delimited.17,29,33,60–63 In the GK structure, the corresponding residues of this region map to the loop that surrounds the glutamate binding site and also to the dimer interface, showing a direct connection between them [Fig. 3(A,B)]. As kinetic assays with E. coli GK have reported,13 proline and glutamate compete for binding while ATP and proline bind independently, indicating that proline binding induces a conformational change of the AAK domain that could modulate the catalytic conversion of glutamate or the inter-subunit signal transmission.
Human P5CS
Among mammals, P5CS, located in the mitochondrial inner membrane, shares a protein sequence identity of over 90%. As far as we are aware, two P5CS isoforms generated by differential display have been described in humans and mice,3 although this exon sliding mechanism is believed to be widespread in mammals. Mammalian P5CS activity was measured for the first time in hamster ovary cells,64 but has also been characterized from rat intestinal mucosa.16
Isoforms and regulation
The human P5CS gene, known as ALDH18A1 (the member 18A1 of the aldehyde dehydrogenase family), is located on chromosome 10,65 spans about 50 kb and comprises 18 exons. The transcript has a length of 3462 bp, encodes a protein of 795 amino acids, and has a molecular weight of 87.3 kDa. The 64 N-terminal residues present characteristic features of a mitochondrial targeting sequence with two potential cleavage sites.3 The P5CS gene undergoes alternative splicing to generate two isoforms which are differentially expressed and differ only by the insertion/deletion of two amino acids at the GK moiety.
The expression of the short isoform, regulated by ornithine, is prominent in the gut, where it directs P5C towards arginine biosynthesis. The long isoform, which is insensitive to ornithine, is ubiquitously expressed and mediates proline biosynthesis. The two amino acid insertion in the long isoform abolishes the feedback inhibition of P5CS activity by ornithine.3 In fact, when these two amino acids are inserted into the E. coli GK, proline inhibition diminishes by around 150-fold. Additionally, the insertion interferes with glutamate binding, thus confirming its key regulatory role (unpublished results from our laboratory). This finding suggests that the location of the catalytic site and structural organization of the inhibitory mechanism of human P5CS are similar to what has been described for bacterial GKs and plant P5CSs.
Arginine synthesis is active in the small intestine, especially in early postnatal life when this amino acid is essential for its critical role in ammonia detoxification. For this reason, the short ornithine-sensitive P5CS isoform is more abundant in the gut and displays an inhibition constant (∼0.4 mM) in a range that is suitable to regulate the enzyme.66 Interestingly, the activation of P5CS by N-acetyl glutamate, as a means of regulating the intestinal synthesis of citruline and arginine, has been reported in mitochondrial preparations of enterocytes of neonatal pigs.67
Conversely, proline biosynthesis is carried out in peripheral tissues with high rates of protein synthesis. In these tissues, proline synthesis is controlled through the second enzyme of the route, pyrroline-5-carboxylate reductase (P5CR), which is inhibited by proline. This allows the differential regulation of both pathways.3
The human P5CS gene is tightly regulated. Its promoter has putative binding sites for p53 and GRE (glucocorticoid response element), and there is a putative binding site for p53 in the first intron. Indeed, it has been demonstrated that P5CS is regulated by growth hormones and p53.2 It has also been shown that P5CS transcription in mouse brain tissues is down-regulated by age, causing hearing loss.68
Domain organization and 3D structure
The human GK moiety has been modeled here using the Swiss Model Workspace program.69 The program selected the E. coli GK structure, which shares a 33% identity, as a suitable template [Fig. 3(C)]. The resulting model showed the common architecture of the amino acid kinase family, a main β sheet of eight strands sandwiched by 3–4 α helices on each side. Interestingly, although E. coli GK has no peripheral sub-domain, a β3-β4 hairpin, similar to that found in N-acetylglutamate kinase (NAGK) structure, was depicted.19 NAGK is the paradigm of the AAK family, and is closely related to GK. It catalyzes the same reaction but uses acetylated glutamate as a substrate. The hairpin, which is composed of exactly the same secondary elements as in NAGK, is apparently involved in glutamate binding. Its hypothetical function would entail opening/closing the binding site to allow the substrate to enter, as predicted in NAGK19 [Fig. 3(C,E)].
In addition, a crystal structure corresponding to the human GPR moiety has been solved (PDB ID 2H5G). It shares the common architecture of the other solved GPRs, with three independent domains and a hinge region between domains II and III [Fig. 3(D,F)]. Based on structure and sequence comparisons, the catalytic cysteine can be identified as Cys612. As with other mammalian enzymes, human GPR moiety contains an extra tail of around 20–35 amino acids which surrounds the oligomerization domain [Fig. 3(F)]. The crystal structure reveals that the human GPR moiety is organized as a dimer with the oligomerization domain from one subunit interacting with the extended region of the hinge between domains II and III of the other monomer. As a result, the oligomerization domains are on one side, and domains I and II from both monomers are located on the other side.
