Abstract
SIRT1 is an NAD-dependent deacetylase critically involved in stress responses, cellular metabolism and, possibly, ageing1–15. The tumour suppressor p53 represents the first non-histone substrate functionally regulated by acetylation and deacetylation16,17; we and others previously found that SIRT1 promotes cell survival by deacetylating p53 (refs 4–6). These results were further supported by the fact that p53 hyperacetylation and increased radiation-induced apoptosis were observed in Sirt1-deficient mice10. Nevertheless, SIRT1-mediated deacetylase function is also implicated in p53-independent pathways under different cellular contexts, and its effects on transcriptional factors such as members of the FOXO family and PGC-1α directly modulate metabolic responses1–15. These studies validate the importance of the deacetylase activity of SIRT1, but how SIRT1 activity is regulated in vivo is not well understood. Here we show that DBC1 (deleted in breast cancer 1) acts as a native inhibitor of SIRT1 in human cells. DBC1-mediated repression of SIRT1 leads to increasing levels of p53 acetylation and upregulation of p53-mediated function. In contrast, depletion of endogenous DBC1 by RNA interference (RNAi) stimulates SIRT1-mediated deacetylation of p53 and inhibits p53-dependent apoptosis. Notably, these effects can be reversed in cells by concomitant knockdown of endogenous SIRT1. Our study demonstrates that DBC1 promotes p53-mediated apoptosis through specific inhibition of SIRT1.
To understand the regulation of SIRT1-mediated deacetylation in vivo, biochemical purification was used to identify cellular factors that stably interact with SIRT1. We isolated physiologically formed protein complexes containing SIRT1 from cell extracts of native HeLa cells by conducting affinity chromatography with affinity-purified antisera raised against the carboxy (C) terminus (amino acids 480–737) of SIRT1 (Supplementary Fig. 1a). As expected, we identified SIRT1 as the major component of the complexes, but several protein bands were also co-purified with SIRT1. Mass spectrometry of a prominent protein band of approximately 130 kilodaltons (kDa) from the SIRT1 complexes revealed peptide sequences corresponding to the DBC1 protein (Supplementary Fig. 1b, Gi: 24432106). The DBC1 gene was initially identified as it is localized to a region of chromosome 8p21 that was homozygously deleted in human breast cancer; however, the molecular function of DBC1 is poorly understood18,19.
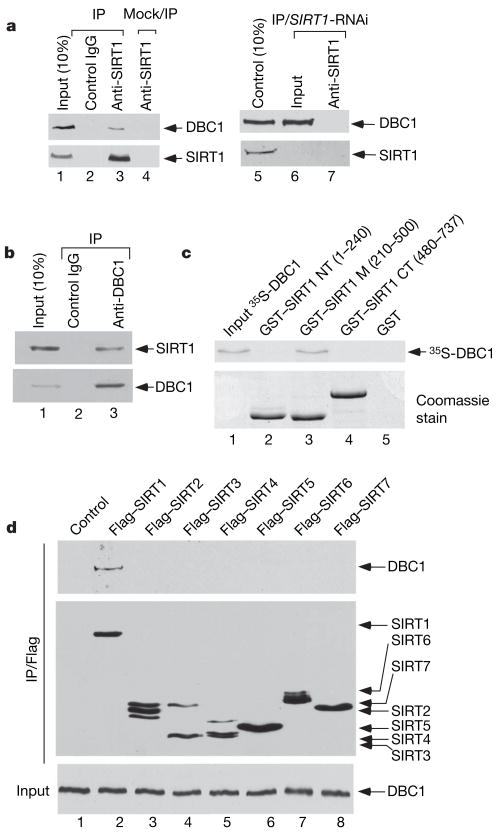
To examine the interaction between endogenous DBC1 and SIRT1, cell extracts from human osteosarcoma U2OS cells were immunoprecipitated with the anti-SIRT1 antibody or with the control IgG. As expected, western blot analysis revealed that DBC1 was clearly detected in the immunoprecipitations obtained with the anti-SIRT1 antiserum (lane 3, Fig. 1a) but not with the control antibody (lane 2). Previous studies indicate that HIC1 can also interact with SIRT1 (ref. 11); nevertheless, we failed to detect a strong interaction between HIC1 and SIRT1 in these cells under the same conditions. To prove the specificity of the SIRT1 antibody, we performed the co-immunoprecipitation in SIRT1-depleted U2OS cells treated with SIRT1-specific RNAi (lanes 6 and 7). Indeed, we failed to detect any DBC1 from the anti-SIRT1 immunoprecipitates with these SIRT1-depleted cells (lane 7). We also performed a reciprocal co-immunoprecipitation assay. As shown in Fig. 1b, endogenous SIRT1 was readily immunoprecipitated with the DBC1-specific antibody (lane 3), but not with a control antibody (lane 2).
