Abstract
Diamond Blackfan anemia (DBA) is a severe congenital failure of erythropoiesis. Despite mutations in one of several ribosome protein genes, including RPS19, the cause of the erythroid specificity is still a mystery. We hypothesize that because the chromatin of late erythroid cells becomes condensed and transcriptionally inactive prior to enucleation, the rapidly proliferating immature cells require very high ribosome synthetic rates. We measured RNA biogenesis in primary mouse fetal liver erythroid progenitor cells; during the first 24 hours, cell number increases 3–4 fold while, remarkably, RNA content increases 6-fold, suggesting an accumulation of an excess of ribosomes during early erythropoiesis. Retrovirus infected siRNA RPS19 knockdown cells show reduced proliferation but normal differentiation, and cell cycle analysis shows a G1/S phase delay. p53 protein is increased in the knockdown cells, and the mRNA level for p21, a transcriptional target of p53, is increased. Furthermore, we show that RPS19 knockdown decreases MYB protein, and KIT mRNA is reduced, as is the amount of cell surface KIT protein. Thus, in this shRNA murine model of DBA, RPS19 insufficient erythroid cells may proliferate poorly because of p53 mediated cell cycle arrest, and also because of decreased expression of the key erythroid signaling protein KIT.
Introduction
Diamond–Blackfan anemia (DBA) is a congenital anemia that develops at or soon after birth. The anemia is due to failure of erythropoiesis, characterized by reticulocytopenia, selective erythroid hypoplasia, and often by macrocytosis and high levels of red cell adenosine deaminase activity. DBA is, however, a broad developmental disease, with congenital abnormalities, particularly short stature, as well as facial, thumb and other abnormalities in 35–47% patients (Ball, et al 1996, Ramenghi, et al 1999 Mar, Willig, et al 1999). DBA is inherited in about 40–45% of cases (Orfali, et al 2004), and genetic studies have identified mutations in ribosomal protein (RP) genes; RPS19, in 25% of cases (Draptchinskaia, et al 1999), RPS24 in 2% of cases (Gazda, et al 2006a), and RPS17 in one case (Cmejla, et al 2007); recent data show evidence for involvement of at least three other RP genes, RPL35a, RPL5 and RPL11 (Farrar, et al 2008, Gazda, et al 2008).
Cellular studies in patients have shown a deficiency of erythroid burst and colony-forming units (BFU-Es and CFU-Es) (Freedman, et al 1976, Nathan, et al 1978), apoptosis of CFU-Es after erythropoietin (EPO) deprivation (Perdahl, et al 1994), and defective expansion and differentiation in liquid culture (Ohene-Abuakwa, et al 2005). Importantly, normal erythropoiesis after transplantation demonstrates that the defect is intrinsic to an erythroid precursor. About 2/3 patients respond to treatment with steroids. Interestingly, and pertinent to our studies, we and others have shown that DBA erythroid colony formation can be corrected in many cases by KIT ligand (stem cell factor (SCF)) (Abkowitz, et al 1991, Olivieri, et al 1991, Sieff, et al 1991).
In yeast and in mammalian cells, including patient cells, RPS19 deletion leads to a block in ribosomal RNA biogenesis (Choesmel, et al 2007, Flygare, et al 2007) (Idol, et al 2007). This important result has led to 2 main hypotheses; first, the block in ribosomal biogenesis leads to nucleolar dysfunction and impaired cell division, and second, a reduction in ribosomes decreases translation and leads to impaired protein synthesis.
A critical question is how haploinsufficiency of ribosomal proteins leads to failure of erythropoiesis. Kinetic considerations may explain why erythroid cells are particularly sensitive to ribosome protein deficiency. During fetal/early development rapid expansion of the erythron requires high proliferation rates and high rates of ribosome synthesis. These unique cells undergo chromatin condensation and enucleate, however, and therefore it is reasonable to propose that translation capacity must be generated early to allow the shift to globin synthesis when cells are becoming smaller and less able to make new ribosomes.
We used primary murine fetal liver erythroid cells to test this hypothesis. Our laboratory developed a flow cytometry assay that allows quantitative evaluation of erythroid differentiation in adult and neonatal hematopoietic tissues (Socolovsky, et al 2001). From E12–E16, mouse fetal liver serves as the primary erythropoietic site for the embryo; erythroid lineage cells comprise >90% of total fetal liver cells, and mouse fetal liver provides a great resource to study erythropoiesis in primary cells. Mouse fetal liver cells were used to develop a series of methods to monitor erythroid differentiation step-by-step both in vivo and in vitro, purify large quantities of primary erythroid progenitors with a very high purity, and culture purified erythroid progenitors in vitro to study their normal terminal proliferation and differentiation. We used purified TER119-negative (Ter119−) cells from fetal livers for our studies; TER119 is a glycophorin A associated protein that is expressed on maturing erythroid cells. CFU-E and proerythroblasts comprise ~70–80% of the TER119− population, which contains essentially no differentiated erythroid cells. These purified E13.5 TER119− cells comprise ~3% B220- positive (CD45R) cells, ~1% CD3- positive cells, essentially no Gr-1- positive (Ly-6G and Ly-6C (granulocytes and some monocytes)) cells and ~9% Mac-1- positive (CD11b) cells. Over 90% of the TER119− cells are KIT positive. The purified TER119− cells are cultured in fibronectin-coated plates in medium with serum and EPO, which is removed from the medium after one day. After one day in culture, early erythroblasts up-regulate the transferrin receptor (CD71) and some differentiate into Ter119+ cells, but most are negative for benzidine (hemoglobin) staining. At the end of two days, these cells further differentiate into benzidine-positive erythroblasts; many of these cells lose their nuclei and form reticulocytes. During this two-day period the number of erythroblasts increases 15–20 fold, corresponding to 4–5 cell divisions and correlating well with the number of terminal cell divisions that a CFU-E goes through to generate terminally differentiated erythrocytes (Gregory, et al 1974, Stephenson, et al 1971). Thus, this culture condition supports both proper terminal proliferation and differentiation of CFU-E progenitors.