A model for the interaction between bacterial GK and GPR, which could also fit the hypothetical arrangement of human P5CS, has been proposed.20 In this model, the molecular face where the active site of GK moiety is located would interact with the catalytic domain of the GPR moiety, and the interaction with the other monomer would be accomplished through the oligomerization domain of the GPR moiety [Supporting Information Fig. S3(A,C)].
Mutations in P5CS and clinical implications
Mutations in human P5CS produce a rare disease (OMIM #612652), characterized by hypoprolinemia and a temporary deficit of urea cycle intermediates, with symptoms that range from joint laxity and skin hyperelasticity to bilateral cataracts, progressive neurodegeneration and peripheral neuropathy.1,70,71 As an amino acid which has no primary amino group,72 proline is implicated in special physiological functions. It is excluded from both the decarboxylation and transamination reactions of amino acid metabolism, and instead acts as chaperone, thereby diminishing protein aggregation. Additionally, proline serves as an antioxidant, fighting ROS and preserving the intracellular glutathione pool, which is the major redox buffer of the cell.73 Hence, some of the symptoms associated with mutations of human P5CS can be explained by the lack of proline. For example, proline is essential for the synthesis of brain polypeptides with neuroprotective and neuromodulator effects68,74 which could account for the neurological disorders observed in humans with P5CS deficiency. Low levels of proline also impair collagen synthesis, causing hyperelasticity. Additionally, proline deficiency could affect the levels of P5C and the redox state of the lens, leading to cataracts.70
The first pathogenic mutation described for P5CS was the missense mutation R84Q observed in two brothers from a consanguineous couple.70 The clinical features, that included hyperelastic skin, hypotonia, chronic vomiting, cataracts, mental retardation, hypoprolinemia, mild hyperammonaemia and complete loss of the ability to walk, correlated with a severe reduction of P5CS enzymatic activity associated with the two isoforms. Moreover, R84Q appeared to destabilize the long isoform1,70 which is ubiquitously expressed for proline biosynthesis. The alignment with other enzymes shows that R84 is highly conserved in the homologous enzymes from mammals. In E. coli, this residue is not conserved (Gly18 is the corresponding residue), but belongs to the β1-αA junction that is located centrally at the site of phosphoryl group transfer. Consequently, in addition to causing structural instability, the R84Q mutation could directly affect ATP binding or/and catalysis, which would also account for the pathophysiological manifestations in these patients [Fig. 3(E)].
A second missense mutation, H784Y, was identified in a New Zealand family with symptoms of a neurocutaneous syndrome, including joint dislocations, lax skin and mental retardation.71 H784 is conserved among the eukaryotic proteins except for V. aconitifolia which has a shorter oligomerization domain. However, a biochemical analysis of metabolites in these patients revealed that the H784Y substitution did not impair proline biosynthesis, suggesting that this might be a milder mutation than R84Q, producing only a partial loss of enzymatic function or that the tissue pathology may occur from the disruption of an unknown function of P5CS. These effects agree to some extent with the 3D structure of the GPR moiety of the human P5CS since the mutation would be located in the last β strand of the oligomerization domain, and would not involve any catalytic function of the enzyme, but would rather alter its quaternary structure [Fig. 3(F)]. The pathological effects of this H784Y mutation could be mediated by reduced cellular pools of P5C that would diminish the ultimate quantity of proline synthesized, hampering additional functions of proline as modulator of redox-dependent metabolic and carcinogenic pathways.73,75
A similar disease known as cutis laxa is produced by mutations in the second enzyme of the proline biosynthetic pathway, pyrroline-5-carboxylate reductase (P5CR),76,77 which converts P5C to proline [Fig. 1]. Patients with cutis laxa also have slightly low but within a normal range of proline levels, and are characterized by the appearance of premature aging, wrinkled and lax skin, joint laxity and mental retardation.76,77 Skin fibroblasts from these patients, which have lower levels of P5CR, display increased sensitivity to oxidative stress and a marked increase in apoptosis, consistent with previous results demonstrating that proline protects against oxidative stress-induced cell death and apoptosis in mammalian cell cultures.73
Recently, a functional genomic study was conducted to analyze the human genes encoding proline metabolic enzymes.4 Nine single nucleotide polymorphisms of P5CS were identified in two cancer cell lines. They comprise two frameshifts, one nonsense and six missense mutations. In addition to R84, polymorphisms in amino acids S266 and T299 were found in the GK moiety. They map near the ATP binding site, thereby confirming its putative clinical importance [Fig. 3(E)]. Interestingly, the E. coli GK contains a residue homologous to S266, which forms a donor hydrogen bond with the β-phosphate from the ATP. Mutations of this residue, Thr169, altered significantly the kinetic parameters for ATP, indicating an important role of the hydroxyl group in ATP binding and catalysis.13 The Q365, S372, and V594 polymorphisms have been reported in the GPR moiety. The first two are contained within the cofactor binding domain, while the last one was located in the catalytic domain [Fig. 3(F)].
