Abstract
Background
The destructive plant disease potato late blight is caused by the oomycete pathogen Phytophthora infestans (Mont.) de Bary. This disease has remained particularly problematic despite intensive breeding efforts to integrate resistance into cultivated potato, largely because of the pathogen's ability to quickly evolve to overcome major resistance genes. The RB gene, identified in the wild potato species S. bulbocastanum, encodes a protein that confers broad-spectrum resistance to most P. infestans isolates through its recognition of highly conserved members of the corresponding pathogen effector family IPI-O. IpiO is a multigene family of effectors and while the majority of IPI-O proteins are recognized by RB to elicit host resistance, some variants exist that are able to elude detection (e.g. IPI-O4).
Methods and Findings
In the present study, analysis of ipiO variants among 40 different P. infestans isolates collected from Guatemala, Thailand, and the United States revealed a high degree of complexity within this gene family. Isolate aggressiveness was correlated with increased ipiO diversity and especially the presence of the ipiO4 variant. Furthermore, isolates expressing IPI-O4 overcame RB-mediated resistance in transgenic potato plants even when the resistance-eliciting IPI-O1 variant was present. In support of this finding, we observed that expression of IPI-O4 via Agrobacterium blocked recognition of IPI-O1, leading to inactivation of RB-mediated programmed cell death in Nicotiana benthamiana.
Conclusions
In this study we definitively demonstrate and provide the first evidence that P. infestans can defeat an R protein through inhibition of recognition of the corresponding effector protein.
Introduction
Late blight, caused by the oomycete pathogen Phytophthora infestans, remains one of the most devastating diseases of potato and tomato, even after many decades of resistance breeding efforts. The oomycetes belong to a diverse group of eukaryotic microorganisms that are closely related to brown algae in the Stramenopiles, one of several major eukaryotic kingdoms. Oomycete plant pathogens, such as P. infestans, secrete many proteins that are important in virulence on the host [1]–[3]. These proteins, termed effectors, are either introduced into the plant extracellular space, where they interact with extracellular targets or surface receptors, or into the plant cell cytoplasm. Deciphering the molecular function of these effectors is key to understanding how these pathogens cause disease. A primary role of pathogen effectors is to suppress host basal defense responses, allowing the pathogen to grow and reproduce [4], [5]. In most cases, host resistance proteins recognize the presence of a single pathogen effector molecule [4]. Activation of resistance proteins elicits a strong resistance response, which includes expression of defense-related proteins, an oxidative burst, and programmed cell death termed the hypersensitive response (HR). Some effectors can suppress resistance responses mediated by R proteins, allowing the pathogen to cause disease even when a corresponding R protein has been activated [6]–[9].
Data mining and functional assays have been very helpful in identifying putative effectors from the P. infestans genome sequence [10], [11]. Using this strategy, the avirulence gene Avr3a was identified in P. infestans and cloned [12]. The Avr3a protein is recognized by resistance protein R3a from S. demissum in the host cytoplasm, triggering plant defense responses. Avr3a is one member of a set of hundreds of P. infestans secreted proteins carrying a highly conserved N-terminal motif RXLR (X denotes any amino acid) [12]. Whisson et al. [13] provided convincing evidence that the RXLR motif acts as a host cell-targeting signal that mediates trafficking into host cells.
That fact that host resistance proteins often recognize the presence of pathogen effectors has made it practicable to screen late blight resistant Solanum species for recognition of putative P. infestans effectors [14]. Using this approach, an RXLR effector ipiO was found to induce hypersensitive resistance in the wild potato species Solanum bulbocastanum, S. stoloniferum, and S. papita [14]. IpiO is only known to be present in P. infestans and the closely related species P. andina, P. ipomoeae, P. phaseoli, and P. mirabilis [15]. Expression of ipiO is induced in planta during the early stage of P. infestans infection [16] but no definitive function for ipiO within the host cytoplasm has been determined. IPI-O variants have been divided into three classes based on diversity of deduced amino acid sequences [15]. Class I and class II variants of IPI-O, which are found in the majority of P. infestans isolates, are recognized by the S. bulbocastanum resistance protein RB (or Rpi-blb1) [15], [17], [18]. Interestingly, class III IPI-O variants (e.g. IPI-O4) are not recognized by RB, suggesting that a P. infestans isolate containing only this variant would be able to overcome RB-mediated resistance [15].
RB confers partial foliar resistance to late blight with no effect on plant performance [17]–[19]. Resistance mediated by RB has been found to be effective against many diverse P. infestans isolates due to the almost ubiquitous presence of the corresponding pathogen effector IPI-O class I and class II variants [15]. This differs from P. infestans immunity derived from the wild potato species S. demissum, where virulent races of the pathogen have overcome a majority of the R genes from this host species likely due to mutation or elimination of the corresponding effector [20]. Based on sequence homology to the previously cloned RB gene, Rpi-stol1 and Rpi-pta1 were cloned from S. stoloniferum and S. papita, respectively, and shown to also recognize IPI-O1 and IPI-O2 [14]. Functional homologs of RB have also been identified in other phylogenetically distant wild potato species, such as S. verrucosum [21], confirming that this gene is likely of ancient origin [18] and suggesting functional conservation of the RB gene throughout the evolution of potato.
In this study we utilized 40 isolates of P. infestans collected from foliar-infected potatoes in Guatemala, Thailand, and the United States to analyze the diversity of IPI-O. Our results support the hypothesis that the ipiO locus is extremely variable between isolates, not only in the presence or absence of specific alleles but also in copy number. We have also found that the class III IPI-O variant IPI-O4 not only eludes detection by RB, but is also capable of inhibiting hypersensitive resistance elicited by the class I variant IPI-O1.
Results
IpiO variants are present among diverse P. infestans isolates
To investigate the diversity of the ipiO gene in P. infestans populations, we collected P. infestans from Central America (Guatemala), South East Asia (Thailand) and North America (United States; Table 1). Amplification of the ipiO genes using PCR resulted in products of 456 base pairs, which lack the region encoding the signal sequence but contain DNA encoding the RXLR motif and the remainder of the protein as well as a portion of the 3′ untranslated region. Although sequences were divergent, all P. infestans isolates yielded amplicons that encode putative protein sequences of 131 amino acids. As expected, each isolate contained multiple copies of ipiO. However, the number of ipiO variants and the presence or absence of specific sequences differed. Where significant variability in the number of ipiO variants was found, as many as 32 additional clones (for a total of 48) were sequenced to ensure that all possible variants had been identified with 99% probability. Interestingly, no isolates were found with the same complement of ipiO gene sequences. Overall, among the 40 P. infestans isolates, we obtained 248 unique deduced IPI-O amino acid sequences. Class I variants (IPI-O1 and IPI-O2 related) were found in all the P. infestans isolates. The presence of class II variants (IPI-O3 related) was only slightly less consistent, with variants of this gene found in 78% of the isolates. An obvious exception was the US isolates, none of which contain class II variants. Class III variants (IPI-O4 related) were much more rare, and were only found in 7 of the 41 isolates (17%).