We show that RNA synthesis of normal fetal liver progenitors is very rapid during the first 24 hours of culture, and exceeds the cell proliferation rate. Although it was shown many years ago that the rate of RNA synthesis is related to cell proliferation rate, our observations are novel in that RNA synthesis actually exceeds cell proliferation rate.
To address the mechanism of erythroid failure in DBA we used small hairpin RNAs (shRNAs) to knockdown Rps19 expression. We show that there is an early defect in cell proliferation, but that differentiation of the residual cells is unimpaired, an observation consistent with the normal differentiation of the surviving erythroid precursors in DBA.
To investigate the molecular mechanism of the erythroid failure we used a modified culture system developed by Mullner, Beug and colleagues (Dolznig, et al 2005). Importantly, this method of serum-free liquid culture in dexamethasone, insulin-like growth factor 1 (IGF-1) and EPO allowed us to expand early erythroid cells for 5–6 days without terminal differentiation. Using this system we show that cells deficient in RPS19 show stabilization of p53 with an increase in p21 and G1 cell cycle delay, and also that MYB, a transcription factor critically important for erythropoiesis, is reduced, leading to reduced expression of one of its critical downstream erythroid transcriptional targets, KIT.
Thus we propose that DBA cells may be unable to produce sufficient RNA during early stages of erythropoiesis, and that the block in ribosome biogenesis leads to nucleolar stress and p53 mediated arrest or apoptosis, as well as a deficiency in KIT and impaired SCF responsiveness.
Methods
Cells
The retrovirus-packaging cell line 293T was maintained in Dulbecco modified Eagle medium (DMEM) containing 10% fetal bovine serum (FBS) (Invitrogen, Carlsbad, CA). Fetal liver cells were isolated from E14.5 Balb/c (Jackson Laboratory, Bar Harbor, ME) or C57BL embryos and mechanically dissociated by pipetting in erythroid-differentiation medium (EDM) (Iscove modified Dulbecco medium [IMDM] containing 20% FBS, 2 mM L-glutamine, and 10−4M β-mercaptoethanol). Single-cell suspensions were prepared by passing the dissociated cells through 70 μm and 25 μm cell strainers. Nucleated cells were counted.
Erythroid culture
When mouse fetal liver cells are double-labeled for erythroid-specific TER119 (Ikuta, et al 1990, Kina, et al 2000), a protein associated with glycophorin A, and non erythroid-specific transferrin receptor (CD71) and analyzed by flow-cytometry, E14.5 fetal livers contain at least five distinct populations of cells, defined by their characteristic staining patterns: CD71medTER119low (R1), CD71highTER119low (R2), CD71highTER119high (R3), CD71medTER119high (R4), and CD71lowTER119high (R5). The R1–R5 cells can be sorted by FACS and further characterized by CFU-E colony assay, surface expression levels of EPOR, and histological staining. R1 cells comprise ≥ 40% CFU-Es. (Zhang, et al 2003a), R2 contain only a few CFU-Es and R3–R5 contain no erythroid progenitors. On average, R1 and R2 cells have ~1,900 cell surface EPOR/cell; later stages have many fewer receptors (Zhang, et al 2003a). When R1–R5 cells are processed by May-Grunwald Giemsa stains, their morphology resembles erythroblasts at different developmental stages (Fawcett 1997); R5 cells have less than 10% the volume of R1 cells, consistent with 4 to 5 cell divisions occurring during erythroid differentiation. The morphologic characteristics generally corresponded to primitive progenitor cells and proerythroblasts in the R1 population, proerythroblasts and early basophilic erythroblasts in R2, early and late basophilic erythroblasts in R3, polychromatophilic and orthochromatophilic erythroblasts in R4, and late orthochromatophilic erythroblasts and reticulocytes in the R5 population.