Figure 1. DBC1 interacts with SIRT1 in vivo and in vitro.
a, Co-immunoprecipitation of DBC1 with SIRT1. Western blot analysis of U2OS whole-cell extracts and SIRT1-antibody, IgG and mock IP (lane 4) immunoprecipitates by DBC1 and SIRT1-antibody (left panel), and after SIRT1 RNAi knockdown (right panel). b, Co-immunoprecipitation of SIRT1 by DBC1 from U2OS whole-cell extracts with DBC1 or IgG analysed by western blot. c, Direct interaction of DBC1 with GST–SIRT1-core (m) domain in in vitro GST pull-down assay detected by autoradiography. d, DBC1 interaction with Sirtuin family in vivo. 293 cells were transfected with Flag–SIRT1–7, and extracts (bottom panel) and M2-immunoprecipitates (upper panels) were analysed by western blot.
Next, we tested whether SIRT1 binds DBC1 in vitro. As shown in Fig. 1c, 35S-labelled in-vitro-translated DBC1 bound the central core domain of SIRT1 (lane 3) but showed no affinity for either its amino (N)-terminal (lane 2) or C-terminal (lane 4) domains. Conversely, we identified the N terminus of DBC1 as the SIRT1-binding domain (Supplementary Fig. 3). Because the enzymatic core sequence represents the most conserved region within the mammalian SIRT protein family, we examined whether DBC1 interacts with other members of this family. Thus, Flag-tagged derivatives of the seven human SIRT polypeptides (SIRT1–7) were each expressed in 293 cells and extracts of the transfected cells were immunoprecipitated with the anti-Flag antibody. Western blots revealed that endogenous DBC1 was clearly detected in the immunoprecipitates of Flag–SIRT1 (lane 2, Fig. 1d). Although similar expression levels for all seven Flag–SIRT polypeptides were detected, none of the other SIRT proteins (SIRT2–7) were able to co-immunoprecipitate DBC1 (lanes 3–8, Fig. 1d). These results demonstrate the specificity of the SIRT1 and DBC1 interaction.
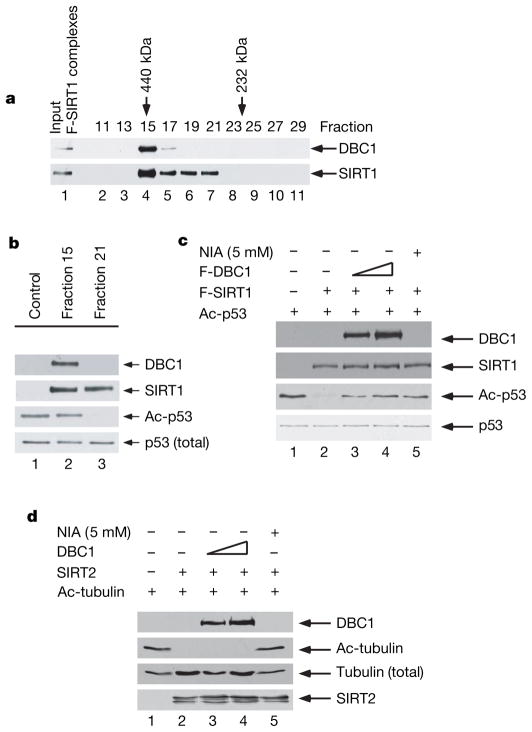
Interestingly, when purified Flag–SIRT1 complexes from human cells were analysed by gel-filtration chromatography on a Superose 12 column, we observed that SIRT1 and DBC1 polypeptides co-eluted in fraction 15 with an apparent molecular mass of 440 kDa (lane 4, Fig. 2a). In contrast, a DBC1-free form of SIRT1 eluted in fractions 19–21 (lanes 6 and 7), suggesting that at least two distinct SIRT1 complexes exist in human cells. As expected, we found that SIRT1 from fraction 21 had a strong NAD-dependent deacetylase activity for p53 (lane 3, Fig. 2b). Surprisingly, however, no activity was detected with fraction 15 (lane 2), raising the notion that SIRT1-mediated deacetylation is inhibited by additional factors in the complexes, such as DBC1. To evaluate a role for DBC1 in regulating SIRT1 function, we examined whether DBC1 can inhibit the deacetylase activity of SIRT1 in a purified system. To this end, Flag-tagged forms of SIRT1 and DBC1 were purified under high-stringency conditions for in vitro deacetylation assays. As indicated in Fig. 2c, deacetylation of p53 was observed when the Flag–SIRT1 protein was incubated with acetylated p53 (lane 2). However, this activity was strongly repressed by Flag–DBC1 in a dose-dependent manner (lanes 3 and 4). DBC-mediated repression is apparently as potent as the effects obtained with 5 mM of nicotinamide (NIA) (lane 5), a known inhibitor of SIRT1-mediated deacetylation4.
