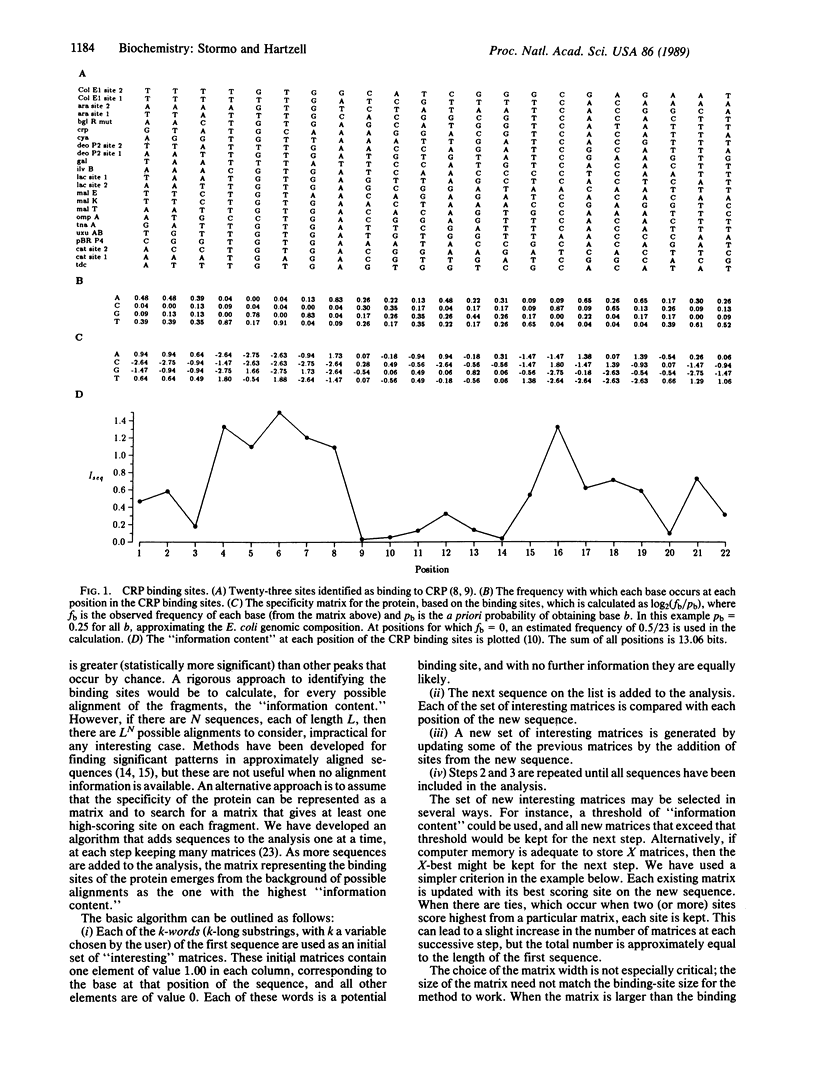
Abstract
The ability to determine important features within DNA sequences from the sequences alone is becoming essential as large-scale sequencing projects are being undertaken. We present a method that can be applied to the problem of identifying the recognition pattern for a DNA-binding protein given only a collection of sequenced DNA fragments, each known to contain somewhere within it a binding site for that protein. Information about the position or orientation of the binding sites within those fragments is not needed. The method compares the "information content" of a large number of possible binding site alignments to arrive at a matrix representation of the binding site pattern. The specificity of the protein is represented as a matrix, rather than a consensus sequence, allowing patterns that are typical of regulatory protein-binding sites to be identified. The reliability of the method improves as the number of sequences increases, but the time required increases only linearly with the number of sequences. An example, using known cAMP receptor protein-binding sites, illustrates the method.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bacon D. J., Anderson W. F. Multiple sequence alignment. J Mol Biol. 1986 Sep 20;191(2):153–161. doi: 10.1016/0022-2836(86)90252-4. [DOI] [PubMed] [Google Scholar]
- Bedouelle H., Hofnung M. A DNA sequence containing the control regions of the malEFG and malK-lamB operons in Escherichia coli K12. Mol Gen Genet. 1982;185(1):82–87. doi: 10.1007/BF00333794. [DOI] [PubMed] [Google Scholar]
- Berg O. G., von Hippel P. H. Selection of DNA binding sites by regulatory proteins. Statistical-mechanical theory and application to operators and promoters. J Mol Biol. 1987 Feb 20;193(4):723–750. doi: 10.1016/0022-2836(87)90354-8. [DOI] [PubMed] [Google Scholar]
- Galas D. J., Eggert M., Waterman M. S. Rigorous pattern-recognition methods for DNA sequences. Analysis of promoter sequences from Escherichia coli. J Mol Biol. 1985 Nov 5;186(1):117–128. doi: 10.1016/0022-2836(85)90262-1. [DOI] [PubMed] [Google Scholar]
- Goss T. J., Datta P. Molecular cloning and expression of the biodegradative threonine dehydratase gene (tdc) of Escherichia coli K12. Mol Gen Genet. 1985;201(2):308–314. doi: 10.1007/BF00425676. [DOI] [PubMed] [Google Scholar]
- Gribskov M., Homyak M., Edenfield J., Eisenberg D. Profile scanning for three-dimensional structural patterns in protein sequences. Comput Appl Biosci. 1988 Mar;4(1):61–66. doi: 10.1093/bioinformatics/4.1.61. [DOI] [PubMed] [Google Scholar]