Future Challenges
Although our knowledge of P5CS has been enhanced during recent years, several important challenges remain. The enzyme is broadly distributed in eukaryotes. As a key metabolic enzyme, it is inhibited by a feedback mechanism and two isoforms frequently exist in the same organism to allow precise regulation of proline metabolism. However, despite the increased information about the structure and function of P5CS, we are far from understanding the biological role of these two isoforms. An activator of the enzyme, N-acetyl glutamate, has even been described in neonatal pig mitochondrial preparations but its universality requires further investigation.67
Proline accumulation, due mainly to an increase in the activity of GK and P5CS, has been reported as an osmoprotective mechanism. However, this mechanism it is still not fully understood. The Ki for proline of the bacterial GKs (and the tomPRO1) is lower than 0.2 mM, while the Ki for most plant P5CSs is over 1 mM. This differential sensitivity to proline as an inhibitor is the reason why bacteria cannot accumulate large amounts of proline. Conversely, however, several plants belonging to the Solanaceae family can accumulate more than 100 mM proline in the cytosol under osmotic stress.38,78
It is somewhat puzzling that plants under stress produce proline in concentrations well above what is needed to inhibit P5CS, yet they preferentially use the glutamate pathway instead of the ornithine pathway, which is not regulated. Several hypotheses can be formulated to explain this paradox. One potential explanation is that the cytosolic concentrations of the different solutes under stress conditions could modify the enzyme kinetics. For instance, it has been reported that when the concentration of glutamate increases in E. coli GK, the sensitivity to proline is diminished.13 Consistent with this, the halophilic bacterium H. halophilus first produces glutamate under conditions of increasing salt to reach a maximal internal concentration of 2 M, which activates the transcription of the proline-producing genes and then it produces proline.28
The exhaustive study of the different duplicated plant P5CSs may also be used to gain insight into the stress response question. One possibility is that the enzyme overexpressed under osmotic stress is not regulated by proline. Among the prokaryotes, B. subtilis has two different GKs, one of which is sensitive to proline and constitutive, while the other (ProJ) is stress-inducible and homologous to that found in H. halophilus. It has been speculated that ProJ is insensitive to proline,79 which represents an interesting issue to investigate.
As discussed earlier, proline and glutamate compete for binding to the GKs and P5CSs which have been characterized, suggesting that the inhibition mechanism could be conserved, with the binding sites for these ligands partially overlapping. However, the numerous mutations which have been demonstrated to effect proline inhibition suggest that proline binding may also influence the hydrogen bond network connecting the active centers from monomers.20 Hence, the identification of the exact location of this binding site will require the crystal resolution of the protein in the presence of the inhibitor.
The crystallographic study has revealed the same protein fold for the GK moiety within the AAK family and the same protein fold for the GPR moiety within the aldehyde dehydrogenase family. The existence of a fully active synthetic polypeptide generated by the fusion of the two prokaryotic proteins from B. subtilis30 implies that the eukaryotic structure could be the result of the two protein folds, one next to the other. To further this notion, we hypothesize that the monomers of human P5CS interact to form the quaternary structure, not only through their oligomerization domains from the GPR moiety, but also through their GK dimer interfaces as seen in other 3D structures since this region seems to play a critical role in regulation [Supporting Information Fig. S3(D)].
The resolution of the crystal structure for P5CS would greatly facilitate our understanding of this enzyme and its regulation. Although the second moiety of the human enzyme has already been solved by crystallography and the first moiety has been modeled, various unknown factors remain to be elucidated including the spatial arrangement of both moieties, the channeling of γ-glutamyl phosphate from one active center to another, the signal transmission between monomers, the oligomerization state, and most importantly, how proline/ornithine binds to the GKs and P5CSs. These parameters represent a bottleneck for understanding the biological significance of P5CS regulation. Strategies to address these important biological questions are currently being developed in our laboratory.
Acknowledgments
The authors thank Deborah Burks (CIPF) for helpful reading of the manuscript.