Figure 2. DBC1 inhibits SIRT1-mediated deacetylation of p53.
a, M2 purified Flag-SIRT1 (F-SIRT1) complexes from 293 cells were fractioned by size-exclusion chromatography and analysed by western blot as indicated. b, In vitro p53 deacetylation by SIRT1 fractions with or without DBC1. c, DBC1 inhibition of SIRT1-mediated deacetylation is dose dependent. Increasing amounts of purified F-DBC1 (lanes 2–4) or nicotinamide (lane 5) can inhibit highly purified F-SIRT1-mediated p53 deacetylation. d, DBC1 does not inhibit SIRT2-mediated tubulin deacetylation in vitro. Increasing amounts of purified F-DBC1 (lanes 2–4) do not inhibit purified F- SIRT2-mediated tubulin deacetylation. Deacetylation assays were analysed by western blot with the indicated antibodies.
Moreover, to prove the specificity of DBC-mediated inhibition of SIRT1 deacetylase activity, we examined the effect of DBC1 on SIRT2-mediated deacetylation of tubulin. As shown in Fig. 2d, deacetylation of tubulin was observed when the purified SIRT2 protein was incubated with acetylated tubulin, as previously reported20. This activity was also inhibited by nicotinamide (lane 5); however, tubulin deacetylation by SIRT2 was not affected by purified DBC1 polypeptides (lanes 3 and 4). Finally, p53 could also be deacetylated by purified HDAC1 complexes, as we have previously shown17 (lane 2, Supplementary Fig. 4); nevertheless, this deacetylase activity was not repressed by DBC1 (lanes 3 and 4). Thus, these data demonstrate that DBC1-mediated inhibitory effects specifically act on SIRT1 deacetylase activity.
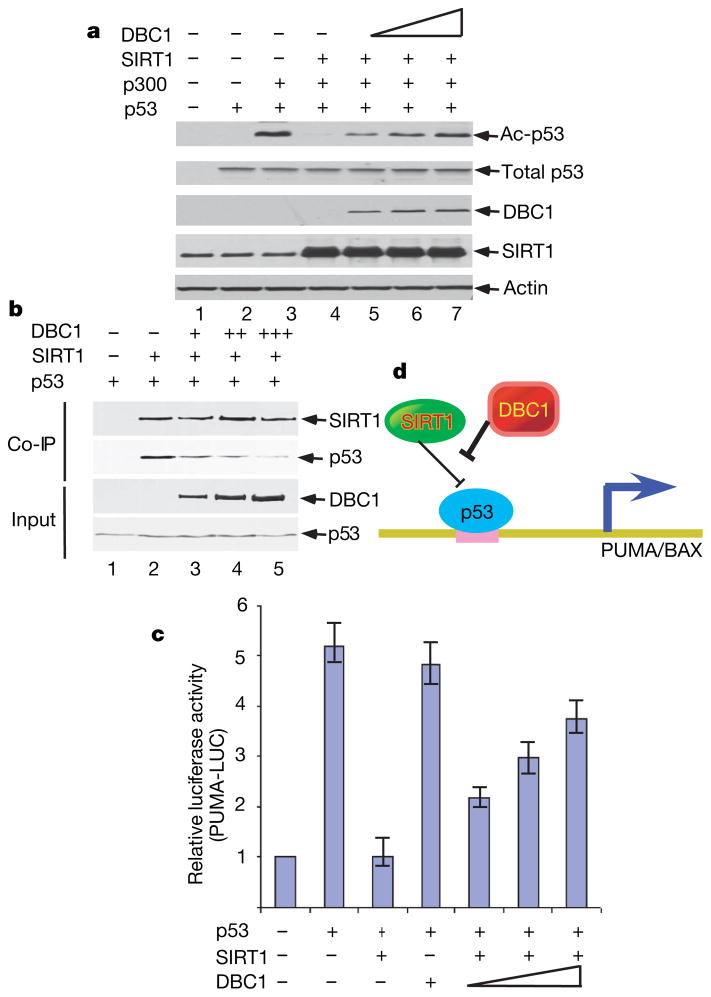
We then tested whether DBC1 expression rescues p53 from SIRT1-mediated deacetylation and repression in human cells. As expected, co-expression of SIRT1 induced p53 deacetylation (lane 4, Fig. 3a); however, the steady-state levels of acetylated p53 were restored by DBC1 expression in a dose-dependent manner (lanes 5–7). To elucidate the mechanism of DBC-mediated effects on SIRT1, we conducted a co-immunoprecipitation assay to test whether the interaction between SIRT1 and p53 is regulated by DBC1. As shown in Fig. 3b, p53 was co-immunoprecipitated with SIRT1 (lane 2). Notably, the p53–SIRT1 interaction was significantly abrogated by DBC1 expression in a dose-dependent fashion (lanes 3–5). These results suggest that DBC1 represses SIRT1 activity in human cells and that these effects may act, in part, through blocking the interactions between SIRT1 and substrates (p53).