Table 1. P. infestans isolates used for IpiO sequencing.
| Isolate Name | Area collected (region, state or province, sub-province, country | Isolate # | Race | Obtained from |
| 19 | Patzicia, Chimaltenango, Guatemala | unknown | direct collection, 2007 | |
| 20 | Patzicia, Chimaltenango, Guatemala | unknown | direct collection, 2007 | |
| 27 | Georginas, Quetzaltenango, Guatemala | unknown | direct collection, 2007 | |
| 34 | Georginas, Quetzaltenango, Guatemala | unknown | direct collection, 2007 | |
| 39 | Georginas, Quetzaltenango, Guatemala | unknown | direct collection, 2007 | |
| 40 | Georginas, Quetzaltenango, Guatemala | unknown | direct collection, 2007 | |
| 44 | Concepción 3, Quetzaltenango, Guatemala | unknown | direct collection, 2007 | |
| 46 | Concepción 3, Quetzaltenango, Guatemala | unknown | direct collection, 2007 | |
| 47 | Aguas Amargas, Quetzaltenango, Guatemala | unknown | direct collection, 2007 | |
| 49 | Aguas Amargas, Quetzaltenango, Guatemala | unknown | direct collection, 2007 | |
| 52 | Aguas Amargas, Quetzaltenango, Guatemala | unknown | direct collection, 2007 | |
| 54 | Aguas Amargas, Quetzaltenango, Guatemala | unknown | direct collection, 2007 | |
| 57 | Aguas Amargas, Quetzaltenango, Guatemala | unknown | direct collection, 2007 | |
| 64 | Cobán, Alta Verapaz, Guatemala | unknown | direct collection, 2007 | |
| 67 | Cobán, Alta Verapaz, Guatemala | unknown | direct collection, 2007 | |
| 68 | Cobán, Alta Verapaz, Guatemala | unknown | direct collection, 2007 | |
| CMPh0-03 | Chiang Mai, Fang, Thailand | unknown | direct collection, 2007 | |
| CMPh0-05 | Chiang Mai, Fang, Thailand | unknown | direct collection, 2007 | |
| CMPh0-07 | Chiang Mai, Fang, Thailand | unknown | direct collection, 2007 | |
| CMSS1-02 | Chiang Mai, San Sai, Chedimaekhrua, Thailand | unknown | direct collection, 2007 | |
| CMSS1-04 | Chiang Mai, San Sai, Chedimaekhrua, Thailand | unknown | direct collection, 2007 | |
| CMSS1-08 | Chiang Mai, San Sai, Chedimaekhrua, Thailand | unknown | direct collection, 2007 | |
| CMSS2-03 | Chiang Mai, San Sai, Mae-Faek-Mai, Thailand | unknown | direct collection, 2007 | |
| CMSS2-10 | Chiang Mai, San Sai, Mae-Faek-Mai, Thailand | unknown | direct collection, 2007 | |
| CMSS2-15 | Chiang Mai, San Sai, Mae-Faek-Mai, Thailand | unknown | direct collection, 2007 | |
| CMSS3-05 | Chiang Mai, San Sai, Nong-Han, Thailand | unknown | direct collection, 2007 | |
| CMSS3-15 | Chiang Mai, San Sai, Nong-Han, Thailand | unknown | direct collection, 2007 | |
| CMSS3-24 | Chiang Mai, San Sai, Nong-Han, Thailand | unknown | direct collection, 2007 | |
| TKPP1-02 | Tak, Phob-Phra, RuamThai-Patana, Thailand | unknown | direct collection, 2007 | |
| TKPP1-05 | Tak, Phob-Phra, RuamThai-Patana, Thailand | unknown | direct collection, 2007 | |
| TKPP1-06 | Tak, Phob-Phra, RuamThai-Patana, Thailand | unknown | direct collection, 2007 | |
| TKPP1-07 | Tak, Phob-Phra, RuamThai-Patana, Thailand | unknown | direct collection, 2007 | |
| US1a | United States | US940501 | 0 | W. Fry, Cornell University |
| US1b | United States | WI 94-1 | unknown | W. Stevenson, University of Wisconsin |
| US11a | United States | S37A1994 | unknown | W. Stevenson, University of Wisconsin |
| US11b | United States | US980008 | unknown | B. Baker, USDA/ARS |
| US8a | United States | US940480 | 0,1,2,3,4,5,6,7,10,11 | B. Baker, USDA/ARS |
| US8b | United States | US930287 | unknown | W. Fry, Cornell University |
| US8c | United States | 693-3 | 0,1,2,3,4,5,6,7,8,10,11 | N. Gudmestad, North Dakota State |
| US8d | United States | 126-C-18 | 0,1,2,3,4,5,6,7,8,9,10,11 | N. Gudmestad, North Dakota State |
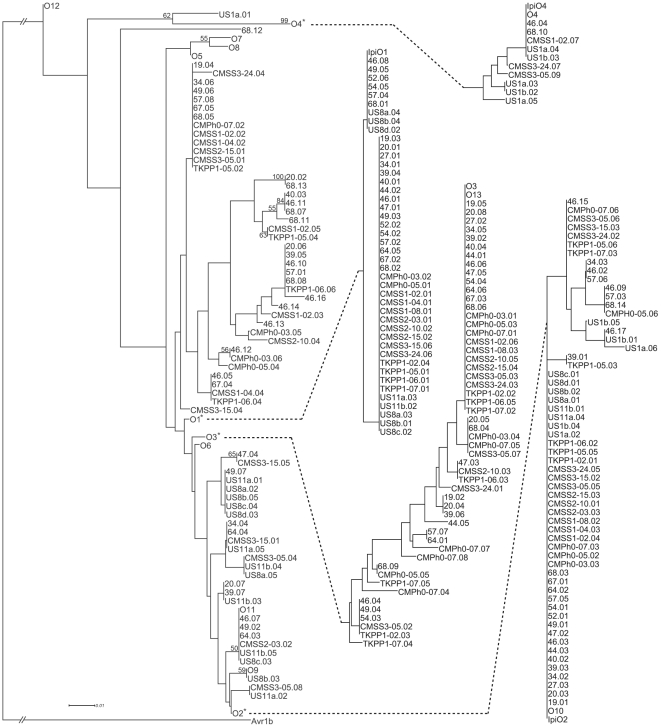
Phylogenetic analysis of deduced IPI-O peptide sequences showed a grouping into multiple clusters (Figure 1). The pattern is similar to another recently published IPI-O diversity analysis [15] with organization into three different classes. A cluster of IPI-O4-related sequences (class III) was clearly distinct from the rest of the IPI-O variants. IPI-O3-related variants represent class II and the remaining IPI-O sequences represent class I. Clustering based on nucleotide sequences yielded similar results. Despite the apparent IPI-O diversity present among isolates, only 28 of the 131 amino acids were polymorphic in the sequences derived from our experiments. The majority (23/28) of the polymorphic sites are located in the 80 amino acids C-terminal to the RSLR-EER motif and 12 are located within the 25 amino acid W-motif, a motif sharing homology between diverse Phytophthora RXLR-EER effectors [11].
Figure 1. Grouping of IPI-O peptide sequences into multiple clusters.
This dendrogram shows hierarchical clustering of deduced amino acid sequences of IPI-O variants identified from 16 P. infestans isolates from Guatemala (beginning with numbers), 16 isolates from Thailand (beginning with CMSS, CMPh, or TKPP), and 8 from the United States (beginning with US). Previously identified IPI-O sequences (O1-O13) were also included. The “*” indicates the IPI-O1, IPI-O2, IPI-O3, and IPI-O4 clusters, which are linked to the dendogram by dotted lines. Branch lengths (solid lines) were not altered and represent the evolutionary distances used to deduce the tree. Bootstrap values ≥50% from 1000 replications are shown at the nodes.
Individual amino acid sites in IPI-O were analyzed for diversifying selection using phylogenetic analysis by maximum likelihood (PAML) [22], [23]. Selection models M2a and M8 both identified the same 14 positively selected amino acids among unique IPI-O sequences (Table 2). A likelihood ratio test of the results of these two models indicated a significant probability that these sites are under positive selection and are not the result of relaxation of selection pressure (p<0.001). Interestingly, six of these sites reside within the conserved W-motif (Figure S1). Of the seven amino acid sites previously identified to be under divergent selection among P. infestans IPI-Os [15], five overlap with the sites identified in our analysis.