Purified TER119− cells fall mostly into the R1 and R2 windows, and when cultured in serum containing medium with EPO proliferate and differentiate into R3, R4 and some R5 cell compartments over a 48 hour culture period. Total fetal liver cells were labeled with biotin-conjugated anti-TER119 antibody (1:100) (BD Pharmingen, San Diego, CA), and TER119 negative (TER119−) cells were purified through a StemSep column as per the manufacturer's instructions (StemCell Technologies, Vancouver, BC, Canada). Purified cells were seeded in fibronectin-coated wells (BD Discovery Labware, Bedford, MA) at a cell density of 1 × 105/mL. On the first day, the purified cells were cultured in Day 1 medium – Iscove modified Dulbecco medium (IMDM) containing 15% fetal bovine serum (FBS), 1% detoxified bovine serum albumin (BSA), 200 μg/mL holo-transferrin (Sigma, St Louis, MO), 10 μg/mL recombinant human insulin (Sigma), 2 mM L-glutamine, 10−4 M β-mercaptoethanol, and 2 U/mL EPO (Amgen, Thousand Oaks, CA). On the second day, this medium was replaced with erythrocyte differentiation medium (EDM) – IMDM containing 20% FBS, 2 mM L-glutamine 10−4 M β-mercaptoethanol. Cells were counted at 24 and 48 hours and aliquots harvested for RNA and protein extraction. Total RNA was measured by absorbance at 260 nm using a Nanodrop device.
A second culture system was introduced to allow amplification of R2 and R3 cells and to measure the longer term effects of RPS19 knockdown. TER119− cells were cultured in 10−6M dexamethasone, 100 ng/mL SCF, 40 ng/mL IGF-1 and 2U/mL EPO in serum free StemPro 34 medium (Dolznig, et al 2005). This culture system allows the amplification of R2 cells with little differentiation into R3 Ter119+ cells over the 5–6 days required for these experiments.
Retroviral constructs
MSCV-pgkGFP-U3-U6P-Bbs vector (murine stem cell retroviral vector-pgk promoter-GFP-U6 promoter shRNA) was a gift from Dr. Biao Luo (Broad Institute, Cambridge MA). The human U6 promoter was inserted at an NheI site in the U3 of the 3' LTR. The U6 promoter is in the opposite orientation to the retroviral LTR. Two back-to-back BbsI sites were used as cloning sites for 4 shRNAs selected to target Rps19.
Generation of retroviral supernatants and infection of primary cells
293T cells were seeded on 100-mm dishes 1 day before transfection. Then, 9 μg retroviral plasmids together with 3μg pCL-Eco vector was used to cotransfect 293T cells with the Lipofectamine Plus reagent (Invitrogen) according to the manufacturer's protocol. The retroviral supernatants were collected 48 hours after transfection and stored in aliquots at −80°C. To titer the packaged viruses, a series of dilutions of the supernatants were used to infect NIH 3T3 cells; titers of 107 to 108 infectious units per milliliter were routinely obtained. For infection of purified TER119− fetal liver cells, 1.2 × 104 to 5 × 105 cells were resuspended in 1 mL thawed viral supernatant containing 10 μg/mL polybrene (Sigma) and centrifuged at 2000 rpm for 1 hour at 37°C.
Immunostaining and flow cytometry analysis of erythroid differentiation
Freshly isolated fetal liver cells were immunostained with phycoerythrin (PE)–conjugated anti-CD71 (1:200) (BD Pharmingen) and allophycocyanin (APC)–conjugated anti-Ter119 (1:200) (BD Pharmingen) antibodies, as previously described (Socolovsky, et al 2001). Propidium iodide was added to exclude dead cells from analysis. GFP+ cells were sorted by a FACS Moflo machine (Cytomation, Fort Collins, CO) with reduced pressure. Flow cytometry was carried out on a FACS LSRII (BD Biosciences, Franklin Lakes, NJ).
Retrovirally transduced TER119− fetal liver cells were cultured in vitro for 6 days in serum-free medium and then incubated at 37°C for 1 hour in the presence of 10 μg/mL Hoechst dye 33342 (Sigma). The cells were placed on ice and labeled with PE-anti-CD71 and APC-anti-Ter119; all incubation and washing steps were carried out carefully at 4°C. Propidium iodide was added to exclude dead cells. Flow cytometry was carried out on an LSR II flow cytometer (BD Biosciences). Collected data were analyzed by ModFit (Verity Software House, Topsham, ME) or FloJo software (Tree Star Inc., Ashland, OR).
Quantitative Reverse Transcription Real Time-Polymerase Chain reaction (qRT-PCR)
For qRT-PCR an 18S rRNA fragment was cloned into the pCR2.1 TOPO vector and used in a dose range 10−1–10−7 ng/well in triplicate with 18S primers and the SYBR green PCR master mix (ABI). The slope and intercept of the standard curve was used to calculate the RNA content of cDNA made from the unknown samples, which were assayed in triplicate or quadruplicate.
Immunoblotting
For most experiments 1×106 cells were used to prepare protein extracts using RIPA buffer supplemented with protease inhibitors. The boiled proteins were electrophoresed on 4–12% Bis Tris NuPAGE precast gels (Invitrogen). The proteins were transferred to nitrocellulose membrane and stained with Ponceau red to monitor transfer and loading, and then blocked in 5% non-fat milk (NFM)/TBS-T. The membranes were incubated overnight in TBS-T with or without 5% NFM in antibodies to MYB (Santa Cruz Biotechnology, Santa Cruz CA or Upstate, Temecula, CA), p53 (Santa Cruz or Novocastra, Newcastle-upon-Tyne NCL-p53–505), GAPDH (Santa Cruz), or RPS19 (Gazda, et al 2004), washed 3 × in TBS-T and incubated for 1 hour at room temperature in the second detection antibody (anti-rabbit, anti-mouse or anti-goat LI-COR fluorescent antibodies. The membrane was scanned on an Odyssey Infrared Imaging System and the density of the bands determined.