Figure 3. DBC1 acts as an inhibitor of SIRT1 in human cells.
a, DBC1 represses SIRT1 deacetylation activity in vivo. Transfected H1299 cell extracts were analysed by western blot using antibodies as shown. b, DBC1 inhibits SIRT1-mediated co-immunoprecipitation of p53 in vivo. H1299 cells were transfected as indicated and Flag–SIRT1 immunoprecipitations were analysed by western blot as shown. c, DBC1 expression rescues SIRT1-mediated repression of p53 transcriptional activation. H1299 cells were transfected with the PUMA-luciferase promoter construct and p53, SIRT1 and DBC1 as indicated. Lysates were assayed for the dual-luciferase activity. Error bars represent s.d., n = 3. d, Model showing DBC1 acting as an inhibitor of SIRT1-mediated repression of p53.
To explore the functional consequences of these interactions further, we tested whether DBC1 can influence SIRT1-mediated repression of p53 transcriptional activation. As shown in Fig. 3c, SIRT1 strongly suppressed p53-mediated transactivation of the PUMA reporter in a luciferase assay. Again, this SIRT1-mediated suppression was abrogated by DBC1 expression in a dose-dependent manner. These data indicate that DBC1 can enhance p53-dependent transactivation of PUMA by inhibiting SIRT1. Because homozygous deletion of the DBC1 gene was reported in human cancers18,21, inactivation of DBC1 may enhance the deacetylase activity of SIRT1 and thereby lead to inhibition of p53 function (Fig. 3d).
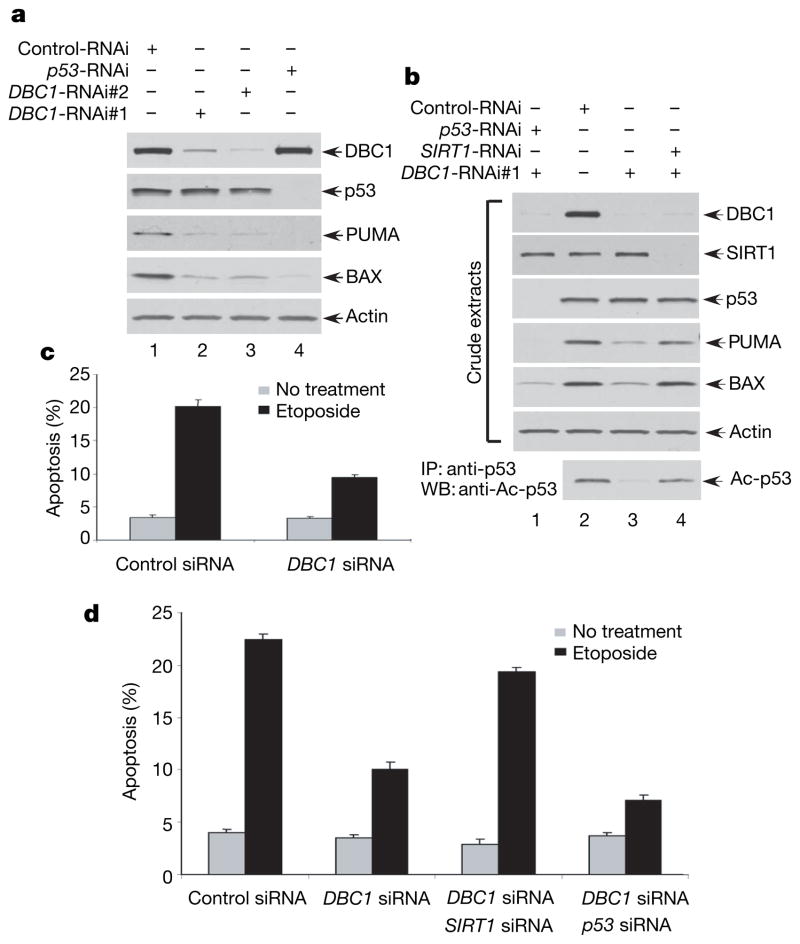
To test the above hypothesis, we first examined whether short interfering RNA (siRNA)-mediated knockdown of endogenous DBC1 has any effect on p53 function. To avoid possible off-target effects caused by the DBC1 RNAi, we used two different RNAi sequences that target different regions of the DBC1 messenger RNA (mRNA). Thus, human osteosarcoma U2OS cells were transfected with the DBC1-specific siRNA#1 (DBC1-RNAi#1), DBC1-specific siRNA#2 (DBC1-RNAi#2) or a control siRNA (Control-RNAi). As shown in Fig. 4a, RNAi-mediated knockdown of DBC1 expression had no obvious effect on p53 stability (lanes 2 and 3) but significantly reduced the expression levels of PUMA and BAX, two major transcriptional targets of p53. As expected, knockdown of p53 expression by p53-specific siRNA (p53-RNAi) completely abolished the expression of both PUMA and BAX (lane 4), validating that expression of these two targets is indeed p53-dependent. These experiments demonstrate that inactivation of endogenous DBC1 leads to down-regulation of p53 activity.
