Abstract
Like phosphorylation, the addition of O-linked β-N-acetylglucosamine (O-GlcNAcylation) is a ubiquitous, reversible process that modifies serine and threonine residues on nuclear and cytoplasmic proteins. Overexpression of the enzyme that adds O-GlcNAc to target proteins, O-GlcNAc transferase (OGT), perturbs cytokinesis and promotes polyploidy, but the molecular targets of OGT that are important for its cell cycle functions are unknown. Here, we identify 141 previously unknown O-GlcNAc sites on proteins that function in spindle assembly and cytokinesis. Many of these O-GlcNAcylation sites are either identical to known phosphorylation sites or in close proximity to them. Furthermore, we found that O-GlcNAcylation altered the phosphorylation of key proteins associated with the mitotic spindle and midbody. Forced overexpression of OGT increased the inhibitory phosphorylation of cyclin-dependent kinase 1 (CDK1) and reduced the phosphorylation of CDK1 target proteins. The increased phosphorylation of CDK1 is explained by increased activation of its upstream kinase, MYT1, and by a concomitant reduction in the transcript for the CDK1 phosphatase, CDC25C. OGT overexpression also caused a reduction in both messenger RNA expression and protein abundance of Polo-like kinase 1, which is upstream of both MYT1 and CDC25C. The data not only illustrate the crosstalk between O-GlcNAcylation and phosphorylation of proteins that are regulators of crucial signaling pathways, but also uncover a mechanism for the role of O-GlcNAcylation in regulation of cell division.
INTRODUCTION
Over the past two decades, extensive work has not only characterized proteins involved in formation of the mitotic spindle, a microtubule-based structure responsible for separating the chromosomes into the two daughter cells, and the midbody, a microtubule-based structure that transiently forms at the site where cytokinesis will occur, but also has defined the signaling cascades regulating these proteins (1–4). Large-scale phosphoproteomic studies on mitotic extracts have identified hundreds of mitotic phosphorylation sites, characterized consensus sequences of mitotic kinases, and defined the timing of phosphorylation events (2, 3, 5). Protein phosphorylation and dephosphorylation cascades created by mitotic kinases, cyclin-dependent kinase 1 (CDK1), Polo-like kinase 1 (PLK1), Aurora kinase B (AURKB), and the phosphatases, protein phosphatase 1 (PP1) and CDC25, control many aspects of mitosis (6–9). However, other common posttranslational modifications may also regulate key steps in mitosis. Although modification of proteins with O-linked β-N-acetylglucosamine (O-GlcNAc) is often overlooked because of the technical difficulty in detection, it rivals phosphorylation in both abundance and distribution of the protein targets for this modification (10–14). Like phosphorylation, O-GlcNAcylation is a reversible modification of nuclear and cytoplasmic proteins and consists of the attachment of a single β-N-acetyl-glucosamine moiety to hydroxyl groups of serine or threonine residues. Modification by O-GlcNAcylation is often competitive with phosphorylation at the same sites or at proximal sites on proteins (10). Furthermore, crosstalk between O-GlcNAcylation and phosphorylation affects the posttranslational state of hundreds of proteins in response to nutrients and stress (15) and plays an important role in chronic diseases of metabolism, such as diabetes and neurodegeneration.
O-GlcNAc transferase (OGT) catalyzes the addition of the sugar moiety from the donor substrate uridine 5′-diphosphate (UDP)–GlcNAc to proteins (10). During M phase, OGT localizes to discrete structures, such as centrosomes (metaphase) and the spindle (anaphase), and then moves to the midbody during cytokinesis (16). OGT, along with O-GlcNAcase (OGA), the enzyme that removes the sugar, dynamically interacts with AURKB and PP1 at the midbody (17). Together, these proteins form a complex regulating M-phase O-GlcNAcylation, which in turn influences the phosphorylation state, of vimentin (17). However, the identity of other OGT mitotic substrates is currently not known.
Peptides modified with O-GlcNAc are difficult to detect by standard mass spectrometric methods (10). The modification is usually present at substoichiometric amounts, modified and unmodified peptides co-elute during high-performance liquid chromatography (HPLC), and ionization of the modified peptide is suppressed in the presence of unmodified peptides. Consequently, sample enrichment is required to successfully detect and characterize O-GlcNAcylated peptides. Enrichment may be achieved through chemoenzymatic approaches that biotinylate O-GlcNAc peptides and capture them by avidin chromatography (12, 18). We improved on this chemoenzymatic approach using a photocleavable biotin-alkyne reagent (PC-biotin-alkyne) tag (19) (fig. S1A). Photocleavage not only allows efficient and quantitative recovery from the affinity column, but also tags the peptide with a charged moiety that facilitates O-GlcNAc site mapping by electron-transfer dissociation (ETD) mass spectrometry (20). This tagging approach also makes it possible to use conventional collision-activated dissociation mass spectrometry (CAD MS) to screen samples for the presence of O-GlcNAc–modified peptides by monitoring for two-signature fragment ions characteristic of the tag (fig. S1B).
Here, we use a combined glycoproteomic and phosphoproteomic approach to characterize O-GlcNAcylation and phosphorylation sites on spindle and midbody proteins of cells collected during cytokinesis. We also quantify changes in the occupancy for these sites that occur during defective cytokinesis caused by overexpression of OGT. Our findings confirm that O-GlcNAcylation rivals phosphorylation in both abundance and distribution of the modified proteins and demonstrate that alterations in O-GlcNAcylation disrupt both the chromosomal passenger complex, containing AURKB, INCENP, PP1, Borealin, and Surviven (21), and the circuits regulating CDK1 activity.
RESULTS
O-GlcNAcylated proteins are abundant at the mitotic spindle and midbody
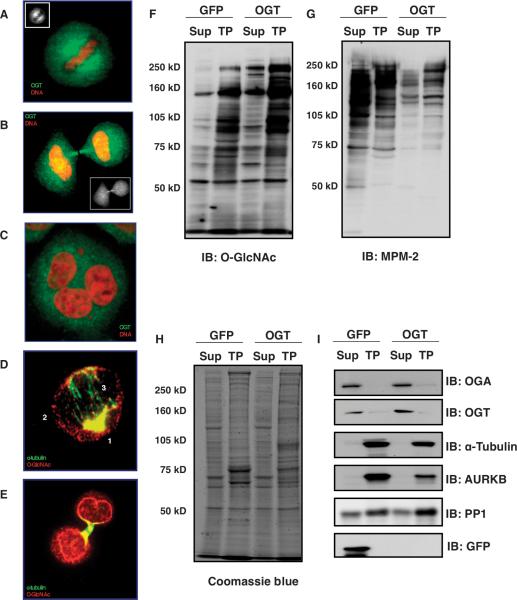
OGT localizes to mitotic spindles and midbodies during M phase (Fig. 1, A and B) (16). HeLa cells forced to overexpress OGT exhibited a substantial increase in polyploidy due to defective cytokinesis (Fig. 1C) (16). To determine how OGT overexpression affected the posttranslational state of mitotic proteins or their function, we used the enhanced O-GlcNAc enrichment and site-mapping method (19) and standard phosphopeptide enrichment methods with stable isotope labeling by amino acids in cell culture (SILAC) on cells synchronized and then harvested at late M phase, which would include cells in anaphase and telophase. With this approach, we identified sites occupied by either O-GlcNAc or phosphate and quantified occupancy changes as a function of cellular perturbation. As briefly outlined in fig. S2A, cells overexpressing OGT were grown in medium with heavy amino acids; cells overexpressing green fluorescent protein (GFP), as a control, were grown in normal medium. After synchronization to G1-S by double thymidine block, cells were released back into SILAC medium. After 6 hours, nocodazole was added to the cells and then, 6 hours later, mitotic cells were collected, washed, and returned into SILAC medium for 1 hour. Cells were harvested, pooled, and then lysed in a Taxol-containing buffer to stabilize microtubes. The pellet was collected by centrifugation, washed, and passed twice through a glycerol gradient. The resulting “Taxol pellet” represents the mitotic spindle and midbody; intact spindles and midbodies were verified by α-tubulin immunostaining (Fig. 1, D and E, and fig. S2B). The supernatant from the lysis in the Taxol buffer step represents the cytosolic fraction. The pellet was then digested with trypsin.
Fig. 1.
OGT is associated with the spindle and midbody. (A and B) OGT localizes to the mitotic spindle, then to the midbody during M phase in HeLa cells as seen by immunostaining (green, OGT; red, DNA). (C) Overexpression of OGT causes polyploidy (green, OGT; red, DNA). (D) Immunostaining of purified spindles shows that O-GlcNAc–modified proteins localize to the periphery of the mitotic spindle–centrosome (labeled as 1), along the spindle (labeled as 2), and a small amount in the spindle midzone (labeled as 3) (green, α-tubulin; red, O-GlcNAc). (E) O-GlcNAc–modified proteins are enriched at the purified midbody and at the nascent nuclear envelope (green, α-tubulin; red, O-GlcNAc). (F) Immunoblot for O-GlcNAc shows enrichment in the mitotic Taxol pellet (TP) and OGT overexpression increases protein O-GlcNAcylation. (G) Mitotic phosphorylation, determined by immunoblot to proline-directed phosphorylation (MPM-2), is disrupted by OGT overexpression. (H) OGT overexpression alters the abundance of a few proteins in the Taxol pellet samples (stained with Coomassie blue dye). All blots were loaded with equal amounts of protein. (I) α-Tubulin and AURKB, markers of Taxol-stabilized spindle midbodies, are enriched in the Taxol pellet. GFP was used as a control for the effects of the overexpression procedure. Experiments shown are representatives of a minimum of three times.