Table 2. IPI-O amino acid sites under diversifying selection.
| Model | Parameter estimates | lnLa | Positively selected sitesb |
| M1a | ω0 = 0, ω1 = 1, p0 = 0.749, p1 = 0.251 | −1368.65 | neutral selection model |
| M2a | ω0 = 0, ω1 = 1, ω2 = 12.141, p0 = 0.674, p1 = 0.200, p2 = 0.125 | −1313.38 | 30V*, 32Y*, 46N*, 68S*, 87L*, 89G*, 92L*, 113A*, 117S*, 122R*, 124L*, 129L, 134A*, 135S, 143N* |
| M7 | p = 0.111, q = 0.039 | −1369.92 | neutral selection model |
| M8 | p0 = 0.874, p1 = 0.006, q = 0.226, ω = 11.682 | −1313.60 | 30V*, 32Y*, 46N*, 68S*, 87L*, 89G*, 92L*, 113A*, 117S*, 122R*, 124L*, 129L, 134A*, 135S, 143N* |
log likelihood value.
p<0.05.
= p<0.01.
P. infestans isolates contain differing numbers of IPI-O variants
The mean/median number of unique deduced amino acid sequences (IPI-O variants) per isolate was 6.8/5.5 in the Guatemalan isolates, 5.6/6.0 in the Thai isolates, and 4.8/5.0 in the U.S. isolates (Table 3). However, in two Guatemalan isolates, named #46 and #68, we found 17 and 14 IPI-O variants, respectively. This level of complexity was not observed in any other isolates although one Thai isolate, CMSS3-05, contained 9 IPI-O variants. In contrast, several isolates, including representatives from Guatemala, Thailand, and the US, contained only 3 IPI-O variants. This variability between isolates is likely not due to amplification-based anomalies since a high-fidelity, high-processivity polymerase and adequate elongation times were used to avoid the possibility of chimera formation during PCR. Additionally, amplification of a majority of the DNA samples produced relatively few variants indicating true copy number variability within the ipiO locus.
Table 3. Number and classification of IPI-O variants in each P. infestans isolate.
| Class | Class | Class | |||||||||||||||
| Isolate | Unique aa sequences | I | II | III | Isolate | Unique aa sequences | I | II | III | Isolate | Unique aa sequences | I | II | III | |||
| Guatemala | 19 | 5 | 3 | 2 | 0 | Thailand | CMPh0-03 | 6 | 4 | 2 | 0 | United States | US1a | 6 | 2 | 0 | 4 |
| 29 | 8 | 5 | 3 | 0 | CMPh0-05 | 6 | 4 | 2 | 0 | US1b | 5 | 3 | 0 | 2 | |||
| 27 | 3 | 2 | 1 | 0 | CMPh0-07 | 8 | 3 | 5 | 0 | US11a | 5 | 5 | 0 | 0 | |||
| 34 | 6 | 5 | 1 | 0 | CMSS1-02 | 7 | 5 | 1 | 1 | US11b | 5 | 5 | 0 | 0 | |||
| 39 | 7 | 5 | 2 | 0 | CMSS1-04 | 4 | 4 | 0 | 0 | US8a | 5 | 5 | 0 | 0 | |||
| 40 | 4 | 3 | 1 | 0 | CMSS1-08 | 3 | 2 | 1 | 0 | US8b | 5 | 5 | 0 | 0 | |||
| 44 | 5 | 2 | 3 | 0 | CMSS2-03 | 3 | 3 | 0 | 0 | US8c | 4 | 4 | 0 | 0 | |||
| 46 | 17 | 15 | 1 | 1 | CMSS2-10 | 5 | 3 | 2 | 0 | US8d | 3 | 3 | 0 | 0 | |||
| 47 | 5 | 3 | 2 | 0 | CMSS2-15 | 4 | 3 | 1 | 0 | Average | 4.8 | ||||||
| 49 | 7 | 6 | 1 | 0 | CMSS3-05 | 9 | 5 | 3 | 1 | Median | 5.0 | ||||||
| 52 | 3 | 3 | 0 | 0 | CMSS3-15 | 6 | 6 | 0 | 0 | ||||||||
| 54 | 5 | 3 | 2 | 0 | CMSS3-24 | 7 | 4 | 2 | 1 | ||||||||
| 57 | 8 | 7 | 1 | 0 | TKPP1-02 | 4 | 2 | 2 | 0 | ||||||||
| 64 | 6 | 4 | 2 | 0 | TKPP1-05 | 6 | 6 | 0 | 0 | ||||||||
| 67 | 5 | 4 | 1 | 0 | TKPP1-06 | 6 | 4 | 2 | 0 | Class | |||||||
| 68 | 14 | 10 | 3 | 1 | TKPP1-07 | 5 | 3 | 2 | 0 | Unique aa sequences | I | II | III | ||||
| Average | 6.8 | Average | 5.6 | All isolates | Average | 5.9 | 4.3 | 1.3 | 0.3 | ||||||||
| Median | 5.5 | Median | 6.0 | Median | 5.0 | 4.0 | 1.0 | 0.0 | |||||||||
However, in order to further verify differences in ipiO copy number, we performed real-time quantitative PCR using genomic DNA from Guatemalan P. infestans isolates #27, #46, #52, and #68 as well as US isolate US8a. Copy number of ipiO was determined through comparison to the P. infestans single copy nuclear gene β-tubulin [24]. Standard curves using RD6F/RD6R and TUB901/TUB1401 primer pairs resulted in correlation coefficients of 0.997 and 0.990, respectively. Amplification efficiency of ipiO was 87.1% and β-tubulin was 86.8% using these primer pairs. The efficiencies were sufficiently similar to compare threshold cycle differences to determine gene copy number. The 2ΔΔCt method was employed using isolate #27 as a calibrator (with 3 copies of IpiO). Using this calibration we estimated that isolate #52 contains 3.5 copies of ipiO, #46 contains 13.4 copies, #68 contains 11.3 copies, and US8a contains 5.4 copies using this method. The correlation coefficient between the number of unique amino acid sequences and the estimated copy number determined by RT-PCR was 0.999.
When comparing variant composition between isolates, we observed a correlation between IPI-O variability and the presence of the class II variant IPI-O4 in Guatemalan and Thai isolates (r = 0.69; p<0.001). Isolates #46 and #68 were the only two Guatemalan isolates containing IPI-O4. Additionally, the presence of IPI-O4 was correlated with increased IPI-O variability in Thai isolates CMSS1-02, CMSS3-05, and CMSS3-24. Among the US isolates, IPI-O4 was found in both US1 strains, both of which contain above average numbers of IPI-O variants.