Figure 4. siRNA-mediated knockdown of DBC1 reduces p53 acetylation and its transcriptional and apoptotic activities.
a, siRNA-mediated DBC1 knockdown inhibits p53 proapoptotic target gene expression. Western blot of U2OS whole-cell extracts treated with siRNA as shown. b, DBC1 siRNA effects on p53-dependent PUMA and BAX expression require SIRT1. Western blot of siRNA-treated U2OS cell extracts. p53 acetylation levels are shown after p53 immunoprecipitation. c, d, DBC1 siRNA inhibits p53-mediated apoptosis after DNA damage. U2OS cells were treated with siRNA as indicated and treated with Etoposide (30 h) before assaying apoptosis by TUNEL staining (c, Supplementary Fig. 11) or Annexin V staining (d, Supplementary Fig. 12). Error bars represent s.d., n = 3 (c, d).
Moreover, to demonstrate that DBC1 acts on p53 by repressing SIRT1 deacetylase activity, we tested whether inactivation of DBC1 indeed reduces acetylation levels of endogenous p53 by SIRT1 and, more importantly, whether these effects are reversed by inactivation of SIRT1 expression. Thus, these cells were transfected with the DBC1-specific siRNA#1 (DBC1-RNAi#1), SIRT1-specific siRNA (SIRT1-RNAi) or a control siRNA (Control-RNAi). As shown in Fig. 4b, RNAi-mediated knockdown of DBC1 expression significantly reduced the acetylation levels of endogenous p53 (Ac-p53, bottom panel, lane 3). Notably, the reduction of p53 acetylation was completely reversed by concomitant knockdown of SIRT1 (Ac-p53, bottom panel, lane 4). Similar results were also observed with DBC1-mediated effects on PUMA and BAX by concomitant knockdown of SIRT1 (PUMA and BAX, middle panel, lanes 3 and 4). Thus our data demonstrate that DBC1-mediated effects on p53 activation act mainly through SIRT1 in vivo.
To investigate the role of DBC1 in the stress response, we tested whether inactivation of DBC1 leads to inhibition of p53-dependent apoptosis on DNA damage. For this, U2OS cells were first transfected with either control or DBC1-specific siRNAs and then exposed to etoposide. Thirty hours later, the cells were stained with DAPI and apoptosis was examined by TUNEL staining. The DBC1-depleted cells were highly resistant to apoptosis, displaying only 8.8% apoptotic cells compared with 20.5% of cells transfected with the control siRNA (Fig. 4c and Supplementary Fig. 11). To confirm further the role of DBC1 in regulating p53-mediated apoptosis, we performed an apoptosis assay by using Annexin V staining followed by flow-cytometry analysis. Again, p53-mediated apoptosis was repressed in DBC1 knockdown cells (Fig. 4d and Supplementary Fig. 12). As expected, inactivation of p53 in these cells completely abolished the apoptotic response by DNA damage. Notably, concomitant knockdown of SIRT1 reversed the inhibitory effects on p53-dependent apoptosis by DBC inactivation. These data demonstrate that DBC1 is critically involved in regulating the p53-mediated apoptotic response by repressing SIRT1 function.
Our findings may have significant implications in the treatment of both metabolic-related disorders and cancer. Small molecule inhibitors of the SIRT1 deacetylase have been proposed as novel anticancer agents22–24, presumably through activating the apoptotic response in cancer cells. On the other hand, activation of SIRT1 in mice also protects them against diet-induced obesity and insulin resistance, mainly through regulating metabolic pathways25,26. Although a role for mammalian SIRT1 in the regulation of lifespan has not been directly determined, the ability of resveratrol, a chemical found in red wine and other foods, to enhance SIRT1 activity and increase lifespan in lower organisms supports the feasibility of this approach in mammals3,27–30. Therefore, both inhibitors and activators of SIRT1 could be therapeutically beneficial by affecting different SIRT1-mediated regulatory pathways. It will be intriguing to know whether DBC1 has differential effects in regulating apoptotic responses versus metabolic pathways and whether manipulations of the DBC1–SIRT1 interaction will help to find more potent activators and/or inhibitors for SIRT1 activity.