Antibody staining for O-GlcNAc in purified mitotic spindle and midbody preparations revealed a concentration of modified proteins at the centrosomes, surrounding the mitotic spindle, and a small amount at the spindle midzone (Fig. 1D). The midbody and the nascent nuclear envelope were also enriched in O-GlcNAc–modified proteins (Fig. 1E). Immunoblots for O-GlcNAc showed that the extent of O-GlcNAcylated proteins increased in cells overexpressing OGT (Fig. 1F) and that, in control (GFP-overexpressing, Fig. 1F; and noninfected cells, fig. S2C) and OGT-overexpressing cells (Fig. 1F), O-GlcNAcylated proteins were enriched in the Taxol pellet relative to the cytosol. Conversely, the cytoplasmic fraction contained a higher amount of phosphorylated protein relative to that in the Taxol pellet as judged by blotting with an antibody specific for proline-directed phosphorylation (Fig. 1G and fig. S2C). Reduced phosphorylation in the cytosol and Taxol pellet was apparent in samples from cells overexpressing OGT (Fig. 1G). The overall amount of protein did not appear substantially different between the samples from the GFP-overexpressing control cells and those from the OGT-overexpressing cells (Fig. 1H). We also checked the quality of our purification by blotting for α-tubulin and AURKB, two proteins that associate with the spindle and midbody (Fig. 1I and fig. S2D). OGT and OGA were mostly found in the supernatant, with a small amount detected in the Taxol pellet (Fig. 1I).
O-GlcNAc–containing peptides generated from tryptic digestion of the proteins contained in the Taxol pellet were chemoenzymatically tagged by PC-biotin-alkyne and enriched by avidin chromatography (19). Tagged O-GlcNAcylated peptides were eluted from the avidin column by photochemical cleavage of the biotin linker, subjected to CAD MS with an LTQ Orbitrap mass spectrometer, and then characterized with a front-end ETD-enabled LTQ Fourier transform ion cyclotron resonance (FT-ICR) mass spectrometer (19). In a separate step, titanium dioxide was used to selectively bind phosphopeptides in the flow-through from the avidin column. The remaining unmodified peptides were analyzed by CAD MS to both identify and quantify more than 700 proteins from the midbody preparation (fig. S3A and table S1).
OGT overexpression reduces the abundance of protein components of the chromosomal passenger complex
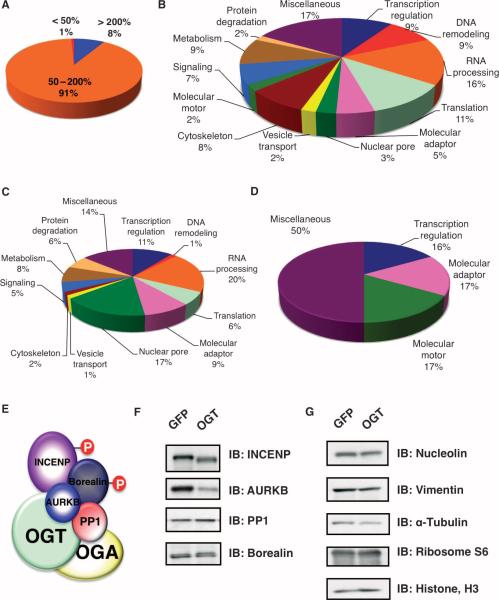
The abundance of most proteins in the spindle and midbody samples from the GFP-overexpressing and OGT-overexpressing cells did not differ substantially (Fig. 2A and fig. S3B). In the spindle and midbody preparation from OGT-overexpressing cells, the abundance of a few proteins (~1%) was decreased (by greater than 50%) or was increased (~8%) by more than 200%. Proteins representing a wide range of functional classes [defined by the UniProt Consortium (The Universal Protein Resource)] were present in the spindle and midbody preparation (Fig. 2B and table S2). Of the proteins with increased abundance as a result of OGT overexpression, one of the largest classes represented nuclear pore proteins (Fig. 2C). In contrast, the largest classes of proteins with decreased abundance in cells overexpressing OGT were the molecular motor and the adaptor proteins (Fig. 2D).
Fig. 2.
OGT overexpression alters the abundance of few proteins that are associated with the spindle and midbody. (A) HeLa cells infected with GFP were cultured in SILAC medium containing no heavy isotope; cells infected with OGT were cultured in SILAC medium containing heavy isotope. After purification and mass spectral analysis, the pie chart displays the ratios of (OGT spindle and midbody/GFP spindle and midbody sample) protein abundance for the more than 700 proteins identified in the MS analysis. Eight percent of identified proteins show increased abundance of more than 200% and 1% show a decrease of more than 50% in abundance. (B)The 700 proteins at the spindle and midbody represent numerous different functional classes as categorized by UniProt. (C) Proteins with a 200% increase in abundance upon OGT overexpression fall into multiple functional classes. (D) Molecular motor proteins and adaptor proteins constitute large proportions of the proteins that decrease in abundance by greater than 50%. (E) Schematic of the chromosomal passenger protein complex that associates with OGT and OGA. (F) Immunoblot (IB) analysis of HeLa synchronized by thymidine-nocodazole from cells overexpressing either GFP or OGT shows that overexpression of OGT causes a decrease in the abundance of several proteins in the chromosomal passenger protein complex. (G) Immunoblotting confirms that the abundance of several proteins is unchanged in cells overexpressing OGT.
Previously, we had shown that OGT interacts with AURKB, but that OGT overexpression did not affect the localization of AURKB (17). Both OGT and OGA interact with AURKB at midbodies (17). AURKB is part of the chromosomal passenger complex, which also includes inner centromere protein (INCENP), Borealin, and PP1 (Fig. 2E) (21). Unexpectedly, the MS analysis showed that in the mitotic spindle and midbody preparations from cells overexpressing OGT, there was an almost 50% decrease in the abundance of the mitotic kinase AURKB (table S1 and fig. S4) and its binding partner INCENP (table S1). These observations were supported by immunoblotting (Fig. 2F). INCENP is essential for the cellular localization and activation of AURKB (22). To determine if overexpression of OGT altered the localization of INCENP, we stained mitotic cells for INCENP and found that, even in cells with aberrant spindles and midbodies, OGT overexpression had no effect on INCENP localization (fig. S5, A and B).
OGT overexpression did not affect the abundance of numerous other proteins, including several involved in mitosis. The lack of change in abundance was confirmed by immunoblotting for vimentin, α-tubulin, ribosome protein S6, and histone H3 (Fig. 2G).
O-GlcNAcylation is nearly as abundant as phosphorylation at spindles and midbodies
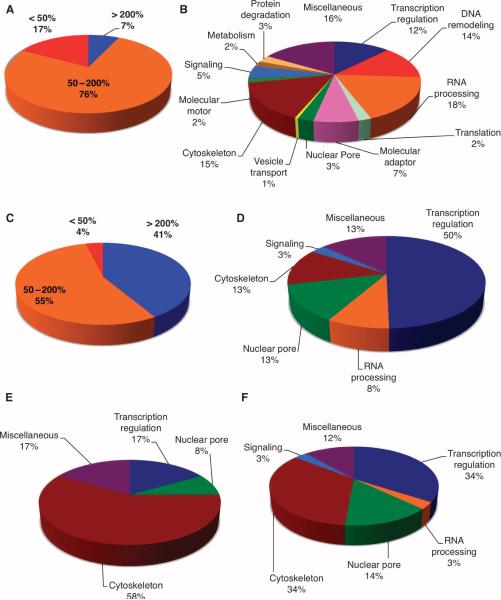
Overexpression of OGT to force cells to have twice the amount of OGT than was present in untransfected cells affected protein phosphorylation (table S3). We identified more than 350 phosphorylation sites in the two spindle and midbody samples (representing GFP-expressing control cells and OGT-overexpressing cells) and quantified changes in the amount of phosphorylation at each site. When OGT activity was increased, phosphorylation was increased by more than 200% for 7% of the observed phosphorylation sites or decreased by more than 50% for 17% of the observed sites (Fig. 3A and fig. S6A). Proteins containing these regulated sites were distributed across numerous functional classes (Fig. 3B, fig. S6, B and C, and table S4). Various sites on these proteins showed large changes in the extent of phosphorylation in spindle and midbodies from cells overexpressing OGT (table S4). We found that 69% of the sites with a lower extent of phosphorylation had the minimum pS/pT-P consensus site for CDK1 (fig. S7A) (23). Other sites exhibiting decreased phosphorylation have sequences similar to those of canonical PLK1 and AURKB phosphorylation motifs (2, 24). We did not detect increased phosphorylation at SQ sites associated with the DNA damage–activated kinases ATM and ATR (24), which suggests that OGT overexpression and the synchronization methods did not induce DNA damage or activate mitotic checkpoints. Proteins with greater than a 200% increase in the amount of phosphorylation did not show any specific sequence (fig. S7B); however, an emphasis for polar amino acids before and after the pS/T site was evident.