IPI-O variant diversity correlates with pathogen aggressiveness
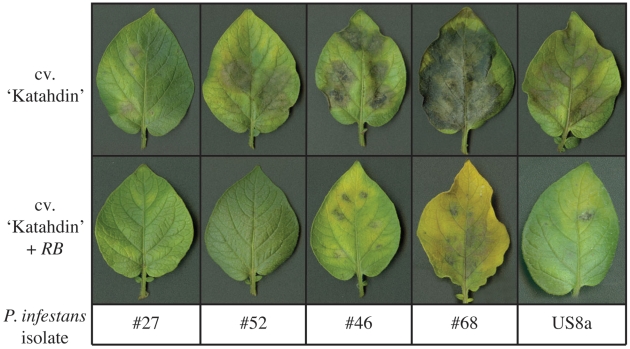
In order to determine whether ipiO genetic variation correlates with isolate aggressiveness, we used detached leaflets of susceptible potato cv. ‘Katahdin’ and resistant cv. ‘Katahdin’ plants containing a single copy of the RB gene (‘SP951’) for inoculation with Guatemalan isolates #46, #68, #27, and #52 along with US8a strain US940480 (5 IPI-O variants; Figure 2; Table 4). Our data show that, based on lesion size, isolate #68, but not #46, was significantly (p<0.01) more aggressive on leaves from cv. ‘Katahdin’ compared to the other isolates. Interestingly, isolates with ipiO4 and multiple variants of other ipiO genes (#46 and #68) were significantly (p<0.01) more aggressive on plants containing the RB resistance gene, indicating their ability to overcome the partial RB-mediated resistance response more effectively than those with fewer alleles and no ipiO4 (#27, #52, and US8a). A comparison of ‘Katahdin’ and ‘SP951’ plants revealed that the presence of the RB gene significantly decreased lesion sizes when inoculated with isolates US8a, #52, and #68. The percentage reduction of lesion area due to the presence of the RB gene was highest with the US8a and #52 isolates. RB had the least effect on isolate #46. This suggested variability among isolates in their ability to overcome RB-mediated resistance.
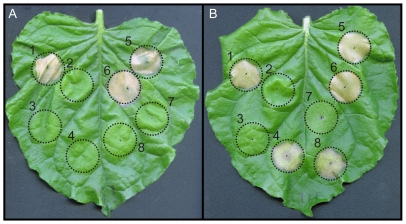
Figure 2. Inoculation of detached leaflets with selected P. infestans isolates.
Detached leaflets were inoculated with P. infestans by placing a 10 µl drop of inoculum at 6 spots on the abaxial surface the leaflets. Photos were taken 5 days after inoculation. Average lesion (necrosis+chlorosis) diameters (in millimeters) of 24 replications are shown in Table 3. Top row: susceptible S. tuberosum cv. ‘Katahdin’; bottom row: transgenic ‘Katahdin’ containing the RB gene.
Table 4. Results of detached leaflet infection assays.
| Average lesion areaa | |||
| P. infestans isolate | cv. ‘Katahdin’ | ‘SP951’ | % reduction due to RB |
| US8a | 62.8±10.7 AY | 1.8±1.1 AZ | 97.1±1.8% |
| #27 | 41.8±13.4 AY | 16.6±7.6 AY | 60.3±22.2% |
| #52 | 69.1±13.1 AY | 1.9±1.8 AZ | 97.3±2.7% |
| #46 | 108.8±15.4 AY | 76.4±17.7 BY | 29.8±18.3% |
| #68 | 388.8±59.1 BY | 67.0±14.1 BZ | 82.8±4.1% |
lesion areas were calculated by averaging 24 inoculation events. Areas followed by different letters indicate they are significantly different within host genotypes (A and B) or between host genotypes (Y and Z) at p<0.01.
IPI-O4 suppresses IPI-O1-induced HR in the presence of RB
The ability of some isolates to overcome RB-mediated resistance raised the possibility that specific IPI-O variants are able to suppress responses induced by activation of RB. In order to test this, we infiltrated RB-transgenic N. benthamiana leaves with A. tumefaciens strains carrying constructs expressing IPI-O4, IPI-O1, INF1, and green fluorescent protein (GFP; Figure 3). INF1 is a P. infestans elicitin that induces an HR in potato and N. benthamiana [25]. Additionally, IPI-O1, INF1 and GFP expressing strains were co-infiltrated with the IPI-O4 expressing strain. Hypersensitive cell death was observed 7 days after infiltration in regions infiltrated with A. tumefaciens expressing INF1 and IPI-O1 alone. No HR was observed in regions exposed to GFP or IPI-O4. Additionally, no HR was observed in areas co-infiltrated with A. tumefaciens expressing IPI-O1 and IPI-O4, indicating a suppression of cell death in the presence of IPI-O4. An HR was observed when IPI-O4 and INF1 were coexpressed, suggesting that IPI-O4 suppression of cell death is IPI-O1-specific. Suppression of the IPI-O1-induced HR was not observed in areas where IPI-O1 and GFP were coexpressed. The suppression phenotype was also observed in overlapping agroinfiltrated regions where IPI-O4-expressing A. tumefaciens was infiltrated three days prior to infiltration with bacteria expressing IPI-O1 (Figure S2). In order to test whether an overabundance of IPI-O1 could overwhelm IPI-O4 suppression, RB-transgenic N. benthamiana plants were infiltrated with a mixture of A. tumefaciens where the ratio of bacteria expressing IPI-O4 to those expressing IPI-O1 was 1∶5 and 1∶10 (Figure 4). Despite the increased amount of A. tumefaciens expressing IPI-O1, no cell death was observed, demonstrating that IPI-O4-mediated suppression of IPI-O1 recognition can take place even with an expected overabundance of the elicitor.
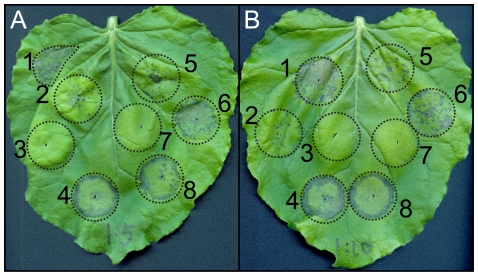
Figure 3. Effect of ipiO4 on the ipiO1-induced hypersensitive response.
Non-transgenic (A) and transgenic N. benthamiana containing the RB gene (B) were infiltrated with A. tumefaciens containing the following constructs: 1) pGR106-INF1; 2) pGR106-GFP; 3) pGR106-ipiO4; 4) pGR106-ipiO1; 5) pGR106-INF1 + pGR106-ipiO4; 6) pGR106-INF1 + pGR106-GFP; 7) pGR106-ipiO1 + pGR106-ipiO4; 8) pGR106-ipiO1 + pGR106-GFP. Co-infiltration was accomplished using a mixture of equal amounts of Agrobacterium. The photograph, which is representative of multiple replications, was taken 7 days after agroinfiltration.
Figure 4. Effect of IPI-O1 overabundance on IPI-O4 mediated HR suppression.
Transgenic N. benthamiana containing the RB gene was infiltrated with A. tumefaciens containing the following constructs: 1) pGR106-INF1; 2) pGR106-GFP; 3) pGR106-ipiO4; 4) pGR106-ipiO1; 5) pGR106-INF1 + pGR106-ipiO4; 6) pGR106-INF1 + pGR106-GFP; 7) pGR106-ipiO1 + pGR106-ipiO4; 8) pGR106-ipiO1 + pGR106-GFP. Co-infiltration was accomplished using a mixture of Agrobacterium strains. A contains 5-fold more and B contains 10-fold more Agrobacterium expressing INF1 or ipiO1 in 5, 6, 7, and 8 compared to the co-infiltrated construct. The photograph was taken 7 days after agroinfiltration.