Results
Early stages of fetal liver erythropoiesis are characterized by very rapid RNA synthesis
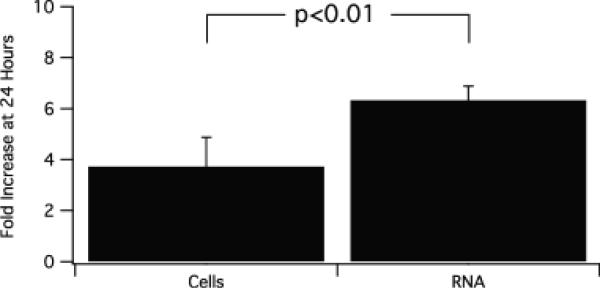
To explore the rate of RNA synthesis during erythropoiesis we enriched E14.5 fetal liver cells for Ter119 negative (Ter119−) cells using biotinylated Ter119 and an immunomagnetic column (StemSep). The cells were cultured on fibronectin coated plates in EPO and serum-containing Day 1 medium supplemented with transferrin and insulin. After 24 hours the cells were centrifuged and resuspended in EDM (IMDM/20% FCS). Cells were counted and total RNA extracted from an aliquot at 24 and 48 hours. Table 1 is an example of an experiment in which the cell and RNA increase in the cultures is monitored. Cell number increases very rapidly, and after 1 and 2 days there is a 4 and 28 fold increase in cell number, representing 4–5 cell divisions. RNA extracted from these cells shows that the greatest fold increase in RNA occurs during the first 24 hours of culture, and exceeds the fold increase in cells. During the later stages of differentiation cell size decreases, RNA yield per cell drops and therefore RNA level plateaus or even decreases slightly. Figure 1 is a summary of 3 experiments showing the marked increase in total RNA (80% of which is ribosomal) during the first 24 hours of culture, significantly greater than the fold increase in cells.
Table 1.
The TER119− Day 0 cells fall into the R1/R2 window (see methods)
| # | Parameter | Day 0 | Day 1 | Day 2 |
|---|---|---|---|---|
| 1 | Cell # (×106) | 2 | 8.3 | 57.6 |
| 2 | Fold Increase in Cells | 1 | 4.15 | 28.8 |
| 3 | RNA/Cell (pg) | 2.59 | 3.84 | 0.44 |
| 4 | Total RNA in Culture (μg) | 5.2 | 31.9 | 25.3 |
| 5 | Fold Increase in RNA | 1 | 6.13 | 4.87 |
Figure 1.
Ter119− fetal liver cells cultured in serum with EPO and transferrin. At 24 hours cell number and total RNA content were measured. N=3, mean +/− 2SD.
Phenotypic consequences of Rps19 depletion in erythroid cells shRNA mediated Rps19 knockdown shows rapid Rps19 mRNA depletion
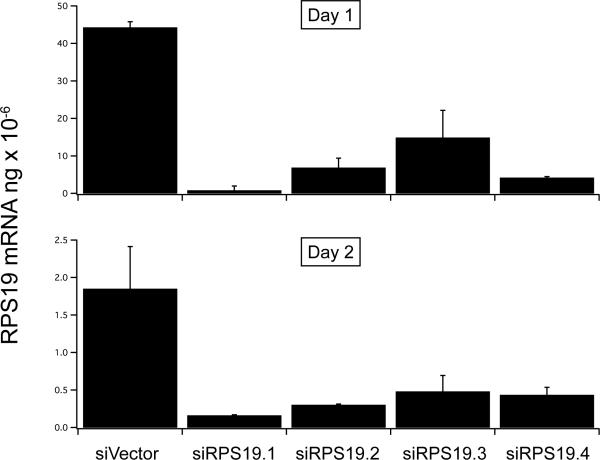
Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) shows that after retrovirus infection of TER119− cells with 4 different shRNAs to Rps19 there is rapid 70–90% depletion of Rps19 mRNA by day 1, with further depletion by day 2 (Figure 2A).
Figure 2.
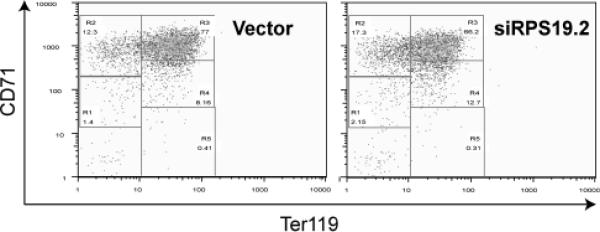
A, purified Ter119 negative fetal liver cells were infected with empty vector or one of 4 shRNA retrovirus constructs designed to knockdown Rps19. At 24 and 48 hours after infection the RNA was extracted and Rps19 mRNA measured by quantitatitive RT-PCR, B, counted, and C, incubated at 48 hours with PE- CD71 and APC-Ter119.