References
- 1.Baumgartner MR, Rabier D, Nassogne MC, Dufier JL, Padovani JP, Kamoun P, Valle D, Saudubray JM. Delta1-pyrroline-5-carboxylate synthase deficiency: neurodegeneration, cataracts and connective tissue manifestations combined with hyperammonaemia and reduced ornithine, citrulline, arginine and proline. Eur J Pediatr. 2005;164:31–36. doi: 10.1007/s00431-004-1545-3. [DOI] [PubMed] [Google Scholar]
- 2.Hu CA, Khalil S, Zhaorigetu S, Liu Z, Tyler M, Wan G, Valle D. Human delta1-pyrroline-5-carboxylate synthase: function and regulation. Amino Acids. 2008;35:665–672. doi: 10.1007/s00726-008-0075-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hu CA, Lin WW, Obie C, Valle D. Molecular enzymology of mammalian delta1-pyrroline-5-carboxylate synthase. Alternative splice donor utilization generates isoforms with different sensitivity to ornithine inhibition. J Biol Chem. 1999;274:6754–6762. doi: 10.1074/jbc.274.10.6754. [DOI] [PubMed] [Google Scholar]
- 4.Hu CA, Bart Williams D, Zhaorigetu S, Khalil S, Wan G, Valle D. Functional genomics and SNP analysis of human genes encoding proline metabolic enzymes. Amino Acids. 2008;35:655–664. doi: 10.1007/s00726-008-0107-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Verbruggen N, Hermans C. Proline accumulation in plants: a review. Amino Acids. 2008;35:753–759. doi: 10.1007/s00726-008-0061-6. [DOI] [PubMed] [Google Scholar]
- 6.Turchetto-Zolet AC, Margis-Pinheiro M, Margis R. The evolution of pyrroline-5-carboxylate synthase in plants: a key enzyme in proline synthesis. Mol Genet Genomics. 2009;281:87–97. doi: 10.1007/s00438-008-0396-4. [DOI] [PubMed] [Google Scholar]
- 7.Zhang CS, Lu Q, Verma DP. Removal of feedback inhibition of delta 1-pyrroline-5-carboxylate synthetase, a bifunctional enzyme catalyzing the first two steps of proline biosynthesis in plants. J Biol Chem. 1995;270:20491–20496. doi: 10.1074/jbc.270.35.20491. [DOI] [PubMed] [Google Scholar]
- 8.Szekely G, Abraham E, Cseplo A, Rigo G, Zsigmond L, Csiszar J, Ayaydin F, Strizhov N, Jasik J, Schmelzer E, Koncz C, Szabados L. Duplicated P5CS genes of Arabidopsis play distinct roles in stress regulation and developmental control of proline biosynthesis. Plant J. 2008;53:11–28. doi: 10.1111/j.1365-313X.2007.03318.x. [DOI] [PubMed] [Google Scholar]
- 9.Ginzberg I, Stein H, Kapulnik Y, Szabados L, Strizhov N, Schell J, Koncz C, Zilberstein A. Isolation and characterization of two different cDNAs of delta1-pyrroline-5-carboxylate synthase in alfalfa, transcriptionally induced upon salt stress. Plant Mol Biol. 1998;38:755–764. doi: 10.1023/a:1006015212391. [DOI] [PubMed] [Google Scholar]
- 10.Kishor P, Hong Z, Miao GH, Hu C, Verma D. Overexpression of δ-pyrroline-5-carboxylate synthetase increases proline production and confers osmotolerance in transgenic plants. Plant Physiol. 1995;108:1387–1394. doi: 10.1104/pp.108.4.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu CA, Delauney AJ, Verma DP. A bifunctional enzyme (delta 1-pyrroline-5-carboxylate synthetase) catalyzes the first two steps in proline biosynthesis in plants. Proc Natl Acad Sci USA. 1992;89:9354–9358. doi: 10.1073/pnas.89.19.9354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Strecker HJ. The interconversion of glutamic acid and proline. I. The formation of delta1-pyrroline-5-carboxylic acid from glutamic acid in Escherichia coli. J Biol Chem. 1957;225:825–834. [PubMed] [Google Scholar]
- 13.Perez-Arellano I, Rubio V, Cervera J. Mapping active site residues in glutamate-5-kinase. The substrate glutamate and the feed-back inhibitor proline bind at overlapping sites. FEBS Lett. 2006;580:6247–6253. doi: 10.1016/j.febslet.2006.10.031. [DOI] [PubMed] [Google Scholar]
- 14.Perez-Arellano I, Rubio V, Cervera J. Dissection of Escherichia coli glutamate 5-kinase: functional impact of the deletion of the PUA domain. FEBS Lett. 2005;579:6903–6908. doi: 10.1016/j.febslet.2005.11.037. [DOI] [PubMed] [Google Scholar]
- 15.Hayzer DJ, Leisinger T. Proline biosynthesis in Escherichia coli. Kinetic and mechanistic properties of glutamate semialdehyde dehydrogenase. Biochim Biophys Acta. 1983;742:391–398. doi: 10.1016/0167-4838(83)90326-6. [DOI] [PubMed] [Google Scholar]
- 16.Wakabayashi Y, Jones ME. Pyrroline-5-carboxylate synthesis from glutamate by rat intestinal mucosa. J Biol Chem. 1983;258:3865–3872. [PubMed] [Google Scholar]