Fig. 3.
OGT overexpression triggers changes in mitotic phosphorylation and O-GlcNAcylation. (A) Phosphopeptides were enriched from pooled spindle-midbody extracts by purification over a titanium dioxide column. OGT overexpression decreased phosphorylation by more than 50% at 17% of the identified phosphopeptides, whereas only 7% of phosphorylation sites showed an increase of more than 200%. (B) Identified phosphorylation sites (397) occur on proteins that fall into similar functional classes. (C) Of the identified O-GlcNAcylation sites, 41% showed increased site modification of more than 200% in preparations from cells overexpressing OGT compared to cells overexpressing GFP, whereas only 4% of the O-GlcNAc sites showed decreased modification by more than 50%. (D) The GlcNAc-modified sites were predominately found on transcriptional regulators; however, RNA processing, nuclear pore, and cytoskeleton proteins also contained a large number of GlcNAcylation sites. (E) Functional classes of proteins whose O-GlcNAc site is also a reciprocal phosphorylation site. (F) Functional class of proteins that have a phosphorylation site within ± 10 amino acids proximal to an O-GlcNAcylation site.
With our enrichment strategy, we detected 450 O-GlcNAc–modified peptide pairs and identified 141 O-GlcNAcylation sites on 64 proteins from less than 15 μg of spindle and midbody preparation (table S5). We also identified more than 350 phosphorylation sites, representing 190 proteins, in the same sample preparation, which suggests that the number of sites modified in the cell by O-GlcNAcylation and phosphorylation is comparable. Not surprisingly, 41% of proteins show greater than a 200% increase in the amount of O-GlcNAcylation (Fig. 3C and fig. S8A) upon OGT overexpression. The largest numbers of O-GlcNAcylated proteins were categorized as involved in regulation of transcription (41%) (25), nuclear pore proteins (13%), and cytoskeletal proteins (13%) (Fig. 3D and table S6) (10).
As a class, cytoskeletal proteins contained the most examples of reciprocally modified sites, that is, sites that may be either phosphorylated or O-GlcNAcylated (Fig. 3E). A number of O-GlcNAcylation sites proximal (within 10 amino acids) to phosphorylation sites were also observed for cytoskeletal and transcriptional regulatory proteins (Fig. 3F). Analysis of the O-GlcNAcylated peptides revealed a preference for hydrophobic and polar amino acids before and after the O-GlcNAcylation site, respectively (fig. S8B). OGT does not recognize a strict consensus sequence, but does appear to prefer the P/V-P/V-V-gS/T-S/T sequence that is present in about 50% of sites mapped to date.
We also mapped 39 asparagine-linked GlcNAc (N-GlcNAc) monosaccharide sites (table S7) in the spindle and midbody preparation. Occupancy at these N-GlcNAcylation sites did not change in response to OGT overexpression, and all of these sites mapped to consensus N-glycan sequences (Asn-X-Ser-Thr). Likely, the proteins containing these N-GlcNAc residues are internalized from the outside of the cell and are degraded by a cytoplasmic endo-N-acetyl-β-D-glucosaminidase that cleaves N-linked oligosaccharides leaving a single N-linked GlcNAc (26), which may contribute to the salvage of oligosaccharides from misfolded proteins (26). Other groups using sensitive methods to detect O-GlcNAc (13) have also reported detection of N-GlcNAcylated sites, and it is likely that as the detection methodologies improve, these rare, low-abundance N-GlcNAcylated sites will become more common. Caution should be taken when interpreting results so as not to confuse N-glycan–derived glycopeptides with O-GlcNAc.
O-GlcNAcylation of three mitotic proteins was validated
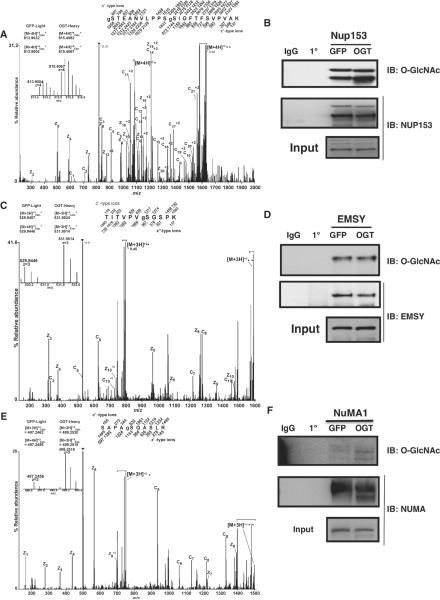
To confirm the glycoproteomic studies, we analyzed the changes in abundance of O-GlcNAcylation on three specific proteins, Nup153, EMSY, and NuMA1. Nup153 is a nuclear pore protein involved in the breakdown of the nuclear envelope at prophase and its reformation at telophase (27), and it contains a zinc finger domain that is critical for the recruitment of COP1 (coatomer complex protein 1) to the nuclear envelope during M phase. Nup153 promotes envelope fusion with the endoplasmic reticulum (28). Phosphorylation of Nup153 by CDK1 at M phase facilitates the breakdown of the nuclear envelope (29). In spindle and midbody preparations from cells overexpressing OGT (compared to those from GFP-expressing cells), we observed increased amounts of O-GlcNAcylation on Nup153 by almost 300% (Fig. 4, A and B). We identified six O-GlcNAcylation sites on Nup153. One site, gSer534 (g represents the GlcNAc) (Fig. 4A), is proximal to a phosphorylation site, pSer529 (p represents phosphate) (3), that is contained in the minimal motif for phosphorylation by CDK1.
Fig. 4.
Validation of O-GlcNAcylation site mapping. (A, C, and E)FETDMS/MS spectra recorded on (A) [M + 4H]+4 ions (m/z 815.6) for the modified peptide, gSTEANVLPPSgSIGFTFSVPVAK, from NUP153; (C) [M + 3H]+3 ions (m/z 531.9) for the modified peptide, TITVPVgSGSPK, from EMSY; (E) [M + 3H]+3 ions (m/z 499.2) for the modified peptide, SAPAgSQASLR, from NUMA. Abbreviations for the amino acid residues are as follows: A, Ala; E, Glu; F, Phe; G, Gly; I, Ile; K, Lys; L, Leu; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; and V, Val. Predicted m/z values for product ions of type c′ and type z′•(monoisotopic and average masses for singly and doubly charged ions, respectively) are listed above and below the peptide sequence. Observed product ions are underlined and also labeled in the spectrum. Ions in the precursor isolation window are labeled with a triangle (▼). Brackets enclose ions corresponding to charge-reduced species, as well as fragments derived from these charge-reduced species. Product ions resulting from loss of an aminomethyl triazole radical are labeled with a circle (●). Insert displays the isotopic distribution, calculated monoisotopic mass, and experimental monoisotopic mass of precursor ions corresponding to the labeled and unlabeled forms of the indicated peptide. (B, D, and F) Nup153, NuMA1, or EMSY were immunopurified from lysates of thymidine-nocodazole synchronized cells overexpressing either GFP or OGT. Precipitates were washed, separated on SDS-PAGE, and transferred to membranes for blotting. Membranes were probed for O-GlcNAc. For Nup153 and NuMA1, the increase in O-GlcNAc in the samples from the OGT-overexpressing cells is evident. For EMSY, the increase is not readily apparent, which may be due to only one of seven O-GlcNAcylation sites exhibiting increased modification.
EMSY is a binding partner to the breast cancer susceptibility protein, BRCA2, which is involved in double-stranded DNA repair (30). BRCA2 is a substrate for PLK1 (31) and interacts with AURKB at the midbody (32). Overexpression of a truncated form of EMSY in an immortalized breast epithelial line results in increased genomic instability (33). Thus, EMSY is believed to play a role in maintaining genomic stability during M phase (30, 33). We identified seven O-GlcNAcylation sites on EMSY; two of those sites map to the BRCA2 binding domain. Although the MS data suggested that overexpression of OGT increased the extent of O-GlcNAcylation on one of these sites (gSer236) by more than twofold in spindle and midbody preparations (Fig. 4, C and D), we failed to detect this increase by immunoblotting with an O-GlcNAc–specific antibody (Fig. 4D), which was likely due to the presence of the other six O-GlcNAcylation sites that were not affected by OGT overexpression.