- Gribskov M., McLachlan A. D., Eisenberg D. Profile analysis: detection of distantly related proteins. Proc Natl Acad Sci U S A. 1987 Jul;84(13):4355–4358. doi: 10.1073/pnas.84.13.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley C. B., Reynolds R. P. Analysis of E. coli promoter sequences. Nucleic Acids Res. 1987 Mar 11;15(5):2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Grice S. F., Matzura H., Marcoli R., Iida S., Bickle T. A. The catabolite-sensitive promoter for the chloramphenicol acetyl transferase gene is preceded by two binding sites for the catabolite gene activator protein. J Bacteriol. 1982 Apr;150(1):312–318. doi: 10.1128/jb.150.1.312-318.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewin R. Genome projects ready to go. Science. 1988 Apr 29;240(4852):602–604. doi: 10.1126/science.3363347. [DOI] [PubMed] [Google Scholar]
- Luo M., Tsao J., Rossmann M. G., Basak S., Compans R. W. Preliminary X-ray crystallographic analysis of canine parvovirus crystals. J Mol Biol. 1988 Mar 5;200(1):209–211. doi: 10.1016/0022-2836(88)90346-4. [DOI] [PubMed] [Google Scholar]
- Mulligan M. E., Hawley D. K., Entriken R., McClure W. R. Escherichia coli promoter sequences predict in vitro RNA polymerase selectivity. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):789–800. doi: 10.1093/nar/12.1part2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler J. R., Sasmor H., Betz J. L. A perfectly symmetric lac operator binds the lac repressor very tightly. Proc Natl Acad Sci U S A. 1983 Nov;80(22):6785–6789. doi: 10.1073/pnas.80.22.6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider T. D., Stormo G. D., Gold L., Ehrenfeucht A. Information content of binding sites on nucleotide sequences. J Mol Biol. 1986 Apr 5;188(3):415–431. doi: 10.1016/0022-2836(86)90165-8. [DOI] [PubMed] [Google Scholar]
- Staden R. Computer methods to locate signals in nucleic acid sequences. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):505–519. doi: 10.1093/nar/12.1part2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. Methods to define and locate patterns of motifs in sequences. Comput Appl Biosci. 1988 Mar;4(1):53–60. doi: 10.1093/bioinformatics/4.1.53. [DOI] [PubMed] [Google Scholar]
- Stormo G. D. Computer methods for analyzing sequence recognition of nucleic acids. Annu Rev Biophys Biophys Chem. 1988;17:241–263. doi: 10.1146/annurev.bb.17.060188.001325. [DOI] [PubMed] [Google Scholar]
- de Crombrugghe B., Busby S., Buc H. Cyclic AMP receptor protein: role in transcription activation. Science. 1984 May 25;224(4651):831–838. doi: 10.1126/science.6372090. [DOI] [PubMed] [Google Scholar]