shRNA knockdown of RPS19 impairs proliferation of erythroid cells
To test the effect of shRNA mediated knockdown of RPS19 on cell proliferation of these erythroid progenitors. cell counts were monitored at 24 and 48 hours (Figure 2B). Although differences in cell number are not apparent on day 1, by day 2 there is a reduction in proliferation of the knockdown cells. To determine the effect on of the knockdown on RPS19 protein levels, we performed Western analysis with cells obtained on day 2; there is a significant reduction in protein with the RPS19.1 knockdown construct that showed the most pronounced reduction in mRNA, but the other constructs show only a small and variable decrease in RPS19 protein by day 2 of culture (not shown).
shRNA knockdown of RPS19 does not affect erythroid differentiation
After purification the Ter119− fetal liver cells do not express Ter119 and are in the R1 and R2 windows (not shown). After 2 days in culture the cells first express more CD71 and then express Ter119 as they move into the R3 window. With further differentiation CD71 is lost and cells move into the R4 and R5 regions (Zhang, et al 2003a). We next determined whether knockdown of RPS19 affected differentiation. Although there are small differences in the number of cells falling into the R2–R5 gates, there was no marked difference in the differentiation capacity of knockdown compared with normal cells (Figure 2C). Thus, even though the number of cells was reduced by RPS19 knockdown, the surviving cells could differentiate normally.
G1 arrest in erythroblasts after siRNA knockdown of Rps19
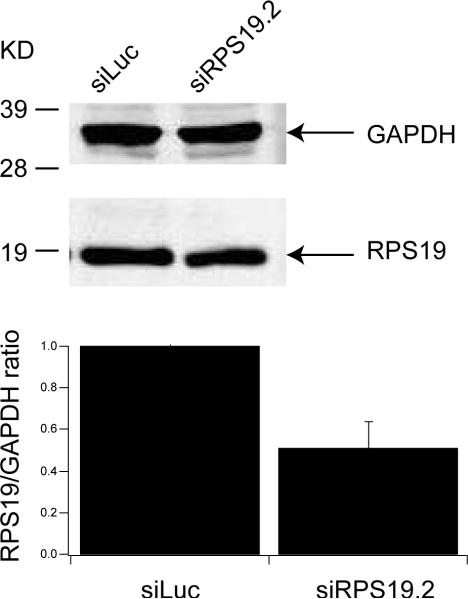
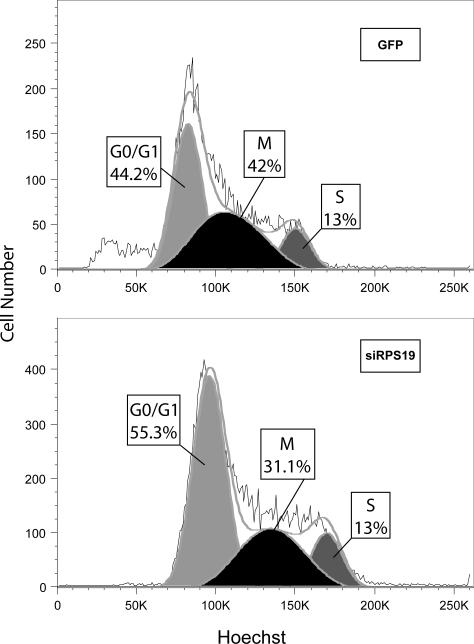
Because the cells differentiate very quickly, by day 2, when the shRNAs are fully expressed, the erythroblasts have already matured and are beginning to exit the cell cycle; this made it difficult to monitor cell cycle status. Furthermore, reduction in RPS19 protein levels in the rapidly proliferating serum cultures were not reproducible from experiment to experiment. To obviate this problem we cultured Ter119− cells in serum-free conditions (StemPro34 medium with nutrients) supplemented with SCF, dexamethasone, IGF-1 and EPO. Under these conditions CD71+, Ter119− (R2) cells can proliferate with little differentiation for up to 14 days (Dolznig, et al 2005). In contrast to the work by Dolznig, Muellner and colleagues we did not remove TER119+ cells at intervals during the culture, and therefore a gradual increase in TER119+ cells was observed. To confirm that these cells are affected by the shRNA knockdown of RPS19, Figure 3A shows growth curves indicating that the RPS19 knockdown cells, similar to the serum containing cultures (Figure 2B, upper panel), proliferate poorly in serum-free conditions; the lower panel confirms that the knockdown cells proliferate more slowly compared to the few residual negative cells left in the culture after FACS sorting on day 2, leading to a more rapid decline of GFP+ knockdown cells compared with the non infected internal control cells or the GFP siLuciferase control. To ensure that this culture system does model DBA, Figure 3B shows that the cellular level of RPS19 protein is decreased to approximately 50% on day 6 of culture, normalized to the control glycolytic enzyme GAPDH. To examine the cell cycle, day 6 erythroblasts were incubated in Hoechst 33342 and cell cycle analysis performed on GFP, CD71, Ter119 positive cells that express either the control siLuciferase or the siRPS19.2 construct. Figure 3C shows that the RPS19 knockdown cells accumulate in G1 to a greater extent than the GFP control cells. These data suggest that RPS19 knockdown is characterized by an increase in G1 cells, consistent with the induction of a p53 response.