Discussion
Late blight remains a worldwide problem for potato production. P. infestans is a particularly destructive pathogen and can lead to significant crop losses due to early defoliation and tuber infection. The pathogen spreads rapidly and can cause total crop loss within days [20]. Race-specific resistance to late blight introgressed from S. demissum has consistently been overcome by new strains of the pathogen. The RB gene, from S. bulbocastanum, confers resistance to most isolates of P. infestans and has therefore been defined as broad-spectrum [17], [18]. The foliar resistance phenotype mediated by RB differs from race-specific immunity derived from the wild species S. demissum since RB-containing plants allow P. infestans to grow and sporulate to a small degree [19], [26]. Separately, we have shown that RB is capable of eliciting the HR and other resistance responses, indicating recognition of P. infestans effectors (Y. Chen and D. Halterman, submitted). However, RB-expressing plants are compromised in their ability to completely stop pathogen spread suggesting that P. infestans is able to suppress some RB-mediated responses before the pathogen can be contained. Recently, Champouret and colleagues [15] categorized IPI-O variants into three classes and concluded that the presence of class I variants determine avirulence on plants with RB and that P. infestans isolates lacking this class are able to overcome resistance mediated by this gene. Our results support these findings and also indicate that the presence of class III variants (IPI-O4) allows P. infestans to overcome the avirulence phenotype even when a class I IPI-O is present. It remains unclear, however, whether IPI-O4 interferes with recognition of products of IPI-O expression or elicitation of the HR after recognition by RB of products of IPI-O1 expression. Preliminary evidence suggests a physical interaction between IPI-O4 and a portion of the RB protein (Z. Liu, Y. Chen, and D. Halterman, unpublished data). Interestingly, we have detected no similar interaction between IPI-O1 and RB, suggesting that IPI-O4 may be directly blocking recognition of IPI-O1 rather than affecting the expression, import, or stability of IPI-O1. The fact that IPI-O1 and IPI-O4 were introduced directly into the cell by Agrobacterium further suggests that inhibition is taking place in the cytoplasm, where RB is presumably located. Whether IPI-O4 is able to inhibit recognition of other class I or II IPI-O variants has yet to be determined and will be a focus of future research.
The ability of IPI-O4 to inhibit programmed cell death mediated by RB is not unique to the co-evolution of P. infestans and its hosts. The I gene from the flax rust pathogen Melampsora lini empowers a normally avirulent strain with the ability to overcome resistance conferred by specific R genes [7], [27], [28]. This activity likely acts at the protein level since the pathogen is able to express the corresponding effector genes normally. Data accumulated from the I effect suggest that inhibition of the R protein is controlled via interactions between different domains within the protein and determined by sequence variation in interacting regions [7], [27], [28]. Similarly, the ATR1NdWsB RXLR effector from the oomycete pathogen Hyaloperonospora parasitica elicits a resistance response mediated by the corresponding Arabidopsis R gene RPP1-WsB when expressed with the R gene via cobombardment [9]. However, the Emco5 isolate containing this effector is virulent on plants with RPP1-WsB [9]. Although the mechanisms involved in allowing Emco5 to overcome resistance are unknown, the authors suggest the possibility that Emco5 is able to evade or suppress recognition of ATR1NdWsB by RPP1-WsB [9]. Effectors of the bacterial pathogen Pseudomonas syringae pv. DC3000 also function to suppress resistance in tomato and Arabidopsis. The Fen gene of tomato is in the same gene family as Pto, which encodes a protein kinase and confers resistance to P. syringae expressing the effectors AvrPto and/or AvrPtoB [29], [30]. Despite 80% amino acid identity, Fen is not able to elicit resistance in the presence of AvrPto or AvrPtoB [31]. Molecular analysis of AvrPtoB has revealed that it encodes a ubiquitin ligase and that Fen is able to elicit resistance to a truncated version of AvrPtoB lacking ligase activity [6], [32]. Full-length AvrPtoB is able to suppress Fen-mediated immunity through ubiquitination of Fen itself leading to degradation of the protein, which demonstrates evolution of the effector to overcome R gene-mediated resistance [6]. Similarly, the P. syringae effector AvrRpt2 disrupts signaling by the resistance protein RPM1 in Arabidopsis by cleaving an essential RPM1-interacting protein, RIN4 [8]. The mechanism by which the P. infestans effector IPI-O4 is able to suppress RB-mediated resistance remains to be fully elucidated. However, results from the work presented here suggest that the phenomenon is specific for RB and that IPI-O4 is not a general suppressor of cell death. It will be interesting to determine whether wild potato species that are able to recognize IPI-O4 to elicit resistance (e.g. S. stoloniferum) contain “defeated” RB-like genes within the locus that allow for recognition of a different suite of IPI-O variants, or whether other factors are involved in IPI-O4 recognition.
Expression of RB is closely correlated with the resistance phenotype and transgenic plants with multiple integration events are typically more resistant than those with fewer copies of RB [33], [34]. ‘SP951’, the transgenic potato plants used in our detached leaflet assays, is transgenic S. tuberosum cv. ‘Katahdin’ containing a single copy of the RB gene. Isolates containing IPI-O4 appeared more aggressive, even when RB was present, possibly due in part to inhibition of IPI-O1 recognition. Interestingly, Guatemalan isolate #27, which does not appear to contain ipiO4, was able to overcome RB-mediated resistance at a similar relative level as isolate #68. This suggests that other factors may be involved, such as ipiO expression or interaction with unidentified host- or pathogen-derived factors. It is possible that an increase in RB expression could overcome IPI-O suppression and restore the resistance phenotype, although further testing of transgenic plants with increased levels of RB expression is necessary to make this determination. The ability to manipulate resistance in this fashion may be completely dependent on the number of IPI-O4 molecules that enter the plant cell and whether RB expression can be increased enough to overcome suppression of IPI-O1 recognition.
The majority of IPI-O diversity in the isolates analyzed in our experiments was restricted to class I, although nearly all isolates also contained at least one class II variant. The median number of total IPI-O variants from all of the isolates was 5, and most of those variants belong to class I, with a median number of 4. However there were obvious exceptions, with Guatemalan isolates #46 and #68 containing 17 and 14 IPI-O variants, respectively. It is unclear at this point what may have driven diversity at the ipiO locus in these isolates. The two strains were collected from separate locations, and other isolates taken from similar locations did not have this level of IPI-O diversity. Isolates #46 and #68 were collected from S. tuberosum plants under field conditions suggesting that the increase in IPI-O diversity is not necessarily due to an interaction with resistant hosts. However, S. bulbocastanum and S. demissum, both of which contain major late blight resistance genes, are present in Guatemala in regions near where the isolates were collected [35]. It is therefore possible that these isolates have had recent contact with resistant or partially resistant wild species, leading to a dramatic increase in IPI-O diversity. This does not explain the increased number of IPI-O variants in some Thai isolates since wild potato species are not common in Thailand. This would suggest that other unknown factors might also influence IPI-O diversity. Previously, two other P. infestans isolates exhibiting virulence on RB-containing plants, PIC99189 and PIC99177, were collected from the wild species S. stoloniferum, from which a functional RB homolog has been identified [15], [36], [37]. However, in the case of PIC99189 and PIC99177, virulence was attributed to the absence of class I IPI-O variants [15], whereas we have found that the presence of certain IPI-O variants, namely IPI-O4, may impact an isolate's ability to overcome resistance.
Based on previous published data [15], [38], we expected to find relatively few IPI-O variants. We were surprised by the level of diversity present within the P. infestans isolates used in this study, particularly those collected recently from infected potato leaves. Incomplete primer extension during the PCR elongation step can result in chimera formation in vitro during the amplification of these products [39]. However, preventative measures (adequate elongation time and high-fidelity polymerase) were used to minimize this possibility. The possibility also exists that IPI-O diversity is maintained in wild populations of P. infestans and may be lost after removal of the pathogen from its natural habitat. This could explain why the majority of Guatemalan and Thai isolates contain increased IPI-O diversity. The US isolates used in this study, which have not been exposed to resistant plants for several years, contained the lowest average and median number of unique IPI-O amino acid sequences. We are currently in the process of passaging isolates through RB-containing hosts to determine whether this interaction can affect IPI-O diversity over multiple generations.