- 17.Fujita T, Maggio A, Garcia-Rios M, Stauffacher C, Bressan RA, Csonka LN. Identification of regions of the tomato gamma-glutamyl kinase that are involved in allosteric regulation by proline. J Biol Chem. 2003;278:14203–14210. doi: 10.1074/jbc.M212177200. [DOI] [PubMed] [Google Scholar]
- 18.Aral B, Kamoun P. The proline biosynthesis in living organisms. Amino Acids. 1997;13:189–217. [Google Scholar]
- 19.Ramon-Maiques S, Marina A, Gil-Ortiz F, Fita I, Rubio V. Structure of acetylglutamate kinase, a key enzyme for arginine biosynthesis and a prototype for the amino acid kinase enzyme family, during catalysis. Structure. 2002;10:329–342. doi: 10.1016/s0969-2126(02)00721-9. [DOI] [PubMed] [Google Scholar]
- 20.Marco-Marin C, Gil-Ortiz F, Perez-Arellano I, Cervera J, Fita I, Rubio V. A novel two-domain architecture within the amino acid kinase enzyme family revealed by the crystal structure of Escherichia coli glutamate 5-kinase. J Mol Biol. 2007;367:1431–1446. doi: 10.1016/j.jmb.2007.01.073. [DOI] [PubMed] [Google Scholar]
- 21.Hayzer DJ, Leisinger T. The gene-enzyme relationships of proline biosynthesis in Escherichia coli. J Gen Microbiol. 1980;118:287–293. doi: 10.1099/00221287-118-2-287. [DOI] [PubMed] [Google Scholar]
- 22.Aravind L, Koonin EV. Novel predicted RNA-binding domains associated with the translation machinery. J Mol Evol. 1999;48:291–302. doi: 10.1007/pl00006472. [DOI] [PubMed] [Google Scholar]
- 23.Perez-Arellano I, Gallego J, Cervera J. The PUA domain—a structural and functional overview. FEBS J. 2007;274:4972–4984. doi: 10.1111/j.1742-4658.2007.06031.x. [DOI] [PubMed] [Google Scholar]
- 24.Seddon AP, Zhao KY, Meister A. Activation of glutamate by gamma-glutamate kinase: formation of gamma-cis-cycloglutamyl phosphate, an analog of gamma-glutamyl phosphate. J Biol Chem. 1989;264:11326–11335. [PubMed] [Google Scholar]
- 25.Baich A. Proline synthesis in Escherichia coli. A proline-inhibitable glutamic acid kinase. Biochim Biophys Acta. 1969;192:462–467. doi: 10.1016/0304-4165(69)90395-x. [DOI] [PubMed] [Google Scholar]
- 26.Fernandez-Murga ML, Rubio V. Basis of arginine sensitivity of microbial N-acetyl-l-glutamate kinases: mutagenesis and protein engineering study with the Pseudomonas aeruginosa and Escherichia coli enzymes. J Bacteriol. 2008;190:3018–3025. doi: 10.1128/JB.01831-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Csonka LN. Proline over-production results in enhanced osmotolerance in Salmonella typhimurium. Mol Gen Genet. 1981;182:82–86. doi: 10.1007/BF00422771. [DOI] [PubMed] [Google Scholar]
- 28.Saum SH, Müller V. Salinity-dependent switching of osmolyte strategies in a moderately halophilic bacterium: glutamate induces proline biosynthesis in Halobacillus halophilus. J Bacteriol. 2007;189:6968–6975. doi: 10.1128/JB.00775-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Csonka LN, Gelvin SB, Goodner BW, Orser CS, Siemieniak D, Slightom JL. Nucleotide sequence of a mutation in the proB gene of Escherichia coli that confers proline overproduction and enhanced tolerance to osmotic stress. Gene. 1988;64:199–205. doi: 10.1016/0378-1119(88)90335-6. [DOI] [PubMed] [Google Scholar]
- 30.Chen M, Cao J, Zheng C, Liu Q. Directed evolution of an artificial bifunctional enzyme, gamma-glutamyl kinase/gamma-glutamyl phosphate reductase, for improved osmotic tolerance of Escherichia coli transformants. FEMS Microbiol Lett. 2006;263:41–47. doi: 10.1111/j.1574-6968.2006.00397.x. [DOI] [PubMed] [Google Scholar]
- 31.Chen M, Wei H, Cao J, Liu R, Wang Y, Zheng C. Expression of Bacillus subtilis proBA genes and reduction of feedback inhibition of proline synthesis increases proline production and confers osmotolerance in transgenic Arabidopsis. J Biochem Mol Biol. 2007;40:396–403. doi: 10.5483/bmbrep.2007.40.3.396. [DOI] [PubMed] [Google Scholar]
- 32.Kaino T, Tateiwa T, Mizukami-Murata S, Shima J, Takagi H. Self-cloning baker's yeasts that accumulate proline enhance freeze tolerance in doughs. Appl Environ Microbiol. 2008;74:5845–5849. doi: 10.1128/AEM.00998-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morita Y, Nakamori S, Takagi H. L-proline accumulation and freeze tolerance of Saccharomyces cerevisiae are caused by a mutation in the PRO1 gene encoding gamma-glutamyl kinase. Appl Environ Microbiol. 2003;69:212–219. doi: 10.1128/AEM.69.1.212-219.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deutch AH, Smith CJ, Rushlow KE. Overproduction and purification of the three enzymes constituting the Escherichia coli proline biosynthetic pathway. Dev Ind Microbiol. 1985;26:437–444. [Google Scholar]