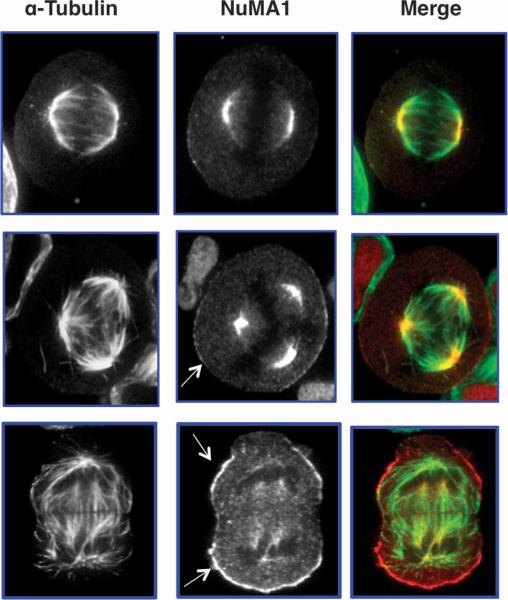
Finally, we found that overexpression of OGT causes a fivefold increase in the extent of O-GlcNAcylation on Ser1844 of NuMA1 (nuclear mitotic apparatus protein 1) (Fig. 4, E and F) in spindle and midbody preparations (compared to those from GFP-overexpressing cells). Cells overexpressing OGT also contained a slightly lower molecular weight version of NuMA1 that appeared extensively O-GlcNAcylated (Fig. 4F). NuMA1 is extensively phosphorylated at M phase (3). Seven phosphorylation sites were found on NuMA1 in the screen and all were decreased after OGT overexpression (table S3). The shift to a more rapidly migrating form of the protein in preparations from cells overexpressing OGT suggests that O-GlcNAcylation interferes with NuMA1 phosphorylation. NuMA1 is an essential protein involved in establishing the spindle poles and in recruiting the cohesion complex to the spindle poles (34, 35). HeLa cells overexpressing OGT exhibit polyploidy (Fig. 1C) and we found that NuMA1 localization was altered in cells with aberrant chromosome number (Fig. 5). Although some NuMA1 was still found at the spindle poles in OGT overexpressing cells with aberrant spindle number, a subset of NuMA1 also localized to the cell periphery in these cells (Fig. 5, bottom panel).
Fig. 5.
NuMA1 localization is disrupted in cells exhibiting aneuploidy. HeLa cells overexpressing OGT were released for 12 hours from double thymidine block, then fixed and stained for α-tubulin (green) or NuMA1 (red). In cells with normal number of mitotic spindles, NuMA1 is at the centromeres (top panel). However, as the number of spindles increase (middle and bottom panel), a subset of NuMA1 appears to localize to the membrane (arrows) of cells and less NuMA1 is associated with the centromeres.
CDK1 activity is decreased in cells overexpressing OGT
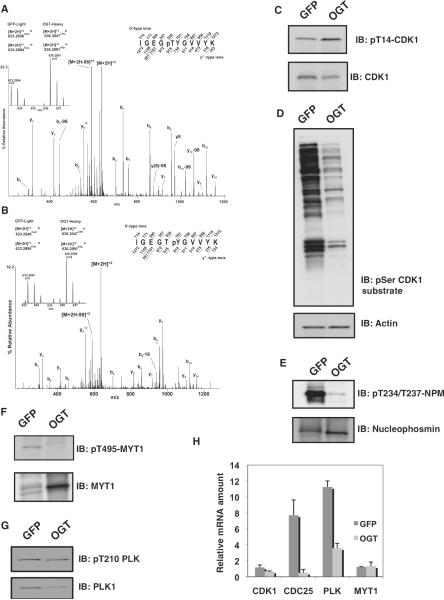
The protein complex containing cyclin B and CDK1 is the master regulator of M-phase progression and is responsible for the phosphorylation of a large number of proteins during M phase (3). This complex accumulates during G2 phase but is inactive as a result of phosphorylation on Thr14 (36) and Tyr15 by the kinases Wee1 and MYT1, respectively (37). Activation of the cyclin B-CDK1 complex results when the phosphatase CDC25c removes the inhibitory phosphate groups, first from Thr14 and then from Tyr15 (fig. S9). We found that overexpression of OGT increased the phosphorylation at both of these inhibitory sites by more than twofold in spindle and midbody preparations (compared to preparations from GFP-expressing cells) (Fig. 6, A to C), suggesting that O-GlcNAcylation is involved in the regulation of CDK1. Furthermore, phosphorylation of CDK1 downstream substrates was diminished as measured by immunoblotting from thymidine-nocodazole synchronized cells with an antibody to phosphorylated serine residues within the CDK1 motif (K/R)(S*)PX(K/R) (Fig. 6D). In the samples from OGT-overexpressing cells, MS data recorded for peptides containing the minimum consensus site for CDK1 (pS/pT-P) (fig. S7A) also showed reduced phosphorylation. Two CDK1 sites on nucleophosmin (Thr234 and Thr237) are of particular interest because they are involved in proper centrosome localization (38). By phosphorylation-specific immunoblotting, we confirmed that the extent of phosphorylation on these sites was substantially reduced in cells overexpressing OGT (Fig. 6E).
Fig. 6.
OGT overexpression disrupts the CDK1 activation circuit. (A and B) CAD MS/MS spectra of phosphopeptides from CDK1 containing pThr14 (A) or pTyr15 (B). Insets show the relative abundances of the precursor ions originating from the two samples. (C) Immunoblotting of thymidine-nocodazole lysates with a phosphorylation-specific antibody to Thr14 on CDK1 (pT14-CDK1) reveals increased phosphorylation at this site in OGT-overexpressing cells. The total amount of CDK1 was slightly lower in OGT-expressing cells compared to GFP-expressing controls. All blots were performed a minimum of three times with different lysates. (D) Immunoblotting with an antibody that recognizes proteins phosphorylated on Ser residue within a CDK1 consensus phosphorylation motif shows that CDK1 activity is decreased in OGT-overexpressing cells. (E) Immunoblotting with an antibody that recognizes nucleophosmin (NPM) phosphorylated on Thr234 and Thr237 shows that phosphorylation of this protein is decreased in OGT-overexpressing cells. (F) Immunoblotting of phosphorylated (pThr495) and total MYT1 shows that phosphorylation of this protein is decreased and its abundance is increased in OGT-overexpressing cells. (G) Immunoblotting of phosphorylated (pThr210) and total PLK1 shows that the abundance of PLK1 is decreased in OGT-overexpressing cells. (H) Relative mRNA amounts at M phase of CDK1, CDC25, PLK1, and MYT1 in cells overexpressing OGT. All transcripts were normalized to actin.
Increased occupancy of the inhibitory phosphorylation sites on CDK1 could result either by increased activity of the upstream inhibitory kinases, MYT1 (36) and Wee1 (37), or through decreased activity of the phosphatase CDC25C (6). The activation circuit for CDK1 involves both inhibitory phosphorylation of MYT1 on Thr495 and an activating phosphorylation on CDC25C, which are both mediated by PLK1 (39). PLK1 is a polo box domain–containing protein that phosphorylates various substrates essential for control of mitotic progression (40). Cells overexpressing OGT exhibited a dramatic reduction in the extent of inhibitory phosphorylation at Thr495 (39, 40) on MYT1 (Fig. 6F). The abundance of PLK1 was also reduced in cells overexpressing OGT (Fig. 6G, table S1, and fig. S10); however, the activating phosphorylation at Thr210 on PLK1 was not altered (Fig. 6G).
Despite altered abundance or phosphorylation, PLK1 and MYT1 did not show altered localization in cells overexpressing OGT. PLK1 is localized to discrete areas of the centromere, spindle midzone, kinetochore, and midbody during Mphase(40). PLK1 localization (detected with an antibody that recognizes all PLK1, as well as an antibody specific for the Thr210 phosphorylated form) was not affected by OGToverexpression (figs. S11A and S12A); PLK1 colocalized with a subset of OGT throughout M phase (fig. S11B). We found MYT1 localized to centrosomal regions in mitotic cells, although in interphase cells the kinase can be found in the ER and golgi (41), and we found that in cells overexpressing OGT, this was unchanged (fig. S12B).
Because we observed that several proteins involved in the CDK1 activation circuit (fig. S9) showed a reduction in protein abundance, changes in phosphorylation, or both in thymidine-nocodazole synchronized cells, we measured the transcript abundance for several of these proteins in synchronized GFP-expressing cells and OGT-overexpressing cells. Little change was evident in the relative amounts of mRNAs for CDK1 and MYT1; however, transcripts for both PLK1 and CDC25 were substantially reduced in the OGT-overexpressing cells (Fig. 6H). Nucleolin, cyclin B, and cyclin E served as internal controls for mitotic genes encoding proteins that did not change in abundance in the MS analysis and for which the transcripts abundance was similar in OGT-overexpressing cells and GFP-expressing cells (fig. S13).
DISCUSSION
We used a sensitive strategy for completing large-scale, O-GlcNAcylation site mapping and showed that O-GlcNAc is nearly as abundant as phosphate on proteins associated with the spindle and midbody. Many of the O-GlcNAcylation sites identified are identical or proximal to known phosphorylation sites. This study suggests O-GlcNAcylation and phosphorylation work together to control complicated mitotic processes, such as spindle formation. For example, OGT overexpression altered the abundance of transcripts and proteins encoded by several mitotic genes, changed the localization of NuMA1, and disrupted the chromosomal passenger complex and the CDK1 activation circuit.