Figure 3.
A, serum free cultures of Ter119− fetal liver cells were monitored daily for cell number (upper panel) and the proportion of GFP+ cells (lower panel) after infection with vector or Rps19 shRNA contructs on day 0 and sorting of positive cells on day 2. B, Western blot of control (siLuciferase) and siRps19 infected cells cultured for 6 days in serum-free conditions and sorted for GFP immediately before lysis. The lower histogram shows the normalized RPS19:GAPDH ratio with standard deviation (n=2) C. Cell cycle analysis of day 6 serum-free cultured erythroblasts after staining with Hoechst 33142 for 1 hour and labeled with CD71 and Ter119. Representative example of 2 experiments.
p53 is increased after siRPS19 knockdown
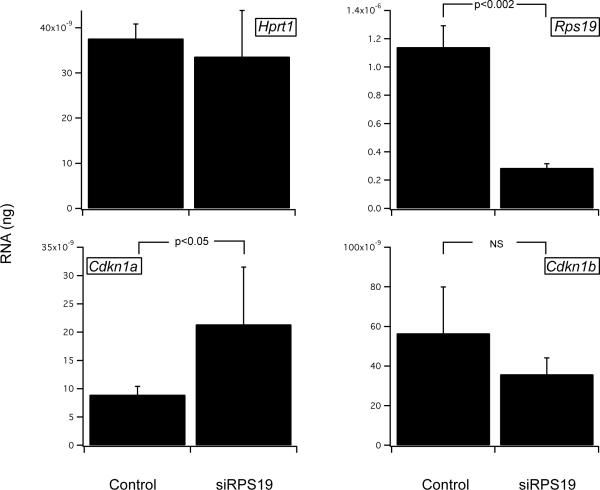
We next compared the levels of p53 in uninfected, GFP-luc and GFP-siRPS19 cells. Figure 4A shows that p53 protein levels are increased in the knockdown cells, and this is confirmed by determining the p53/GAPDH ratio, normalized to the siLuciferase vector control in 3 independent experiments (Figure 4B). Stabilization of p53 can lead to apoptosis or cell cycle arrest. Figure 5A shows that the mRNA level of p21, a cyclin/cyclin dependent kinase (CDK) inhibitor that is a known target of p53, is increased. In contrast, there is little change in the level of p27, another cell cycle inhibitor that is not induced by p53. We did not find evidence of apoptosis by annexin V staining in these cells (data not shown).
Figure 4.
A, Ter119− fetal liver cells cultured without (control) or following infection with siLuciferase (GFP) or siRps19 for 6 days; Western analysis of protein expression using antibodies to p53, c-Myb or GAPDH. B, normalized ratio of p53:GAPDH, showing a modest 20% increase in p53 in siRPS19.2 compared with the siLuciferase empty vector control (n=3). C, normalized ratio of MYB:GAPDH, showing a marked decrease in MYB in siRPS19.2 compared with the siLuciferase control (n=3).
Figure 5.
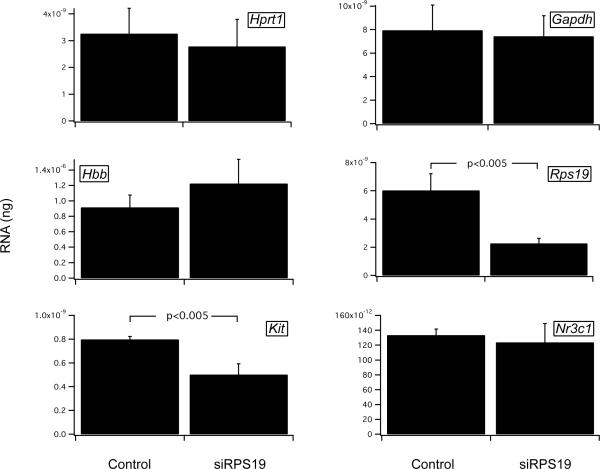
Control or siRNA Rps19 infected Ter119− fetal liver cells cultured for 6 days in serum free conditions and FACS sorted before analysis of mRNA levels by quantitative RT-PCR. A, HPRT (control) RPS19, p21 and p27. B, HPRT and GAPDH (controls), globin (Hbb), c-Kit and glucocorticoid receptor (GR).
MYB is depleted after siRps19 knockdown
We have shown previously in human gene expression microarray studies that MYB is one of the most highly under-expressed genes in DBA RPS19 mutant compared with normal CD34+ cells (Gazda, et al 2006b). To determine whether the same is true of the RPS19 knockdown cells, we compared MYB protein by Western blot and found that MYB levels are decreased in the knockdown cells (Figure 4A). Figure 4C shows the results of 3 independent experiments normalized to the siLuciferase vector control.