- 35.Page R, Nelson MS, von Delft F, Elsliger MA, Canaves JM, Brinen LS, Dai X, Deacon AM, Floyd R, Godzik A, Grittini C, Grzechnik SK, Jaroszewski L, Klock HE, Koesema E, Kovarik JS, Kreusch A, Kuhn P, Lesley SA, McMullan D, McPhillips TM, Miller MD, Morse A, Moy K, Ouyang J, Robb A, Rodrigues K, Schwarzenbacher R, Spraggon G, Stevens RC, van den Bedem H, Velasquez J, Vincent J, Wang X, West B, Wolf G, Hodgson KO, Wooley J, Wilson IA. Crystal structure of gamma-glutamyl phosphate reductase (TM0293) from Thermotoga maritima at 2.0 A resolution. Proteins. 2004;54:157–161. doi: 10.1002/prot.10562. [DOI] [PubMed] [Google Scholar]
- 36.Liu ZJ, Sun YJ, Rose J, Chung YJ, Hsiao CD, Chang WR, Kuo I, Perozich J, Lindahl R, Hempel J, Wang BC. The first structure of an aldehyde dehydrogenase reveals novel interactions between NAD and the Rossmann fold. Nat Struct Biol. 1997;4:317–326. doi: 10.1038/nsb0497-317. [DOI] [PubMed] [Google Scholar]
- 37.Spector D, Labarre J, Toledano MB. A genetic investigation of the essential role of glutathione: mutations in the proline biosynthesis pathway are the only suppressors of glutathione auxotrophy in yeast. J Biol Chem. 2001;276:7011–7016. doi: 10.1074/jbc.M009814200. [DOI] [PubMed] [Google Scholar]
- 38.Delauney AJ, Verma DPS. Proline biosynthesis and osmoregulation in plants. Plant J. 1993;4:215–223. [Google Scholar]
- 39.Yoshiba Y, Kiyosue T, Katagiri T, Ueda H, Mizoguchi T, Yamaguchi-Shinozaki K, Wada K, Harada Y, Shinozaki K. Correlation between the induction of a gene for delta 1-pyrroline-5-carboxylate synthetase and the accumulation of proline in Arabidopsis thaliana under osmotic stress. Plant J. 1995;7:751–760. doi: 10.1046/j.1365-313x.1995.07050751.x. [DOI] [PubMed] [Google Scholar]
- 40.Xue X, Liu A, Hua X. Proline accumulation and transcriptional regulation of proline biosynthesis and degradation in Brassica napus. BMB Rep. 2009;42:28–34. doi: 10.5483/bmbrep.2009.42.1.028. [DOI] [PubMed] [Google Scholar]
- 41.Hmida-Sayari A, Gargouri-Bouzid R, Bidani A, Jaoua L, Savoure A, Jaoua S. Overexpression of Δ1-pyrroline-5-carboxylate synthetase increases proline production and confers salt tolerance in transgenic potato plants. Plant Sci. 2005;169:746–752. [Google Scholar]
- 42.Pavlikova D, Pavlik M, Staszkova L, Motyka V, Szakova J, Tlustos P, Balik J. Glutamate kinase as a potential biomarker of heavy metal stress in plants. Ecotoxicol Environ Saf. 2008;70:223–230. doi: 10.1016/j.ecoenv.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 43.Fujita T, Maggio A, Garcia-Rios M, Bressan RA, Csonka LN. Comparative analysis of the regulation of expression and structures of two evolutionarily divergent genes for delta1-pyrroline-5-carboxylate synthetase from tomato. Plant Physiol. 1998;118:661–674. doi: 10.1104/pp.118.2.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Silva-Ortega CO, Ochoa-Alfaro AE, Reyes-Aguero JA, Aguado-Santacruz GA, Jimenez-Bremont JF. Salt stress increases the expression of p5cs gene and induces proline accumulation in cactus pear. Plant Physiol Biochem. 2008;46:82–92. doi: 10.1016/j.plaphy.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 45.Farzaneh M, Jazi FR, Motamed N. Application of dotbloting for detecting the expression of p5cs gene in transgenic olive plantlets. FEBS J. 2005;272:547. [Google Scholar]
- 46.Mattioli R, Falasca G, Sabatini S, Altamura MM, Costantino P, Trovato M. The proline biosynthetic genes P5CS1 and P5CS2 play overlapping roles in Arabidopsis flower transition but not in embryo development. Physiol Plant. 2009 doi: 10.1111/j.1399-3054.2009.01261.x. [DOI] [PubMed] [Google Scholar]
- 47.Islam MM, Hoque MA, Okuma E, Jannat R, Banu MN, Jahan MS, Nakamura Y, Murata Y. Proline and glycinebetaine confer cadmium tolerance on tobacco bright yellow-2 cells by increasing ascorbate-glutathione cycle enzyme activities. Biosci Biotechnol Biochem. 2009;73:2320–2323. doi: 10.1271/bbb.90305. [DOI] [PubMed] [Google Scholar]