Up until the early 1990s, US1 was the most prevalent strain of P. infestans in the US, but was replaced by US8 and US6 isolates within a period of a few years [40], [41]. The US1 isolate used in this study has been used previously to test for virulence on RB-containing plants with no notable increase in aggressiveness despite the fact that it contains ipiO4 [17, D. Halterman, unpublished data]. This would suggest that factors contributing to virulence on plants with the RB gene remain to be found. Despite the fact that RB confers broad spectrum resistance, isolates that can overcome resistance have already been identified [15]. Therefore, it is essential to identify germplasm with resistance sources that will allow recognition of diverse IPI-O variants. Pyramiding of R genes that recognize different P. infestans effectors, whether they be multiple variants of ipiO or unrelated effectors such as avrRpi-blb2, will be important in battling this disease at the breeding level. The RB gene remains a valuable trait because it confers broad-spectrum resistance to P. infestans with no impact on yield in tested transgenic cultivars [19]. The accumulated costs of control efforts and losses due to late blight alone are estimated at more than $3 billion/year worldwide [20]. Therefore, the deployment of plants that require little or no fungicide input and remain resistant even under conditions ideal for disease development should significantly impact costs associated with growing potato everywhere the crop is grown.
Materials and Methods
Collection and maintenance of P. infestans
Infected leaf samples were collected from areas of major potato production during the rainy seasons in Guatemala and Thailand (August and September 2007 in Guatemala; February and December in Thailand). Guatemalan and Thai isolates were collected from S. tuberosum plants of varying cultivars. Leaves with a single lesion of late blight were collected and the process of isolation was started the same day of collection. Late blight-infected lesions were excised from the leaf and placed on clarified V8 (cV8) agar (15% clarified V8 juice, 1.5% CaCO3; and 1.5% agar in distilled water) and incubated in the dark at 15°C. Subsamples of P. infestans hyphal tips were collected and placed on fresh cV8 media and stored in the dark at 15°C.
Isolation of ipiO alleles from Phytophthora
P. infestans isolates were grown in liquid pea broth media and DNA was isolated using a previously published protocol [42]. Fifty nanograms of Phytophthora DNA were used as a template for polymerase chain reaction (PCR). Primers RD6F (5′ - CGCATCGATGGTTTCATCCAATCTCAACACCGCCG - 3′) and RD6R (5′ - GATGCGGCCGCTATACGATGTCATAGCATGACA - 3′) were used to amplify ipiO alleles using the following parameters: 94° for 1 min; 40 cycles of 94° for 15 s, 52° for 30 s, 68° for 1.5 min; 68° for 15 minutes. All amplifications were carried out using Platinum® PCR SuperMix High Fidelity (Invitrogen, Carlsbad, CA) with 5 nmoles of each primer. PCR products were cloned into pGEM-T Easy vector (Promega, Madison, WI) according to the manufacturers instructions.
Sequencing and analysis
At least 16 plasmid clones containing ipiO variants from each isolate were sequenced in both directions using vector-specific primers. Poisson distribution was used to ensure that the cumulative probability of finding all alleles was greater than 99%. Double-strand sequencing of DNA was carried out at the University of Wisconsin-Madison Biotechnology Center sequencing facility. Vector sequence removal was performed using Vector NTI software (Invitrogen, CA). Duplicate identical sequences from the same P. infestans isolate were removed from the analysis. Sequence alignments were done using ClustalX [43] using the P. infestans effector Avr1b as an outgroup. Trees were visualized using Dendroscope [44]. Bootstrap values in percentage (≥50) from 1000 repetitions are shown at the nodes.
Identification of positively selected sites
The 78 unique IpiO DNA sequences were used to identify amino acid codons under divergent selection using the codeml program within the PAML v. 4 package [23]. Maximum-likelihood models M0, M1a, M2a, M7 and M8 were used for the analysis. The log-likelihood values from the M1a (neutral selection) and M2a (positive selection) models and the M7 (neutral selection) and M8 (positive selection models were used to perform a likelihood ratio test (LRT). The LRT statistic was calculated as twice the difference in log-likelihood values for the two models, and the cumulative distribution of test statistics for each set of simulated data was plotted against a chi-square distribution with two degrees of freedom.
Estimation of ipiO copy number using real time PCR
Real-time PCR reactions were carried out using a Bio-Rad MyiQ thermocycler (Bio-Rad, CA). All reactions were run in triplicate. For each reaction, 1 µl of each primer and 50 ng of P. infestans genomic DNA was added to 22.5 µl of iQ SYBR Green Supermix (Invitrogen, CA). Primers RD6F and RD6R were used to amplify ipiO and primers TUB901 (5′-TACGACATTTGCTTCCG-3′) and TUB1401 (5′- CGCTTGAACATCTCCTGG-3′) were used to amplify β-tubulin [24]. PCR was performed as follows: 3 min. at 94°C; 40 cycles of 15 sec at 94°C, 30 sec at 56°C, and 30 sec at 68°C. Reactions were followed by melt curve analysis to verify the presence of only one amplicon. The Ct values were determined using the instrument's software. We calculated ipiO copy number using the 2ΔΔCt method [45] where ΔCt = Ct (ipiO) - Ct (β-tubulin) and ΔΔCt = ΔCt (unknown isolate) - ΔCt (isolate #27). Estimated copy number was determined using the 2ΔΔCt and calibrating the results so that the reference isolate #27 contained three copies of ipiO.
Agroexpression of IPI-O variants in Nicotiana benthamiana
N. benthamiana plants were transformed with the RB gene at the University of Wisconsin-Madison Biotechnology Center plant transformation facility. The RB gene construct used for transformation was the same as previously published [17]. Agroexpression of ipiO variants was performed using vector pGR106 [46] in Agrobacterium tumefaciens strain GV3101 [47] as previously described [48]. A. tumefaciens strains were diluted to a concentration of OD600 = 0.3 before infiltration with a needleless syringe. Inoculated N. benthamiana plants were incubated in a growth chamber (22°C day/18°C night temperatures with 16 h of light). Photographs were taken 7 days after inoculation.
Detached leaf infection assays
All potatoes were propagated from cuttings and maintained in the greenhouse, which was set for 18 h of daylight, a daytime temperature between 17 and 19°C, and a nighttime temperature between 13 and 15°C. Leaflets from six to eight week old cv. ‘Katahdin’ and ‘SP951’ (cv. ‘Katahdin’ with one copy of RB) plants were collected. Petioles were trimmed and the leaflets were inserted into plastic boxes containing 0.67% water agar 2 to 24 hours before inoculating with 2 leaflets per cube. P. infestans cultures (16–22 days old) were flooded with sterile, distilled water and washes were combined to obtain a suspension of approximately 45,000 sporangia/ml. Sporangial suspensions were placed at 12°C for 3 hours to induce zoospore release. Leaflets were then inoculated with 6 evenly spaced 10 µl drops per leaflet, for a total of 24 independent inoculations per isolate/genotype interaction. Inoculations with each P. infestans isolate were repeated twice. Cubes were covered and placed in a 15°C incubator for 6 days before reading results. Six days after inoculation, necrotic plus chlorotic lesion diameters were measured in millimeters. The diameters were used to calculate average lesion areas per inoculation from the 24 inoculation events.