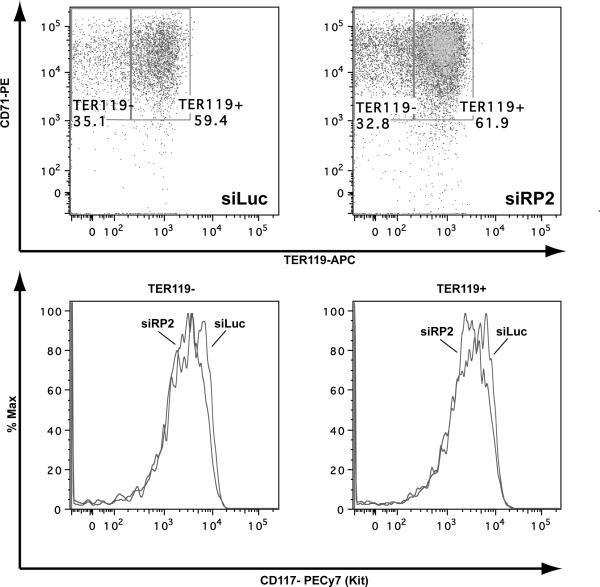
Two transcriptional targets of MYB are KIT and the glucocorticoid receptor. We found that in comparison with GAPDH and HRPT housekeeping messages, which are unchanged in the RPS19 knockdown cells, KIT mRNA levels are significantly reduced. In contrast, there is no significant difference in glucocorticoid receptor mRNA levels (Figure 5B). We next measured cell surface KIT (CD117) protein level by labeling cells with fluorescent antibodies to CD117. Figure 6 shows that there is an siRPS19 induced reduction in cell surface expression of KIT on day 6 of serum-free culture (note that this is a log scale) for both the less differentiated TER119− and the more differentiated TER119+ cells.
Figure 6.
Purified Ter119− fetal liver cells were infected with siLuciferase conrol (GFP) or an shRNA retrovirus construct designed to knockdown Rps19 (RP2). On day 6 the cells were labeled with antibodies to CD71, Ter119 and CD117 (c-Kit) and the GFP+ cells analyzed for differentiation stage (upper panels) and c-Kit expression (lower panel). The results show decreased KIT expression in siRPS19.2 compared with siLuciferase control for both the less differentiated TER119− gate (mean fluorescence values 2394 and 3521, respectively) and the more differentiated TER119+ gate (mean fluorescence values 3893 and 4144, respectively). Representative example of 2 experiments.
Discussion
In this paper we ask why the erythroid lineage is selectively affected by RPS19 deficiency in DBA. Our data raise two interesting possibilities that are not mutually exclusive. First, during the first 24 hours of normal erythropoiesis, the rate of RNA production exceeds the cell doubling rate. This indicates that these early erythroid precursors require very high rates of RNA synthesis and may therefore be particularly sensitive to the block in ribosomal biogenesis that is a feature of DBA cells. And second, we provide novel evidence in a murine shRNA model of DBA that KIT, a critical regulator of erythropoiesis, is not optimally expressed in cells that contain reduced amounts of RPS19.
To test RNA kinetics we measured RNA production in normal primary fetal liver erythroid cells. In this system erythroid differentiation starts with the early committed erythroid progenitor (the CFU-E), which expresses the highest level of EPO receptors and is critically dependent on EPO, particularly for the early differentiation steps. This cell rapidly goes through about 5 divisions in 48 hours as it matures. During the first 24 hours RNA production exceeds the cell doubling rate. In contrast, during the next 24 hours, despite an equally rapid cell proliferation rate, RNA production rate plateaus or declines (Table 1). It was shown many years ago that RNA synthesis correlates with cell growth rate (Levine, et al 1965, Todaro, et al 1965, Ward and Plagemann 1969). This regulation affects rRNA synthesis, as cells placed in media with low or high concentrations of amino acids show low or high incorporation of labeled precursors into RNA, respectively, which is blocked by antibiotics that prevent rRNA synthesis (Franze-Fernandez and Pogo 1971). In contrast, our data show that there is a discordance in RNA production during erythropoiesis; despite a continuous rapid proliferation rate over 48 hours, the major phase of RNA production, which contains ~80% ribosomal RNA, is at the earliest stage of erythropoiesis. This suggests that these cells could be particularly sensitive to a block in ribosome biogenesis, and is consistent with clinical laboratory descriptions in DBA patients of a block or absence at the proerythroblast stage of differentiation.
Our data are consistent with RNA and protein expression studies in murine erythroblasts that show the highest levels of RPS19 in the most primitive cells (Da Costa, et al 2003). Human microarray data confirm these findings at the RNA level and show that expression of RPS19 mRNA declines in freshly isolated human CD34 positive cells induced to differentiate over 2 weeks by stem cell factor and erythropoietin (Ebert, et al 2005). Furthermore, reduced protein level in DBA patients with nonsense RPS19 mutations compared with normal control cells was only detectable in CD34 positive bone marrow cells, but the reduction was not demonstrable in DBA blood mononuclear cells (Gazda, et al 2004). An explanation for this discrepancy might be that the differences are easier to detect in highly proliferative early erythroid cells (contained in the CD34 fraction) compared with slowly proliferating longer lived monocytes and lymphocytes in the mononuclear fraction that express much less RPS19. Thus, we suggest that haploinsufficiency of RPS19 might be particularly limiting in cell populations with high RPS19 expression, such as erythroid progenitors, which have an enormous requirement for RNA and protein synthesis.