We find a complex interplay exists between O-GlcNAcylation and phosphorylation for several protein classes, most noticeably transcriptional regulators and cytoskeletal proteins. Many of the O-GlcNAcylation and phosphorylation sites are located in the regulatory head domains of intermediate filament proteins. Phosphorylation of these sites causes filament disassociation during M phase (17). For example, vimentin is phosphorylated at multiple sites during M phase and thereisan O-GlcNAcylation site that is also a mitotic phosphorylation site (Ser55)(16, 17, 42, 43). Here, we characterized three additional O-GlcNAcylation sites on vimentin at Ser7,Thr33,and Ser34 (tables S5 and S6), all of which are in the regulatory head domain of the protein. Two of these, Ser7 and Ser34, are also phosphorylation sites (3, 43). It seems likely that signaling pathways involving cytoskeletal proteins are regulated by reciprocal occupancy on specific sites by phosphate and O-GlcNAc. These data suggest that in these classes of molecules, areas of multiple phosphorylation are also likely to be targeted for O-GlcNAcylation.
Furthermore, the data show that OGT overexpression profoundly affects multiple mitotic signaling circuits. Although overexpression of OGT does not interfere with the formation of the midbody complex or localization of AURKB, previously we showed that AURKB activity is altered toward the cytoskeletal protein, vimentin (17). Potentially, the reduction in the abundance of AURKB or INCENP might dampen kinase activity to a point that retards mitotic progression especially during anaphase and telephase (8). Furthermore, OGToverexpression reduced phosphorylation of INCENP and borealin, but to what extent this alters the function of the midbody complex is unclear.
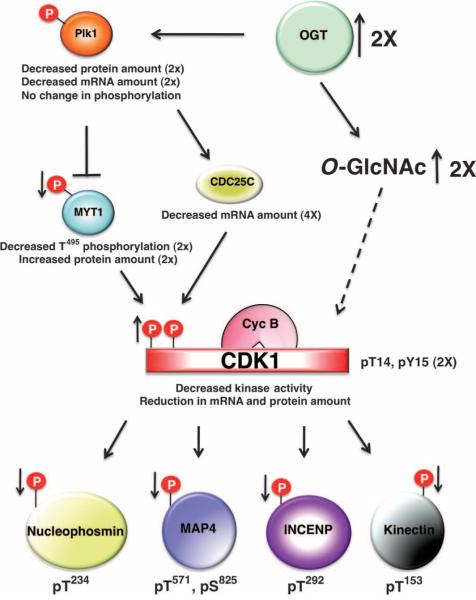
Multiple components of the cyclin B-CDK1 activation circuit were disrupted by the overexpression of OGT. The loss of PLK1 inhibitory phosphorylation on MYT1 and the increase in theabundanceof MYT1 are likely contributors to the loss in cyclin B-CDK1 activity observed in OGT-overexpressing cells (Fig. 7). However, the reduction in cyclin B-CDK1 activity is likely only partially due to the increase in MYT1 activity, because the mRNA for CDC25C, the key CDK1 dual-specific phosphatase, is substantially reduced. The “on” switch for CDK1 activation, the reduction of MYT1 and the increase in CDC25C activity, is pushed toward “off ” by OGT overexpression. Both MYT1 and CDC25C are substrates for PLK1. The protein and transcript abundance of PLK1 is substantially reduced in response to OGT overexpression, but there is little change in the extent of activating phosphorylation of PLK1. Whether the decrease in MYT1 inhibitory phosphorylation in OGT-overexpressing cells is solely due to the decreased abundance of PLK1 is not known.
Fig. 7.
Model illustrating how OGT overexpression affects cell cycle progression. OGT overexpression causes a decrease in the protein and mRNA abundance of PLK1, although the amount of PLK1's activating phosphorylation is not changed. MYT1, a target of PLK1, displays an increase in protein amount but a reduction in phosphorylation, whereas the other PLK1 target, CDC25C, shows a decrease in mRNA amount. MYT1 phosphorylates CDK1, inhibiting CDK1 activity, and leading to a decrease in CDK1 mitotic phosphorylation on numerous substrates. Thus, the increase in CDK1 inhibitory phosphorylation leads to a reduction in the phosphorylation of CDK1 substrates.
Because O-GlcNAcylation is directly coupled to nutrient uptake and metabolism, the sugar residue is an ideal metabolic sensor for regulating mitotic progression. Whereas phosphorylation might act as a master switch initiating the mitotic process, O-GlcNAcylation may act as an adjuster of signals to make these processes more responsive to environmental cues. How O-GlcNAcylation exerts control on specific mitotic proteins and how O-GlcNAcylation will integrate into well-known signaling pathways represent another layer of cellular regulation.
MATERIALS AND METHODS
Reagents
Antibodies used in the study are the following: antibodies against AURKB (1:1000, AIM-1 611083, BD Transduction Laboratories), PP1c (1:1000, sc-7482 and sc-443, Santa Cruz Biotechnology), GFP (1:5000, sc-9996 Santa Cruz Biotechnology), α-tubulin (1:1000, T5168, Sigma), MPM-2 (1:1000, 05-368, Upstate Biotechnology, Inc), INCENP (1:1000, ab36453, Abcam), PLK1 (1:1000, Zymed 37-7000, and 1:1000, ab14209, Abcam), pThr210 PLK1 (1:1000, ab12157, Abcam), Borealin (1:500, PAB3096, Abnova), Nucleolin (1:1000, sc-8031, Santa Cruz), ribosome S6 (1:1000, 2217, Cell Signaling), Histone H3 (1:1000, 9715, Cell Signaling), CDK1 (1:1000, ab18, Abcam), phosphorylated T14 CDK1 (1:1000, ab4824, Abcam), phosphorylated T234 and T237 Nucleophosmin (1:1000, 619101, Biolegend), nucleophosmin (1:1000, 32-5200, Zymed), PhosphoSer CDK1 substrate (1:1000, 2324, Cell Signaling), Nup153 (1:1000, ab24700, Abcam), EMSY (1:1000, ab123, Abcam), NuMA (1:1000, ab36999, Abcam), Vimentin (1:5000, V 6630, Sigma), and O-GlcNAc (1:10000, 110.6 (44), available from Sigma or Covance). Polyclonal antibodies for OGT (1:1000, AL-28) and OGA (1:1000, 345) were raised in rabbit and chicken, respectively. GammaBind G Sepharose beads were from GE Healthcare (17-0885-01). Horseradish peroxidase–conjugated secondary antibodies were purchased from GE Healthcare (rabbit, NA934V; mouse, NA931V; rat, NA935V) or Sigma (chicken, A 9046; antibody against mouse immunoglobulin (Ig) M, A 8786). Fluorescently labeled secondary antibodies, Alexa Fluor 647 and Alexa Fluor 488 (A11029, A21245), and propidium iodide (P1304MP) were from Invitrogen-Molecular Probes. Blots were developed with ECL reagent (GE Healthcare) on Hyper Film (GE Healthcare). Nocodazole, thymidine, paclitaxel, and phalloidin were from Sigma (M1404, T1895, T7402, P2141). Dimethyl sulfoxide (DMSO) was from Sigma (D8418).
Cell culture and synchronization
HeLa cells were grown in SILAC Dulbecco's modified Eagle's medium (89985, Thermo Scientific) supplemented with 10% dialyzed fetal bovine serum (26400044, Invitrogen) and 5% penicillin-streptomycin (30-001-CI, Cellgro). For light SILAC medium, the medium was supplemented with tyrosine (T1145, Sigma), lysine (L5626, Sigma), and arginine (BP2505500, Fisher Scientific), and for SILAC medium, the medium was supplemented with tyrosine, [13C6]lysine (CLM-2247, Cambridge Isotopes), and [13C6]arginine (CLM-2265, Cambridge Isotopes). After five passages in SILAC medium, cells were synchronized by double thymidine block as previously described (16). Cells were infected at an MOI (multiplicity of infection) of 100 with either GFP adenovirus (Baylor Vector Laboratories) or OGT adenovirus (16) at the first thymidine synchronization. After 6 hours into the second thymidine release, nocodazole was added at 0.8 ng/ml, and then mitotic cells were harvested by mitotic shake-off (gently shaking the plate to dislodge mitotic cells) 6 hours later. The mitotic cells were washed twice in warm SILAC medium and replated for 1 hour. Next, Taxol and phalloidin (5 μg/ml) were added for 5 min. Cells were harvested by shake-off, combined, and washed in warm phosphate-buffered saline (PBS). HeLa cells used for validation of the SILAC results were treated as above, except grown in normal medium and lysed in NP-40 buffer (45) after harvesting.
Mitotic spindle and midbody preparation
Spindles and midbodies were prepared as described previously (1). Briefly, cells were lysed in a hypotonic buffer [2 mM Pipes (pH 6.9), 0.25% Triton X-100, phosphatase inhibitor cocktail (P2850 and P5726, Sigma-Aldrich), and Taxol (20 μg/ml) at 37°] for 20 min. Lysates were centrifuged for 20 min at 325 relative centrifugal force (RCF), and the pellet was collected. The pellet was resuspended in MES buffer (50 mM, pH 6.3, 4°C), and an aliquot of samples was plated for confocal microscopy. The resuspended pellet was washed twice and then spun through a 40% glycerol cushion for 20 min at 16,000 RCF. The pellet was washed with MES and spun through the glycerol cushion for a second time. The pellet was washed and resuspended in either ammonium bicarbonate buffer (50 mM, pH 7.8) or tris-EDTA buffer [25 mM tris (pH 8.5), 1 mM EDTA] (T6066 and E6758, Sigma) and digested overnight at 37°C in trypsin (V5280, Promega) or endoproteinase Lys-C (11-420-429-001, Roche Applied Science), respectively.