METHODS SUMMARY
H1299, U2OS, 293 and HeLa cells were maintained in DMEM medium supplemented with 10% fetal bovine serum. H1299 and 293 cells were transfected with plasmid DNA by the calcium phosphate protocol or the siRNA RNA duplex, as indicated in the relevant figures, by Lipofectamine2000 according to the manufacturer’s protocol. In vitro deacetylation assays were performed as previously described. Purified acetylated p53 was incubated with purified SIRT1 and DBC1 as indicated at 30 °C for 1 h in the presence of 50 μM NAD, and reactions were resolved by SDS–polyacrylamide gel electrophoresis (SDS–PAGE) and analysed by western blot. To immunoprecipitate the ectopically expressed Flag-tagged proteins, transfected cells were lysed 24 h post-transfection in Flag-lysis buffer and precipitated using anti-Flag monoclonal antibody-conjugated M2 agarose beads. The eluted material was resolved by SDS–PAGE and detected by antibodies as indicated. The endogenous SIRT1 complex was purified from HeLa cells by using the SIRT1-CT-specific antibody for immunoprecipitation. Interacting proteins were identified by mass spectrometry. To analyse the SIRT1 complex, 50 μl of M2-eluted Flag-SIRT1 containing approximately 12.5 μg of total purified Flag-SIRT1 was fractionated by size-exclusion chromatography. GST pull-downs were performed as previously described, using recombinant fragments of GST–SIRT1 or GST–control to pull-down 35S-methionine-labelled DBC1. Direct in vitro interaction was detected by autoradiation after the reactions were resolved by SDS–PAGE. Dual luciferase reporter assays to determine transcriptional activity were performed in H1299 cells according to the manufacturer’s guidelines 24 h post-transfection. The ablation of DBC1 was performed by transfection of the U2OS cells with either of two siRNA duplex oligonucleotides which covered mRNA regions of nucleotides 582–602 (amino acids 55–61) and nucleotides 2097–2115 (amino acids 560–565) of DBC1, respectively, by using Lipofectamine2000 according to the manufacturer’s protocol. SIRT1 RNAi, p53 RNAi and Control RNAi were used and transfected according to the manufacturer’s guidelines. Annexin V-FITC and TUNNEL staining were performed according to the manufacturer’s guidelines.
METHODS
Cell culture and transfections
H1299, U2OS, 293 and HeLa cells were maintained in DMEM medium supplemented with 10% fetal bovine serum. H1299 and 293 cells were transfected with the plasmid DNA by the calcium phosphate protocol. U2OS cells were transfected with siRNA duplexes by Lipofectamine2000 (Invitrogen) according to the manufacturer’s protocol.
In vitro deacetylation assays
In vitro deacetylation assays were performed as previously described. Purified acetylated p53 was incubated with purified SIRT1 and DBC1 as indicated at 30 °C for 1 h in the presence of 50 μM NAD. Reactions were performed in a buffer containing 50 mM Tris-HCl (pH 9.0), 50 mM NaCl, 4 mM MgCl2, 0.5 mM DTT, 0.2 mM PMSF, 0.02% NP-40 and 5% glycerol. Nicotinamide (Sigma) was added to 5 mM as indicated to inhibit all deacetylase activity of the Sirtuins. The reactions were resolved on SDS–PAGE and analysed by western blot using antibodies specific for acetylated p53, total p53 (DO-1, sc-126, Santa Cruz), Sir2-CT and DBC1 (Bethyl, BL1924).
Immunoprecipitation and immunoblot
Cells were transfected with Flag-tagged expression constructs for p53, SIRT1 and DBC1 using the calcium phosphate method as previously described. To immunoprecipitate the ectopically expressed Flag-tagged proteins, transfected cells were lysed 24 h post-transfection in Flag-lysis buffer (50 mM Tris-HCl pH 7.9, 137 mM NaCl, 10 mM NaF, 1 mM EDTA, 1% Triton X-100, 0.2% Sarkosyl, 10% glycerol and fresh proteinase inhibitor cocktail (Sigma)) or for high stringency in BC500 (20 mM Tris pH 7.9, 500 mM NaCl, 10% glycerol, 0.2 mM EDTA, 0.5% Triton X-100 and fresh proteinase inhibitor cocktail). The whole-cell extracts were immunoprecipitated with the monoclonal anti-Flag antibody-conjugated M2 agarose beads (Sigma) at 4 °C overnight. After three washes with either BC500 or Flag-lysis buffer, followed by two washes with BC100 (20 mM Tris pH 7.9, 100 mM NaCl, 10% glycerol, 0.2 mM EDTA, 0.1% Triton X-100), the bound proteins were eluted using Flag-Peptide (Sigma)/BC100 for 3 h at 4 °C. The eluted material was resolved by SDS–PAGE and detected by antibodies as indicated. The endogenous SIRT1 complex was purified from HeLa cells with the SIRT1-CT-specific antibody for immunoprecipitation. Interacting proteins were identified by mass spectrometry. For analysis of the SIRT1 complex, 50 μl of M2-eluted Flag-SIRT1 containing approximately 12.5 μg of total purified Flag-SIRT1 was fractionated by size exclusion chromatography on a Sepharose 12 Column on the SMART System (GE Healthcare) according to the manufacturer’s protocol. To co-immunoprecipitate SIRT1 and DBC1 with specific antibodies, cells were lysed in BC100 and cell lysates were first incubated with 20 μl protein G agarose beads for 2 h with gentle rotation. The cleared supernatants were incubated with DBC1- or SIRT1-specific antibodies overnight before addition of 20 μl of protein G agarose beads for 4 h. After four washes with the lysis buffer, the immunoprecipitated materials were eluted with the SDS sample buffer with boiling, resolved by SDS–PAGE and detected with antibodies as indicated.