Supporting Information
Amino acid alignment of IPI-O sequences. For simplicity, duplicate sequences were removed. IPI-O1 sequence is shown along the top. Identical amino acids were replaced with “.” while polymorphic amino acids are shown. A “*” denotes amino acids determined to be under selection for divergence (see Table 2). Lines above the sequence show the RXLR/RGD, DEER, and predicted W motifs from left to right, respectively. Secondary structure prediction (shown at bottom) was done using the PSIPRED protein structure prediction server (http://www.psipred.net/psiform.html). C = coil, E = strand, H = helix. Confidence values (0 = low, 9 = high) are shown below each structure prediction.
(0.26 MB DOC)
Transgenic N. benthamiana containing the RB gene was infiltrated with A. tumefaciens containing the following constructs: A), C), and F) pGR106-IpiO1; B) and D) pGR106-GFP; E) and G) pGR106-ipiO4. C) and F) were infiltrated three days after the other constructs. The photograph was taken 5 days after agroinfiltration of the final constructs.
(2.29 MB DOC)
Acknowledgments
We would like to thank Gail Middleton for expert technical assistance and Dr. Pipat Chiampiriyakul, Maejo University, Chiang Mai, Thailand for supplying P. infestans DNA. This article reports the results of research only. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U. S. Department of Agriculture.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: Research funding was primarily provided by base funds from the United States Department of Agriculture/Agricultural Research Service. Salary support for Yu Chen was provided by the Department of Plant Pathology at the University of Wisconsin-Madison. Salary support for Julio Berduo and Amilcar Sanchez-Perez was provided by the University of San Carlos. Salary support for Jiraphan Sopee was provided by Kasetsart University. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Birch PRJ, Rehmany AP, Pritchard L, Kamoun S, Beynon JL. Trafficking arms: oomycete effectors enter host plant cells. Trends in Microbiology. 2006;14:8–11. doi: 10.1016/j.tim.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 2.Kamoun S. A catalogue of the effector secretome of plant pathogenic Oomycetes. Annual Review of Phytopathology. 2006;44:41. doi: 10.1146/annurev.phyto.44.070505.143436. [DOI] [PubMed] [Google Scholar]
- 3.O'Connell RJ, Panstruga R. Tête á tête inside a plant cell: establishing compatibility between plants and biotrophic fungi and oomycetes. New Phytologist. 2006;171:699–718. doi: 10.1111/j.1469-8137.2006.01829.x. [DOI] [PubMed] [Google Scholar]
- 4.Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 5.Tyler BM, Tripathy S, Zhang X, Dehal P, Jiang RHY, et al. Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science. 2006;313:1261–1266. doi: 10.1126/science.1128796. [DOI] [PubMed] [Google Scholar]
- 6.Janjusevic R, Abramovitch RB, Martin GB, Stebbins CE. A bacterial inhibitor of host programmed cell death defenses is an E3 ubiquitin ligase. Science. 2006;311:222–226. doi: 10.1126/science.1120131. [DOI] [PubMed] [Google Scholar]
- 7.Ellis JG, Dodds PN, Lawrence GJ. Flax rust resistance gene specificity is based on direct resistance-avirulence protein interactions. Annual Review of Phytopathology. 2007;45:289–306. doi: 10.1146/annurev.phyto.45.062806.094331. [DOI] [PubMed] [Google Scholar]
- 8.Kim H-S, Desveaux D, Singer AU, Patel P, Sondek J, et al. The Pseudomonas syringae effector AvrRpt2 cleaves its C-terminally acylated target, RIN4, from Arabidopsis membranes to block RPM1 activation. Proceedings of the National Academy of Sciences. 2005;102:6496–6501. doi: 10.1073/pnas.0500792102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rehmany AP, Gordon A, Rose LE, Allen RL, Armstrong MR, et al. Differential recognition of highly divergent downy mildew avirulence gene alleles by RPP1 resistance genes from two Arabidopsis lines. Plant Cell. 2005;17:1839–1850. doi: 10.1105/tpc.105.031807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooke DEL, Drenth A, Duncan JM, Wagels G, Brasier CM. A molecular phylogeny of Phytophthora and related Oomycetes. Fungal Genetics and Biology. 2000;30:17–32. doi: 10.1006/fgbi.2000.1202. [DOI] [PubMed] [Google Scholar]
- 11.Jiang RHY, Tripathy S, Govers F, Tyler BM. RXLR effector reservoir in two Phytophthora species is dominated by a single rapidly evolving superfamily with more than 700 members. Proceedings of the National Academy of Sciences. 2008;105:4874–4879. doi: 10.1073/pnas.0709303105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Armstrong MR, Whisson SC, Pritchard L, Bos JIB, Venter E, et al. An ancestral oomycete locus contains late blight avirulence gene Avr3a, encoding a protein that is recognized in the host cytoplasm. Proceedings of the National Academy of Sciences. 2005;102:7766–7771. doi: 10.1073/pnas.0500113102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whisson SC, Boevink PC, Moleleki L, Avrova AO, Morales JG, et al. A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature. 2007;450:115–118. doi: 10.1038/nature06203. [DOI] [PubMed] [Google Scholar]
- 14.Vleeshouwers VG, Rietman H, Krenek P, Champouret N, Young C, et al. Effector genomics accelerates discovery and functional profiling of potato disease resistance and Phytophthora infestans avirulence genes. PLoS ONE. 2008;3:e2875. doi: 10.1371/journal.pone.0002875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Champouret N, Bouwmeester K, Rietman H, van der Lee T, Maliepaard C, et al. Phytophthora infestans isolates lacking class I ipiO variants are virulent on Rpi-blb1 potato. Molecular Plant-Microbe Interactions. 2009;22:1535–1545. doi: 10.1094/MPMI-22-12-1535. [DOI] [PubMed] [Google Scholar]
- 16.van West P, de Jong AJ, Judelson HS, Emons AMC, Govers F. The ipiO gene of Phytophthora infestans is highly expressed in invading hyphae during infection. Fungal Genetics and Biology. 1998;23:126–138. doi: 10.1006/fgbi.1998.1036. [DOI] [PubMed] [Google Scholar]
- 17.Song J, Bradeen JM, Naess SK, Raasch JA, Wielgus SM, et al. Gene RB cloned from Solanum bulbocastanum confers broad spectrum resistance to potato late blight. Proceedings of the National Academy of Sciences. 2003;100:9128–9133. doi: 10.1073/pnas.1533501100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van der Vossen E, Sikkema A, Hekkert BtL, Gros J, Stevens P, et al. An ancient R gene from the wild potato species Solanum bulbocastanum confers broad-spectrum resistance to Phytophthora infestans in cultivated potato and tomato. Plant Journal. 2003;36:867–882. doi: 10.1046/j.1365-313x.2003.01934.x. [DOI] [PubMed] [Google Scholar]
- 19.Halterman D, Kramer LC, Weilgus S, Jiang J. Performance of transgenic potato containing the late blight resistance gene RB. Plant Disease. 2008;92:339–343. doi: 10.1094/PDIS-92-3-0339. [DOI] [PubMed] [Google Scholar]
- 20.Fry W. Phytophthora infestans: the plant (and R gene) destroyer. Molecular Plant Pathology. 2008;9:385–402. doi: 10.1111/j.1364-3703.2007.00465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Z, Halterman D. Identification and characterization of RB orthologous genes from the late blight resistant wild potato species Solanum verrucosum. Physiological and Molecular Plant Pathology. 2006;69:230–239. [Google Scholar]