Peptide enrichment
Solutions containing 15 μg of digested peptides were filtered through a 10-kD molecular mass cutoff membrane (Millipore) to remove excess proteases and lyophilized. The peptides were resuspended in 150 μl of Hepes (pH 7.9) containing 5 mM MnCl2, ~5× UDP-GalNAz and mutant GalT1 (C33368, Invitrogen), and 5 U PNGase F (P0705, New England Biolabs) and incubated overnight at 4°C and at room temperature for additional hours with rotation. Excess UDP-GalNAz was removed with a C18 spin column (Nestgroup). The labeled peptides were again lyophilized and resuspended in a 30-μl solution containing PC-PEG-biotin-alkyne (~4× excess in methanol) (19), 10 mM sodium ascorbate (A7631, Sigma-Aldrich), 1 mM tris [(1-benzyl-1H-1, 2, 3-triazol-4-yl) methyl] amine (TBTA, in 4:1 t-butanol–DMSO), and 2mMCuSO4. The cycloaddition proceeded at room temperature overnight with light protection. Then, the mixture was diluted into strong cation exchange (SCX) loading buffer [5 mM KH2PO4, 25% acetonitrile (pH 3.0)] and passed through a SCX spin column (Nestgroup). Peptides were washed with loading buffer and then eluted with a series of high-salt buffers containing 50, 100, 200, and 400 mM KCl. After adjusting pH to around 7, each fraction was allowed to bind to high-capacity avidin beads (29202, Pierce) for 2 hours at room temperature. The beads were extensively washed with PBS followed by a final wash with 20% methanol. The beads were then resuspended in 70% methanol, transferred to clear thin-walled polymerase chain reaction (PCR) tubes, and illuminated by 365 nm UV (Spectroline ENF-240C) for 25 min. After photocleavage, the supernatant from each fraction was collected, lyophilized, and stored at −20°C. The flow-through of each fraction from the avidin chromatography was collected, lyophilized, and resuspended in 80% acetonitrile containing 1 M glycolic acid (124737, Sigma-Aldrich) and 4% trifluoroacetic acid (TFA) (T6508, Sigma-Aldrich). The solution was passed through TiO2 columns (TT2TIO, Glycogen). After extensive washing with 80% acetonitrile–4% TFA, bound peptides were eluted by NH4OH (pH 11.5). The elutant was immediately neutralized, desalted, and lyophilized. Flow-through and the initial wash from the TiO2 was also collected, desalted, and lyophilized.
CAD tandem mass spectrometry analysis
An aliquot of the enriched glycol-peptides was resuspended by 0.1% formic acid and analyzed by CAD on an LTQ-Orbitrap XL (Thermo) mass spectrometer coupled with a nano-LC system (Eksigent). Peptides were desalted on a precolumn [75-μm inside diameter (ID), 3-cm length, packed with irregular size particles 5 to 15 μm, 120 Å], and separated on an RF analytical column packed with 10 cm of C18 beads (5 μm, 120 Å, YMC ODS-AQ, Wather). The main HPLC gradient was 5 to 40% solvent B (A, 0.1% formic acid; B, 90% acetonitrile, 0.1% formic acid) in 60 min at a flow rate of 200 nl/min. The scan was set in data-dependent mode with each survey scan [Fourier transform, 30,000 resolution at a mass to charge ratio (m/z)of400] followed by CAD tandem mass spectrometry (MS/MS) (ion trap) of up to five most intense precursor ions. Normalized collision energy was set at 35.0. Activation time was 30 ms with activation Q at 0.25. Phosphorylated and unmodified peptides were similarly analyzed by CAD. Technical duplicates were performed for all CAD analysis.
ETD MS/MS analysis
Lyophilized samples were reconstituted in 0.1% acetic acid (338826, Sigma-Aldrich). For all MS analyses, an aliquot of sample was pressure-loaded onto a 360-μm outside diameter (OD) by 75-μm ID capillary precolumn (2000019, Polymicro Technologies) packed with C18 resin (5 to 20 μm diameter, 120 Å pore size, YMC) (46, 47). To remove salts, the precolumn was rinsed with ~20 column volumes of 0.1% acetic acid. The precolumn was connected to a 360-μm OD by 50-μm ID capillary (2000017, Polymicro Technologies) analytical column packed with C18 resin (5 μm diameter, 120 Å pore size, YMC) and integrated with an electrospray emitter tip (46, 48). All samples were analyzed by nanoflow (60 nl/min) HPLC (HP 1100, Agilent Technologies) interfaced to an LTQ FT hybrid mass spectrometer (Thermo Scientific) (46, 48). A 60-min gradient [0 to 60% solvent B; A, 0.1 M acetic acid; B, 0.1 M acetic acid in 70% acetonitrile (9853, Mallinckrodt Baker)] was used for all experiments.
All samples were analyzed on an LTQ FT mass spectrometer (Thermo Scientific) equipped with a new ion source that facilitates simultaneous generation of positively charged peptides by electrospray ionization and fluoranthene radical anion reagents for ETD, both from the front side of the instrument (FETD). The FT-ICR analyzer was used to record high-resolution MS1 spectra (resolving power of 25,000 at m/z 400). FETD MS/MS spectra were acquired with a quadrupole linear ion trap analyzer operating in the data-dependent mode (reaction time, 60 ms; reagent AGC target, 2 × 105 ion counts; full AGC target, 1 × 106 ion counts; MSn AGC target, 1 × 104 ion counts; isolation window, 3 m/z; repeat count, 2; repeat duration, 20 s; exclusion duration, 30 s).
Mass spectrometric data analysis
CAD MS/MS spectra of potential phosphopeptides were searched against the human RefSeq database using the Mascot algorithm with the following parameters: trypsin or Lys-C as enzyme with up to two missed cleavages; no fixed modification; oxidation (M), deamidated (NQ), phospho (STY) (for phosphopeptides only), label-13C(6) (K), and label-13C(6)(R)asvariable modifications. Mass tolerance was set to 8 parts per million (ppm) for precursors and 0.8 atomic mass units for fragment ions. Spectra assigned by the algorithm to phosphopeptides with a significance threshold less than 0.05 were accepted and then validated manually. Assigned sequences were deemed to be correct if the spectra contained fragment ions corresponding to the loss of phosphoric acid from the precursor ion, displayed b and y fragment ions that matched those predicted for the theoretical peptide sequence, and were devoid of abundant ions that failed to correlate with the assigned sequence. For database searches of raw data corresponding to unmodified peptides, a false-positive rate of 0.25% was estimated by searching against a reverse database. Relative abundances of isotopically labeled and unlabeled phosphopeptides were determined with the MSQuant algorithm (http://www.msquant.sourceforge.net) and manually inspected to ensure quality. Relative occupancy ratios were calculated as previously described (18).
ETD MS/MS spectra were searched against all human proteins in the National Center for Biotechnology Information RefSeq database with the Open Mass Spectrometry Search Algorithm (OMSSA) (49). Parameters for the search include the following: precursor mass tolerance, ± 0.01 dalton; product ion mass tolerance, ± 0.35 dalton; and differential modifications, +15.9949 on methionine residues, +502.2024 on serine, threonine, and asparagine residues. Spectra assigned to O-GlcNAcylated peptides, regardless of score, were interpreted manually by two of the following three authors: N.D.U., J.S., and D.F.H. A protocol similar to that described in Nichols and White (50) was used for this process. The 12C isotope of the precursor ion was measured with a mass accuracy of less than 3 ppm. Singly and doubly charged fragments of type c and z plus charge-reduced ions and their corresponding neutral loss species were annotated and then used to confirm the peptide sequence and define the specific site of modification (19). Peptide sequence assignments were deemed to be correct if the observed c- and z-type fragment ions plus the measured mass of the precursor were consistent with only one sequence in the RefSeq database. Relative ion abundances of isotopically labeled and unlabeled O-GlcNAcylated peptides were determined from extracted ion chromatograms (0.01-dalton window around the calculated m/z values) by summing signal intensities (area under the curve) corresponding to the 12C isotopes of both peptide species.
Real-time PCR primer design
Primers were designed with oligoperfect program available at Invitrogen (http://www.invitrogen.com). Primer sequences are the following: actin: forward (F), ctcttccagccttccttcct, reverse (R), agcactgtgttggcgtacag; PLK1, F, gcccctcacagtcctcaata, R, ctgcagcatgtcactgaggt; Nucleolin: F, cgttcgggcaaggatagtta, R, agccaccttcacccttaggt; cyclin B: F, ttggtgtcactgccatgttt, R, ccgacccagaccaaagttta; cyclin E: F, atcctccaaagttgcaccag, R, aggggacttaaacgccactt; cdc2C: F, gaacaggccaagactgaagc, R, gcccctggttagaatcttcc; MYT1: F, agcctagggccttcactctc, R, tcacttaggtccagggcatc; CDK1: F, ctggggtcagctcgttactc, R, agtgcccaaagctctgaaaa.