GST pull-down
pCIN4-DBC1 or pCIN4-SIRT1 were labelled by incorporation of 35S-labelled methionine during in vitro translation (TNT Coupled Reticulocyte Lysate System, Promega Corporation).
Five microlitres of 35S-labelled protein was incubated with 3 μg of the purified GST protein fragments as indicated in the presence of 0.2% BSA in BC100 on a rotator overnight at 4 °C. The proteins were pulled down using GST beads, and the beads were washed five times with BC100 before elution with 50 μl of BC100 plus 20 mM reduced glutathione for 2 h with gentle rotation. Eluted materials were resolved on SDS–PAGE. The presence of 35S-labelled protein was detected by autoradiography and the levels of the GST proteins by Coomassie blue stain.
siRNA-mediated ablation of DBC1, SIRT1 and p5
The ablation of DBC1 was performed by transfection of the U2OS cells with either of two siRNA duplex oligonucleotides (DBC1-RNAi#1: 5′-CAGCGGGUCUUCACUGGUAUU-3′; or DBC1-RNAi#2: 5′-CUACUGAGCCUUCCUGAAUU-3′ (synthesized by Dharmacon)), which covered mRNA regions of nucleotides 582–602 (amino acids 55–61) and nucleotides 2097–2115 (amino acids 560–565) of DBC1, respectively, by using Lipofectamine2000 according to the manufacturer’s protocol. SIRT1 RNAi (SiGenome Smartpool M-003540-01 (Dharmacon)), p53 RNAi (SiGenome Smartpool M-003329-01-0010 (Dharmacon)) and Control RNAi (On-target plus siControl non-targeting pool D-001810-10-20 (Dharmacon)) were used and transfected according to the manufacturer’s guidelines.
Luciferase reporter gene assay
H1299 cells were transfected at 70% confluence in six-well plates with plasmid DNA as indicated in the relevant figures. After incubation for 24 h, cells were harvested and the luciferase activity was measured by the Dual Luciferase Reporter Assay System Kit from Promega according the manufacturer’s protocol. Activity was assayed in three separate experiments and shown as the mean ± s.d.
In vitro acetylation assay
In vitro acetylation assays were performed as described previously16. Recombinant GST–p53 was incubated with Flag-purified SIRT1 and DBC1 as indicated in the presence of 14C-labelled acetyl-CoA in reaction buffer containing 50 mM HEPES (pH 8.0), 10% glycerol, 1 mM DTT, 1 mM PMSF and 10 mM sodium butyrate. Reactions were incubated at 30 °C for 1 h and resolved on SDS–PAGE. Protein levels were detected by Coomassie blue staining, and in vitro acetylation was detected by autoradiography.
Immunofluorescent staining
Cells were fixed with 4% paraformaldehyde for 20 min on ice, rehydrated for 5 min in serum-free DMEM, and permeabilized with 0.2% Triton X-100 (Fisher) for 10 min on ice. Cells were incubated in 1% bovine serum albumin (BSA) (Sigma)/phosphate buffered salt solution (PBS) (Cellgro) for 30 min. Primary antibodies (as indicated) were added in 1% BSA/PBS for 45 min at room temperature. After washing with 1% BSA/PBS, secondary antibodies were added and incubated for 30 min at room temperature. Finally, cells were counterstained with DAPI to visualize the nuclei essentially as described before.
Annexin V-FITC staining
The apopotosis assay was performed with the BD-Bioscience Annexin V-FITC staining kit according to the manufacturer’s protocol. After siRNA-mediated knockdown, U2OS cells were treated with Etoposide (30 h) before fixing and TUNEL staining or Annexin V assays. Apoptoses observed in TUNEL and Annexin V assays were quantified for three separate experiments and presented as the mean ± s.d.
Supplementary Material
Acknowledgments
We thank R. Baer for suggestions on this manuscript. We also thank E. White, M. Wigler and E. Verdin for reagents. This work was supported in part by Ellison Medical Foundation and grants from the National Institutes of Health/National Cancer Institute to J.Q. and W.G.
Footnotes
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Information Reprints and permissions information is available at www.nature.com/reprints.