- 48.Rhodes D, Handa S, Bressan RA. Metabolic changes associated with adaptation of plant cells to water stress. Plant Physiol. 1986;82:890–903. doi: 10.1104/pp.82.4.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boggess SF, Stewart CR. Effect of water stress on proline synthesis from radioactive precursors. Plant Physiol. 1976;58:398–401. doi: 10.1104/pp.58.3.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parvanova D, Ivanov S, Konstantinova T, Karanov E, Atanassov A, Tsvetkov T, Alexieva V, Djilianov D. Transgenic tobacco plants accumulating osmolytes show reduced oxidative damage under freezing stress. Plant Physiol Biochem. 2004;42:57–63. doi: 10.1016/j.plaphy.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 51.Goring H, Thien BH. Influence of nutrient deficiency on proline accumulation in the cytoplasm of Zea mays seedlings. Biochem Physiol Pflanz. 1979;17:9–16. [Google Scholar]
- 52.Siripornadulsil S, Traina S, Verma DP, Sayre RT. Molecular mechanisms of proline-mediated tolerance to toxic heavy metals in transgenic microalgae. Plant Cell. 2002;14:2837–2847. doi: 10.1105/tpc.004853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saradhi PP, Alia, Arora S, Prasad KV. Proline accumulates in plants exposed to UV radiation and protects them against UV induced peroxidation. Biochem Biophys Res Commun. 1995;209:1–5. doi: 10.1006/bbrc.1995.1461. [DOI] [PubMed] [Google Scholar]
- 54.Haudecoeur E, Planamente S, Cirou A, Tannieres M, Shelp BJ, Morera S, Faure D. Proline antagonizes GABA-induced quenching of quorum-sensing in Agrobacterium tumefaciens. Proc Natl Acad Sci U S A. 2009;106:14587–14592. doi: 10.1073/pnas.0808005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Savoure A, Jaoua S, Hua XJ, Ardiles W, Van Montagu M, Verbruggen N. Isolation, characterization, and chromosomal location of a gene encoding the delta 1-pyrroline-5-carboxylate synthetase in Arabidopsis thaliana. FEBS Lett. 1995;372:13–19. doi: 10.1016/0014-5793(95)00935-3. [DOI] [PubMed] [Google Scholar]
- 56.Venekamp JH, Koot JTM. The distribution of free amino acids, especially proline, in the organs of field bean plants, Vicia faba L, during development in the field. J Plant Physiol. 1984;116:343–349. doi: 10.1016/S0176-1617(84)80113-3. [DOI] [PubMed] [Google Scholar]
- 57.Stines AP, Naylor DJ, Hoj PB, van Heeswijck R. Proline accumulation in developing grapevine fruit occurs independently of changes in the levels of delta1-pyrroline-5-carboxylate synthetase mRNA or protein. Plant Physiol. 1999;120:923. doi: 10.1104/pp.120.3.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rayapati PJ, Stewart CR. Solubilization of a proline dehydrogenase from maize (Zea mays L.) mitochondria. Plant Physiol. 1991;95:787–791. doi: 10.1104/pp.95.3.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Deuschle K, Funck D, Forlani G, Stransky H, Biehl A, Leister D, van der Graaff E, Kunze R, Frommer WB. The role of Δ1-pyrroline-5-carboxylate dehydrogenase in proline degradation. Plant Cell. 2004;16:3413–3425. doi: 10.1105/tpc.104.023622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rushlow KE, Deutch AH, Smith CJ. Identification of a mutation that relieves gamma-glutamyl kinase from allosteric feedback inhibition by proline. Gene. 1985;39:109–112. doi: 10.1016/0378-1119(85)90115-5. [DOI] [PubMed] [Google Scholar]
- 61.Omori K, Suzuki S, Imai Y, Komatsubara S. Analysis of the mutant proBA operon from a proline-producing strain of Serratia marcescens. J Gen Microbiol. 1992;138:693–699. doi: 10.1099/00221287-138-4-693. [DOI] [PubMed] [Google Scholar]
- 62.Sleator RD, Gahan CG, Hill C. Mutations in the listerial proB gene leading to proline overproduction: effects on salt tolerance and murine infection. Appl Environ Microbiol. 2001;67:4560–4565. doi: 10.1128/AEM.67.10.4560-4565.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kosuge T, Hoshino T. Construction of a proline-producing mutant of the extremely thermophilic eubacterium Thermus thermophilus HB27. Appl Environ Microbiol. 1998;64:4328–4332. doi: 10.1128/aem.64.11.4328-4332.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smith RJ, Downing SJ, Phang JM, Lodato RF, Aoki TT. Pyrroline-5-carboxylate synthase activity in mammalian cells. Proc Natl Acad Sci USA. 1980;77:5221–5225. doi: 10.1073/pnas.77.9.5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu G, Maunoury C, Kamoun P, Aral B. Assignment of the human gene encoding the delta 1-pyrroline-5-carboxylate synthetase (P5CS) to 10q24.3 by in situ hybridization. Genomics. 1996;37:145–146. doi: 10.1006/geno.1996.0535. [DOI] [PubMed] [Google Scholar]