Real-time PCR
RNAwas isolated from cells using Trizol (Invitrogen) according to the manufacturer's instructions. Complementary DNA was prepared with oligo(dT) with SuperScript II (Invitrogen) according to the manufacturer's instructions. mRNA concentrations were detected by quantitative PCR (qPCR) with Platinum SYBR Green qPCR SuperMix (Invitrogen) on a MX3000P qPCR machine (Stratagene). Primers were designed with Oligoperfect from Invitrogen. Relative mRNAwas calculated after normalization to actin mRNA concentrations.
Immunopurification, Western blotting, and immunofluorescence
Proteins were immunoprecipitated from whole-cell lysates of thymidine-nocodazole synchronized cells as described previously (17). After separation on SDS–polyacrylamide gel electrophoresis (SDS-PAGE) and transfer to polyvinylidene difluoride membrane (IPVH0010, Millipore), membranes were blotted against either phosphorylation- or O-GlcNAcylation–specific antibodies, stripped, and reprobed with antibodies that detected the total protein (17).
HeLa cells were plated and fixed for staining as previously described (16). After fixation, cells were washed twice for 10 min each wash in PBS-Mg+2 containing 100 mM glycine (pH 7.4) (BP381–5, Fisher Scientific). Next, cells were permeabilized in PBS-Mg+2 containing 1% (v/v) Triton X-100 (BP151–500, Fisher Scientific) for 20 min, washed, and then blocked for 1 hour in tris-buffered saline with 0.05% Tween 20 (T2700, Sigma) containing 3% bovine serum albumin (A9647, Sigma). Slides were sequentially incubated overnight at 4°C with primary antibody (1:200 PLK1, INCENP, NuMA, 1:1000 OGT, 1:1000 α-tubulin, and 1:10,000 O-GlcNAc antibody 110.6), washed as described above, incubated with secondary antibody (1:1000) for 1 hour at 25°C, and washed. Before mounting, some slides were incubated in PBS-Mg+2 containing 0.1% Triton X-100 and propidium iodide (1 μg/ml) for 1 min and washed as described above. Fluorescent images were obtained on a 3i Spinning Disk Confocal microscope with Olympus Slidebook software at the Johns Hopkins University School of Medicine Core Microscopy Facility.
Supplementary Material
Acknowledgements
We thank the Johns Hopkins microscope core facility group for their assistance and A. Skop and M. K. Bonner from the University of Wisconsin for their help with the spindle/midbody protocol. This work was supported by the National Cancer Institute, NIH grants CA42486 and DK61671 (to G.W.H.), and NIH grant GM 37537 (to D.F.H). The use of the orbitrap was supported in part by grant S10RR023025-01 (R. J. Cotter, PI) from the High End Instrumentation Program of the NIH. Additionally, we thank the Hart laboratory members for critical reading of the manuscript.
Footnotes
REFERENCES AND NOTES
- 1.Skop AR, Liu H, Yates J, III, Meyer BJ, Heald R. Dissection of the mammalian midbody proteome reveals conserved cytokinesis mechanisms. Science. 2004;305:61–66. doi: 10.1126/science.1097931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nousiainen M, Sillje HH, Sauer G, Nigg EA, Korner R. Phosphoproteome analysis of the human mitotic spindle. Proc. Natl. Acad. Sci. U.S.A. 2006;103:5391–5396. doi: 10.1073/pnas.0507066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dephoure N, Zhou C, Villen J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP. A quantitative atlas of mitotic phosphorylation. Proc.Natl.Acad.Sci.U.S.A. 2008;105:10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glotzer M. The 3Ms of central spindle assembly: Microtubules, motors and MAPs. Nat. Rev. Mol. Cell Biol. 2009;10:9–20. doi: 10.1038/nrm2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daub H, Olsen JV, Bairlein M, Gnad F, Oppermann FS, Korner R, Greff Z, Keri G, Stemmann O, Mann M. Kinase-selective enrichment enables quantitative phospho-proteomics of the kinome across the cell cycle. Mol. Cell. 2008;31:438–448. doi: 10.1016/j.molcel.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 6.Karlsson-Rosenthal C, Millar JB. Cdc25: Mechanisms of checkpoint inhibition and recovery. Trends Cell Biol. 2006;16:285–292. doi: 10.1016/j.tcb.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 7.Rahal R, Amon A. Mitotic CDKs control the metaphase-anaphase transition and trigger spindle elongation. Genes Dev. 2008;22:1534–1548. doi: 10.1101/gad.1638308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuller BG, Lampson MA, Foley EA, Rosasco-Nitcher S, Le KV, Tobelmann P, Brautigan DL, Stukenberg PT, Kapoor TM. Midzone activation of aurora B in anaphase produces an intracellular phosphorylation gradient. Nature. 2008;453:1132–1136. doi: 10.1038/nature06923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trinkle-Mulcahy L, Lamond AI. Mitotic phosphatases: No longer silent partners. Curr. Opin. Cell Biol. 2006;18:623–631. doi: 10.1016/j.ceb.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 10.Hart GW, Housley MP, Slawson C. Cycling of O-linked β-N-acetylglucosamine on nucleocytoplasmic proteins. Nature. 2007;446:1017–1022. doi: 10.1038/nature05815. [DOI] [PubMed] [Google Scholar]
- 11.Vosseller K, Trinidad JC, Chalkley RJ, Specht CG, Thalhammer A, Lynn AJ, Snedecor JO, Guan S, Medzihradszky KF, Maltby DA, Schoepfer R, Burlingame AL. O-Linked N-acetylglucosamine proteomics of postsynaptic density preparations using lectin weak affinity chromatography and mass spectrometry. Mol. Cell. Proteomics. 2006;5:923–934. doi: 10.1074/mcp.T500040-MCP200. [DOI] [PubMed] [Google Scholar]
- 12.Khidekel N, Ficarro SB, Clark PM, Bryan MC, Swaney DL, Rexach JE, Sun YE, Coon JJ, Peters EC, Hsieh-Wilson LC. Probing the dynamics of O-GlcNAc glycosylation in the brain using quantitative proteomics. Nat. Chem. Biol. 2007;3:339–348. doi: 10.1038/nchembio881. [DOI] [PubMed] [Google Scholar]
- 13.Chalkley RJ, Thalhammer A, Schoepfer R, Burlingame AL. Identification of protein O-GlcNAcylation sites using electron transfer dissociation mass spectrometry on native peptides. Proc. Natl. Acad. Sci. U.S.A. 2009;106:8894–8899. doi: 10.1073/pnas.0900288106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khidekel N, Ficarro SB, Peters EC, Hsieh-Wilson LC. Exploring the O-GlcNAc proteome: Direct identification of O-GlcNAc-modified proteins from the brain. Proc. Natl. Acad. Sci. U.S.A. 2004;101:13132–13137. doi: 10.1073/pnas.0403471101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Z, Gucek M, Hart GW. Cross-talk between GlcNAcylation and phosphorylation: Site-specific phosphorylation dynamics in response to globally elevated O-GlcNAc. Proc. Natl. Acad. Sci. U.S.A. 2008;105:13793–13798. doi: 10.1073/pnas.0806216105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Slawson C, Zachara NE, Vosseller K, Cheung WD, Lane MD, Hart GW. Perturbations in O-linked β-N-acetylglucosamine protein modification cause severe defects in mitotic progression and cytokinesis. J. Biol. Chem. 2005;280:32944–32956. doi: 10.1074/jbc.M503396200. [DOI] [PubMed] [Google Scholar]
- 17.Slawson C, Lakshmanan T, Knapp S, Hart GW. A mitotic GlcNAcylation/phosphorylation signaling complex alters the posttranslational state of the cytoskeletal protein vimentin. Mol. Biol. Cell. 2008;19:4130–4140. doi: 10.1091/mbc.E07-11-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Z, Park K, Comer F, Hsieh-Wilson LC, Saudek CD, Hart GW. Site-specific GlcNAcylation of human erythrocyte proteins: Potential biomarker(s) for diabetes. Diabetes. 2009;58:309–317. doi: 10.2337/db08-0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Z, Udeshi ND, O'Malley M, Shabanowitz J, Hunt DF, Hart GW. Enrichment and site-mapping of O-linked N-acetylglucosamine by a combination of chemical/enzymatic tagging, photochemical cleavage, and electron transfer dissociation (ETD) mass spectrometry. Mol. Cell. Proteomics. 2009:M900268–MCP200. doi: 10.1074/mcp.M900268-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carmena M, Ruchaud S, Earnshaw WC. Making the Auroras glow: Regulation of Aurora A and B kinase function by interacting proteins. Curr. Opin. Cell Biol. 2009;21:796–805. doi: 10.1016/j.ceb.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adams RR, Maiato H, Earnshaw WC, Carmena M. Essential roles of Drosophila inner centromere protein (INCENP) and aurora B in histone H3 phosphorylation, metaphase chromosome alignment, kinetochore disjunction, and chromosome segregation. J. Cell Biol. 2001;153:865–880. doi: 10.1083/jcb.153.4.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Songyang Z, Blechner S, Hoagland N, Hoekstra MF, Piwnica-Worms H, Cantley LC. Use of an oriented peptide library to determine the optimal substrates of protein kinases. Curr. Biol. 1994;4:973–982. doi: 10.1016/s0960-9822(00)00221-9. [DOI] [PubMed] [Google Scholar]