References
- 1.Bordone L, Guarente L. Calorie restriction, SIRT1 and metabolism: understanding longevity. Nature Rev Mol Cell Biol. 2005;6:298–305. doi: 10.1038/nrm1616. [DOI] [PubMed] [Google Scholar]
- 2.North BJ, Verdin E. Sirtuins: Sir2-related NAD-dependent protein deacetylases. Genome Biol. 2004;5:224.1–224.12. doi: 10.1186/gb-2004-5-5-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baur JA, Sinclair DA. Therapeutic potential of resveratrol: the in vivo evidence. Nature Rev Drug Discov. 2006;5:493–506. doi: 10.1038/nrd2060. [DOI] [PubMed] [Google Scholar]
- 4.Luo J, et al. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 5.Langley E, et al. Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. EMBO J. 2002;21:2383–2396. doi: 10.1093/emboj/21.10.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vaziri H, et al. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 7.Motta MC, et al. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116:551–563. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- 8.Brunet A, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 9.Kitamura YI, et al. FoxO1 protects against pancreatic beta cell failure through NeuroD and MafA induction. Cell Metab. 2005;2:153–163. doi: 10.1016/j.cmet.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 10.Cheng HL, et al. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci USA. 2003;100:10794–10799. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen WY, et al. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell. 2005;123:437–448. doi: 10.1016/j.cell.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 12.Yeung F, et al. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004;23:2369–2380. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greene WC, Chen LF. Regulation of NF-kappaB action by reversible acetylation. Novartis Found Symp. 2004;259:208–217. discussion 218–225. [PubMed] [Google Scholar]
- 14.Rodgers JT, et al. Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 15.Cohen HY, et al. Acetylation of the C terminus of Ku70 by CBP and PCAF controls Bax-mediated apoptosis. Mol Cell. 2004;13:627–638. doi: 10.1016/s1097-2765(04)00094-2. [DOI] [PubMed] [Google Scholar]
- 16.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 17.Luo J, Su F, Chen D, Shiloh A, Gu W. Deacetylation of p53 modulates its effect on cell growth and apoptosis. Nature. 2000;408:377–381. doi: 10.1038/35042612. [DOI] [PubMed] [Google Scholar]
- 18.Hamaguchi M, et al. DBC2, a candidate for a tumor suppressor gene involved in breast cancer. Proc Natl Acad Sci USA. 2002;99:13647–13652. doi: 10.1073/pnas.212516099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sundararajan R, Chen G, Mukherjee C, White E. Caspase-dependent processing activates the proapoptotic activity of deleted in breast cancer-1 during tumor necrosis factor-alpha-mediated death signaling. Oncogene. 2005;24:4908–4920. doi: 10.1038/sj.onc.1208681. [DOI] [PubMed] [Google Scholar]
- 20.North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell. 2003;11:437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- 21.Knowles MA, Aveyard JS, Taylor CF, Harnden P, Bass S. Mutation analysis of the 8p candidate tumour suppressor genes DBC2 (RHOBTB2) and LZTS1 in bladder cancer. Cancer Lett. 2005;225:121–130. doi: 10.1016/j.canlet.2004.10.047. [DOI] [PubMed] [Google Scholar]
- 22.Heltweg B, et al. Antitumor activity of a small-molecule inhibitor of human silent information regulator 2 enzymes. Cancer Res. 2006;66:4368–4377. doi: 10.1158/0008-5472.CAN-05-3617. [DOI] [PubMed] [Google Scholar]
- 23.Olaharski AJ, et al. The flavoring agent dihydrocoumarin reverses epigenetic silencing and inhibits sirtuin deacetylases. PLoS Genet. 2005;1:e77. doi: 10.1371/journal.pgen.0010077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mai A, et al. Design, synthesis, and biological evaluation of sirtinol analogues as class III histone/protein deacetylase (Sirtuin) inhibitors. J Med Chem. 2005;48:7789–7795. doi: 10.1021/jm050100l. [DOI] [PubMed] [Google Scholar]
- 25.Lagouge M, et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 26.Baur JA, et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–342. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Howitz KT, et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 28.Kaeberlein M, et al. Substrate-specific activation of sirtuins by resveratrol. J Biol Chem. 2005;280:17038–17045. doi: 10.1074/jbc.M500655200. [DOI] [PubMed] [Google Scholar]
- 29.Borra MT, Smith BC, Denu JM. Mechanism of human SIRT1 activation by resveratrol. J Biol Chem. 2005;280:17187–17195. doi: 10.1074/jbc.M501250200. [DOI] [PubMed] [Google Scholar]
- 30.Viswanathan M, Kim SK, Berdichevsky A, Guarente L. A role for SIR-2.1 regulation of ER stress response genes in determining C. elegans life span. Dev Cell. 2005;9:605–615. doi: 10.1016/j.devcel.2005.09.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.