- 22.Yang Z, Wong WSW, Nielsen R. Bayes empirical bayes inference of amino acid sites under positive selection. Molecular Biology and Evolution. 2005;22:1107–1118. doi: 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]
- 23.Yang Z. PAML 4: Phylogenetic analysis by paximum likelihood. Molecular Biology and Evolution. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 24.Gómez-Alpizar L, Carbone I, Ristaino JB. An Andean origin of Phytophthora infestans inferred from mitochondrial and nuclear gene genealogies. Proceedings of the National Academy of Sciences. 2007;104:3306–3311. doi: 10.1073/pnas.0611479104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kamoun S, van West P, Vleeshouwers V, de Groot KE, Govers F. Resistance of Nicotiana benthamiana to Phytophthora infestans is mediated by the recognition of the elicitor protein INF1. Plant Cell. 1998;10:1413–1425. doi: 10.1105/tpc.10.9.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Y, Halterman D. Phenotypic characterization of potato late blight resistance mediated by the broad-spectrum resistance gene RB. Phytopathology submitted. 2010 doi: 10.1094/PHYTO-04-10-0119. [DOI] [PubMed] [Google Scholar]
- 27.Lawrence GJ, Mayo GME, Shepherd KW. Interactions between genes controlling pathogenicity in the flax rust fungus. Phytopathology. 1981;71:12–19. [Google Scholar]
- 28.Jones DA. Genetic properties of inhibitor genes in flax rust that alter avirulence to virulence on flax. Phytopathology. 1988;78:342–344. [Google Scholar]
- 29.Martin GB, Frary A, Wu T, Brommonschenkel S, Chunwongse J, et al. A member of the tomato Pto gene family confers sensitivity to fenthion resulting in rapid cell death. Plant Cell. 1994;6:1543–1552. doi: 10.1105/tpc.6.11.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pedley KF, Martin GB. Molecular Basis of Pto-Mediated resistance to Bacterial Speck Disease in Tomato. Annual Review of Phytopathology. 2003;41:215–243. doi: 10.1146/annurev.phyto.41.121602.143032. [DOI] [PubMed] [Google Scholar]
- 31.Kim YJ, Lin N-C, Martin GB. Two distinct Pseudomonas effector proteins interact with the Pto kinase and activate plant immunity. Cell. 2002;109:589–598. doi: 10.1016/s0092-8674(02)00743-2. [DOI] [PubMed] [Google Scholar]
- 32.Rosebrock TR, Zeng L, Brady JJ, Abramovitch RB, Xiao F, et al. A bacterial E3 ubiquitin ligase targets a host protein kinase to disrupt plant immunity. Nature. 2007;448:370–374. doi: 10.1038/nature05966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kramer LC, Choudoir MJ, Wielgus SM, Bhaskar PB, Jiang J. Correlation between transcript abundance of the RB gene and the level of the RB-mediated late blight resistance in potato. Molecular Plant-Microbe Interactions. 2009;22:447–455. doi: 10.1094/MPMI-22-4-0447. [DOI] [PubMed] [Google Scholar]
- 34.Bradeen JM, Iorizzo M, Mollov DS, Raasch J, Kramer LC, et al. Higher copy numbers of the potato RB transgene correspond to enhanced transcript and late blight resistance levels. Molecular Plant-Microbe Interactions. 2009;22:437–446. doi: 10.1094/MPMI-22-4-0437. [DOI] [PubMed] [Google Scholar]
- 35.Spooner D, Hoekstra R, Van den Berg R, Martínez V. Solanum sect. petota in Guatemala; Taxonomy and genetic resources. American Journal of Potato Research. 1998;75:3–17. [Google Scholar]
- 36.Flier WG, Grünwald NJ, Kroon LPNM, Van Den Bosch TBM, Garay-Serrano E, et al. Phytophthora ipomoeae sp. nov., a new homothallic species causing leaf blight on Ipomoea longipedunculata in the Toluca Valley of central Mexico. Mycological Research. 2002;106:848–856. [Google Scholar]
- 37.Wang M, Allefs S, van den Berg R, Vleeshouwers V, van der Vossen E, et al. Allele mining in Solanum: conserved homologues of Rpi-blb1 are identified in Solanum stoloniferum. Theoretical and Applied Genetics. 2008;116:933–943. doi: 10.1007/s00122-008-0725-3. [DOI] [PubMed] [Google Scholar]
- 38.Pieterse CM, van West P, Verbakel HM, Brassé PW, van den Berg-Velthuis GC, et al. Structure and genomic organization of the ipiB and ipiO gene clusters of Phytophthora infestans. Gene. 1994;138:67–77. doi: 10.1016/0378-1119(94)90784-6. [DOI] [PubMed] [Google Scholar]
- 39.Judo M, Wedel A, Wilson C. Stimulation and suppression of PCR-mediated recombination. Nucleic Acids Research. 1998;26:1819–1825. doi: 10.1093/nar/26.7.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goodwin SB, Cohen BA, Deahl KL, Fry WE. Migration from Northern Mexico as the probable cause of recent genetic changes in populations of Phytophthora infestans in the United States and Canada. Phytopathology. 1994;84:553–558. doi: 10.1094/PHYTO.1998.88.9.939. [DOI] [PubMed] [Google Scholar]
- 41.Marshall-Farrar KD, McGrath M, James RV, Stevenson WR. Characterization of Phytophthora infestans in Wisconsin from 1993 to 1995. Plant Disease. 1998;82:434–436. doi: 10.1094/PDIS.1998.82.4.434. [DOI] [PubMed] [Google Scholar]
- 42.Goodwin SB, Drenth A, Fry WE. Cloning and genetic analyses of two highly polymorphic, moderately repetitive nuclear DNAs from Phytophthora infestans. Current Genetics. 1992;22:107–115. doi: 10.1007/BF00351469. [DOI] [PubMed] [Google Scholar]
- 43.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 44.Huson D, Richter D, Rausch C, Dezulian T, Franz M, et al. Dendroscope: An interactive viewer for large phylogenetic trees. BMC Bioinformatics. 2007;8:460. doi: 10.1186/1471-2105-8-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 46.Lu R, Martin-Hernandez AM, Peart JR, Malcuit I, Baulcombe DC. Virus-induced gene silencing in plants. Methods. 2003;30:296–303. doi: 10.1016/s1046-2023(03)00037-9. [DOI] [PubMed] [Google Scholar]
- 47.Koncz C, Schell J. The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Molecular and General Genetics. 1986;204:383–396. [Google Scholar]
- 48.Vleeshouwers VGAA, Rietman H, Krenek P, Champouret N, Young C, et al. Effector genomics accelerates discovery and functional profiling of potato disease resistance and Phytophthora Infestans avirulence genes. PLoS ONE. 2006;3:e2875. doi: 10.1371/journal.pone.0002875. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Amino acid alignment of IPI-O sequences. For simplicity, duplicate sequences were removed. IPI-O1 sequence is shown along the top. Identical amino acids were replaced with “.” while polymorphic amino acids are shown. A “*” denotes amino acids determined to be under selection for divergence (see Table 2). Lines above the sequence show the RXLR/RGD, DEER, and predicted W motifs from left to right, respectively. Secondary structure prediction (shown at bottom) was done using the PSIPRED protein structure prediction server (http://www.psipred.net/psiform.html). C = coil, E = strand, H = helix. Confidence values (0 = low, 9 = high) are shown below each structure prediction.
(0.26 MB DOC)
Transgenic N. benthamiana containing the RB gene was infiltrated with A. tumefaciens containing the following constructs: A), C), and F) pGR106-IpiO1; B) and D) pGR106-GFP; E) and G) pGR106-ipiO4. C) and F) were infiltrated three days after the other constructs. The photograph was taken 5 days after agroinfiltration of the final constructs.
(2.29 MB DOC)