- 66.Valle D, Simell O. Disorders of proline and hydroxyproline metabolism. In: Scriver C, Beaudet A, Sly W, Valle D, editors. The Metabolic and Molecular Bases of Inherited Disease. New York: McGraw Hill; 1995. pp. 1147–1185. [Google Scholar]
- 67.Wu G, Knabe DA, Kim SW. Arginine nutrition in neonatal pigs. J Nutr. 2004;134:2783S–2790S. doi: 10.1093/jn/134.10.2783S. discussion 2796S–2797S. [DOI] [PubMed] [Google Scholar]
- 68.Tadros SF, D'Souza M, Zettel ML, Zhu X, Waxmonsky NC, Frisina RD. Glutamate-related gene expression changes with age in the mouse auditory midbrain. Brain Res. 2007;1127:1–9. doi: 10.1016/j.brainres.2006.09.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 70.Baumgartner MR, Hu CA, Almashanu S, Steel G, Obie C, Aral B, Rabier D, Kamoun P, Saudubray JM, Valle D. Hyperammonemia with reduced ornithine, citrulline, arginine and proline: a new inborn error caused by a mutation in the gene encoding delta(1)-pyrroline-5-carboxylate synthase. Hum Mol Genet. 2000;9:2853–2858. doi: 10.1093/hmg/9.19.2853. [DOI] [PubMed] [Google Scholar]
- 71.Bicknell LS, Pitt J, Aftimos S, Ramadas R, Maw MA, Robertson SP. A missense mutation in ALDH18A1, encoding Delta1-pyrroline-5-carboxylate synthase (P5CS), causes an autosomal recessive neurocutaneous syndrome. Eur J Hum Genet. 2008;16:1176–1186. doi: 10.1038/ejhg.2008.91. [DOI] [PubMed] [Google Scholar]
- 72.Phang JM, Hu CA, Valle D. Disorders of proline and hydroxyproline metabolism. In: Scriver CR, Beaudet AL, Sly WS, Valle D, editors. The Metabolic and Molecular Bases of Inherited Disease. New York: McGraw-Hill; 2001. pp. 1821–1838. [Google Scholar]
- 73.Krishnan N, Dickman MB, Becker DF. Proline modulates the intracellular redox environment and protects mammalian cells against oxidative stress. Free Radic Biol Med. 2008;44:671–681. doi: 10.1016/j.freeradbiomed.2007.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shafqat S, Velaz-Faircloth M, Henzi VA, Whitney KD, Yang-Feng TL, Seldin MF, Fremeau RT., Jr Human brain-specific l-proline transporter: molecular cloning, functional expression, and chromosomal localization of the gene in human and mouse genomes. Mol Pharmacol. 1995;48:219–229. [PubMed] [Google Scholar]
- 75.Phang JM, Donald SP, Pandhare J, Liu Y. The metabolism of proline, a stress substrate, modulates carcinogenic pathways. Amino Acids. 2008;35:681–690. doi: 10.1007/s00726-008-0063-4. [DOI] [PubMed] [Google Scholar]
- 76.Reversade B, Escande-Beillard N, Dimopoulou A, Fischer B, Chng SC, Li Y, Shboul M, Tham PY, Kayserili H, Al-Gazali L, Shahwan M, Brancati F, Lee H, O'Connor BD, Schmidt-von Kegler M, Merriman B, Nelson SF, Masri A, Alkazaleh F, Guerra D, Ferrari P, Nanda A, Rajab A, Markie D, Gray M, Nelson J, Grix A, Sommer A, Savarirayan R, Janecke AR, Steichen E, Sillence D, Hausser I, Budde B, Nürnberg G, Nürnberg P, Seemann P, Kunkel D, Zambruno G, Dallapiccola B, Schuelke M, Robertson S, Hamamy H, Wollnik B, Van Maldergem L, Mundlos S, Kornak U. Mutations in PYCR1 cause cutis laxa with progeroid features. Nat Genet. 2009;41:1016–1021. doi: 10.1038/ng.413. [DOI] [PubMed] [Google Scholar]
- 77.Guernsey DL, Jiang H, Evans SC, Ferguson M, Matsuoka M, Nightingale M, Rideout AL, Provost S, Bedard K, Orr A, Dubé MP, Ludman M, Samuels ME. Mutation in pyrroline-5-carboxylate reductase 1 gene in families with cutis laxa type 2. Am J Hum Genet. 2009;85:120–129. doi: 10.1016/j.ajhg.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Binzel ML, Hasegawa PM, Rhodes D, Handa S, Handa AK, Bressan RA. Solute accumulation in tobacco cells adapted to NaCl. Plant Physiol. 1987;84:1408–1415. doi: 10.1104/pp.84.4.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bremer E. Osmoregulation through accumulation and release of compatible solutes in bacteria. In: Storz G, Hengge-Aronis R, Reinhard K, editors. bacterial Stress Responses. Washington, DC: American Society for Microbiology; 2000. pp. 79–97. [Google Scholar]