- 24.Amanchy R, Periaswamy B, Mathivanan S, Reddy R, Tattikota SG, Pandey A. A curated compendium of phosphorylation motifs. Nat. Biotechnol. 2007;25:285–286. doi: 10.1038/nbt0307-285. [DOI] [PubMed] [Google Scholar]
- 25.Copeland RJ, Bullen JW, Hart GW. Cross-talk between GlcNAcylation and phosphorylation: Roles in insulin resistance and glucose toxicity. Am. J. Physiol. Endocrinol. Metab. 2008;295:E17–E28. doi: 10.1152/ajpendo.90281.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chantret I, Moore SE. Free oligosaccharide regulation during mammalian protein N-glycosylation. Glycobiology. 2008;18:210–224. doi: 10.1093/glycob/cwn003. [DOI] [PubMed] [Google Scholar]
- 27.Mackay DR, Elgort SW, Ullman KS. The nucleoporin Nup153 has separable roles in both early mitotic progression and the resolution of mitosis. Mol. Biol. Cell. 2009;20:1652–1660. doi: 10.1091/mbc.E08-08-0883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prunuske AJ, Liu J, Elgort S, Joseph J, Dasso M, Ullman KS. Nuclear envelope breakdown is coordinated by both Nup358/RanBP2 and Nup153, two nucleoporins with zinc finger modules. Mol. Biol. Cell. 2006;17:760–769. doi: 10.1091/mbc.E05-06-0485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Favreau C, Worman HJ, Wozniak RW, Frappier T, Courvalin JC. Cell cycle-dependent phosphorylation of nucleoporins and nuclear pore membrane protein Gp210. Biochemistry. 1996;35:8035–8044. doi: 10.1021/bi9600660. [DOI] [PubMed] [Google Scholar]
- 30.Hughes-Davies L, Huntsman D, Ruas M, Fuks F, Bye J, Chin SF, Milner J, Brown LA, Hsu F, Gilks B, Nielsen T, Schulzer M, Chia S, Ragaz J, Cahn A, Linger L, Ozdag H, Cattaneo E, Jordanova ES, Schuuring E, Yu DS, Venkitaraman A, Ponder B, Doherty A, Aparicio S, Bentley D, Theillet C, Ponting CP, Caldas C, Kouzarides T. EMSY links the BRCA2 pathway to sporadic breast and ovarian cancer. Cell. 2003;115:523–535. doi: 10.1016/s0092-8674(03)00930-9. [DOI] [PubMed] [Google Scholar]
- 31.Lin HR, Ting NS, Qin J, Lee WH. M phase-specific phosphorylation of BRCA2 by Polo-like kinase 1 correlates with the dissociation of the BRCA2-P/CAF complex. J. Biol. Chem. 2003;278:35979–35987. doi: 10.1074/jbc.M210659200. [DOI] [PubMed] [Google Scholar]
- 32.Daniels MJ, Wang Y, Lee M, Venkitaraman AR. Abnormal cytokinesis in cells deficient in the breast cancer susceptibility protein BRCA2. Science. 2004;306:876–879. doi: 10.1126/science.1102574. [DOI] [PubMed] [Google Scholar]
- 33.Raouf A, Brown L, Vrcelj N, To K, Kwok W, Huntsman D, Eaves CJ. Genomic instability of human mammary epithelial cells overexpressing a truncated form of EMSY. J. Natl. Cancer Inst. 2005;97:1302–1306. doi: 10.1093/jnci/dji254. [DOI] [PubMed] [Google Scholar]
- 34.Silk AD, Holland AJ, Cleveland DW. Requirements for NuMA in maintenance and establishment of mammalian spindle poles. J. Cell Biol. 2009;184:677–690. doi: 10.1083/jcb.200810091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kong X, Ball AR, Jr., Sonoda E, Feng J, Takeda S, Fukagawa T, Yen TJ, Yokomori K. Cohesin associates with spindle poles in a mitosis-specific manner and functions in spindle assembly in vertebrate cells. Mol. Biol. Cell. 2009;20:1289–1301. doi: 10.1091/mbc.E08-04-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mueller PR, Coleman TR, Kumagai A, Dunphy WG. Myt1: A membrane-associated inhibitory kinase that phosphorylates Cdc2 on both threonine-14 and tyrosine-15. Science. 1995;270:86–90. doi: 10.1126/science.270.5233.86. [DOI] [PubMed] [Google Scholar]
- 37.Parker LL, Piwnica-Worms H. Inactivation of the p34cdc2-cyclin B complex by the human WEE1 tyrosine kinase. Science. 1992;257:1955–1957. doi: 10.1126/science.1384126. [DOI] [PubMed] [Google Scholar]
- 38.Cha H, Hancock C, Dangi S, Maiguel D, Carrier F, Shapiro P. Phosphorylation regulates nucleophosmin targeting to the centrosome during mitosis as detected by cross-reactive phosphorylation-specific MKK1/MKK2 antibodies. Biochem. J. 2004;378:857–865. doi: 10.1042/BJ20031173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakajima H, Toyoshima-Morimoto F, Taniguchi E, Nishida E. Identification of a consensus motif for Plk (Polo-like kinase) phosphorylation reveals Myt1 as a Plk1 substrate. J. Biol. Chem. 2003;278:25277–25280. doi: 10.1074/jbc.C300126200. [DOI] [PubMed] [Google Scholar]
- 40.Strebhardt K, Ullrich A. Targeting polo-like kinase 1 for cancer therapy. Nat. Rev. Cancer. 2006;6:321–330. doi: 10.1038/nrc1841. [DOI] [PubMed] [Google Scholar]
- 41.Liu F, Stanton JJ, Wu Z, Piwnica-Worms H. The human Myt1 kinase preferentially phosphorylates Cdc2 on threonine 14 and localizes to the endoplasmic reticulum and Golgi complex. Mol. Cell. Biol. 1997;17:571–583. doi: 10.1128/mcb.17.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Z, Pandey A, Hart GW. Dynamic interplay between O-linked N-acetylglucosaminylation and glycogen synthase kinase-3-dependent phosphorylation. Mol. Cell. Proteomics. 2007;6:1365–1379. doi: 10.1074/mcp.M600453-MCP200. [DOI] [PubMed] [Google Scholar]
- 43.Molina H, Horn DM, Tang N, Mathivanan S, Pandey A. Global proteomic profiling of phosphopeptides using electron transfer dissociation tandem mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 2007;104:2199–2204. doi: 10.1073/pnas.0611217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Comer FI, Vosseller K, Wells L, Accavitti MA, Hart GW. Characterization of a mouse monoclonal antibody specific for O-linked N-acetylglucosamine. Anal. Biochem. 2001;293:169–177. doi: 10.1006/abio.2001.5132. [DOI] [PubMed] [Google Scholar]
- 45.Zachara NE, O'Donnell N, Cheung WD, Mercer JJ, Marth JD, Hart GW. Dynamic O-GlcNAc modification of nucleocytoplasmic proteins in response to stress. A survival response of mammalian cells. J. Biol. Chem. 2004;279:30133–30142. doi: 10.1074/jbc.M403773200. [DOI] [PubMed] [Google Scholar]
- 46.Udeshi ND, Compton PD, Shabanowitz J, Hunt DF, Rose KL. Methods for analyzing peptides and proteins on a chromatographic timescale by electron-transfer dissociation mass spectrometry. Nat. Protoc. 2008;3:1709–1717. doi: 10.1038/nprot.2008.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schroeder MJ, Shabanowitz J, Schwartz JC, Hunt DF, Coon JJ. A neutral loss activation method for improved phosphopeptide sequence analysis by quadrupole ion trap mass spectrometry. Anal. Chem. 2004;76:3590–3598. doi: 10.1021/ac0497104. [DOI] [PubMed] [Google Scholar]
- 48.Martin SE, Shabanowitz J, Hunt DF, Marto JA. Subfemtomole MS and MS/MS peptide sequence analysis using nano-HPLC micro-ESI fourier transform ion cyclotron resonance mass spectrometry. Anal. Chem. 2000;72:4266–4274. doi: 10.1021/ac000497v. [DOI] [PubMed] [Google Scholar]
- 49.Geer LY, Markey SP, Kowalak JA, Wagner L, Xu M, Maynard DM, Yang X, Shi W, Bryant SH. Open mass spectrometry search algorithm. J. Proteome Res. 2004;3:958–964. doi: 10.1021/pr0499491. [DOI] [PubMed] [Google Scholar]
- 50.Nichols AM, White FM. Manual validation of peptide sequence and sites of tyrosine phosphorylation from MS/MS spectra. In: Lipton MS, Pasa-Tolic L, editors. Mass Spectrometry of Proteins and Peptides. Humana Press; Totowa, NJ: 2009. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.