Our data show that the cell cycle is delayed but that the surviving cells differentiated normally. We did not observe an increase in the number of apoptotic cells in the knockdown cells (not shown). The data showing normal differentiation are consistent with observations in patients, where surviving erythroid cells synthesize normal amounts of hemoglobin and are not hypochromic. An increase in apoptotic cells has been described in DBA patient cells and in cell lines by some (Farrar, et al 2008, Miyake, et al 2008, Perdahl, et al 1994) but not by others (Kuramitsu, et al 2008), and an increase in apoptotic cells has been observed in the bone marrow cells of a dark skinned mouse with a mutation in Rps19 and a very mild erythroid phenotype (McGowan, et al 2008). It is possible that apoptosis or cell cycle arrest depend on the degree of ribosomal stress and the effect on p53, and our studies in primary knockdown cells, which have a limited time course for experimentation, may not allow sufficient time for apoptosis to become evident.
To explore the molecular mechanism by which lack of ribosome proteins could trigger cell cycle arrest and apoptosis, we used a serum-free culture system that allows expansion of primitive erythroid progenitors in dexamethasone, SCF, erythropoietin and IGF-1 (Dolznig, et al 2005). This modification of the culture system allows one to evaluate the effect of RPS19 knockdown on the cell cycle as well as gene and protein expression associated with such a knockdown. Cell cycle analysis shows a G1/S phase block, suggesting a delay in the cell cycle. The oncoprotein MDM2 is a crucial regulator of the tumor suppressor p53, targeting it for ubiquitin-mediated destruction (Haupt, et al 1997, Honda, et al 1997, Oliner, et al 1993). Nucleolar or ribosomal stress-associated pathways can be activated to sequester MDM2 and thereby stabilize p53 (Pestov, et al 2001, Rubbi and Milner 2003), and the free ribosomal proteins L5, L11 and L23 can mediate this process by binding to MDM2 (Dai and Lu 2004, Jin, et al 2004, Lohrum, et al 2003, Marechal, et al 1994, Zhang, et al 2003b). We found that p53 is increased in RPS19 knockdown cells and that a transcriptional target of p53 that inhibits cyclin/CDK and the cell cycle, p21, is increased. Although p53 levels are increased the increase is modest
We showed previously in gene expression microarray studies that in comparison with normal CD34+ cells, MYB message levels are much lower in DBA RPS19 mutant cells (Gazda, et al 2006b). Embryonic erythropoiesis in Myb knockout mice is normal, but there is complete failure of definitive erythropoiesis in fetal liver.(Mucenski, et al 1991) Progenitors of other lineages, but not megakaryocytes, were also decreased, indicating that MYB is required for early definitive cellular expansion. Furthermore, a knockdown allele of Myb shows that suboptimal levels of MYB favor macrophage and megakaryocyte differentiation while higher levels are particularly important for erythropoiesis and lymphopoiesis (Emambokus, et al 2003). Moreover, in primary murine fetal liver erythroid cells, study of Myb hypomorphs showed that MYB is a critical regulator of KIT expression (Vegiopoulos, et al 2006). There is evidence in chicken and mouse cells that Myb transcription is regulated by glucocorticoids (Ganguli, et al 2002, Wessely, et al 1997), and this may contribute to the potent role of glucocorticoids in the dexamethasone mediated proliferation and blocked differentiation of immature erythroid progenitors.(Dolznig, et al 2001) Interestingly, MYB interacts physically with nucleolin, a nucleolar protein important in rRNA biogenesis; it is translocated from the nucleolus after nucleolar stress induced by heat shock or ionizing radiation, and nucleolin binds to MYB and inhibits its transcriptional activity (Daniely, et al 2002, Ying, et al 2000).
Here we show that, like DBA patient cells, MYB protein is decreased after shRNA mediated knockdown of RPS19. Furthermore, KIT, a transcriptional target of MYB, is reduced at the mRNA and protein levels. Support for a role of KIT comes from studies of the dark skin mice and parallel studies done in Rps6 conditional loxP mice (McGowan, et al 2008, Volarevic, et al 2000). The dark skin in the Rps19 mutant mice appears to result from an indirect SCF-mediated effect on melanocytes in the epidermis, and this phenotype is recapitulated by hemizygosity of Rps6 in keratinocytes (McGowan, et al 2008). However, both the dark skin Dsk3 (Rps19) or Dsk4 (Rps20) mutant mice and deletion of Rps6 in melanocytes show a reduction in the number and/or proliferative capacity of melanocytes; this results in lighter skin and impaired migration of melanocytes, with a white belly spot in 10% of the Dsk3 animals that is reminiscent of the white spot in mice with Kit mutations; indeed, the size of the white spot is enhanced when placed on a KitWv genetic background (McGowan, et al 2008).
In conclusion, we propose that the erythroid defect in DBA results from an inability of early erythroid progenitors to make sufficient ribosomal RNA due to a block in ribosome biogenesis; these early progenitors are particularly susceptible to deficiency of ribosomal proteins as their rate of ribosomal biogenesis is very rapid. This is accompanied by nucleolar stress with p53 stabilization and depletion of a critical erythroid transcription factor MYB, which results in impaired expression of KIT.
Acknowledgements
We are grateful for financial support from the March of Dimes, the Daniella Maria Arturi Foundation and the Diamond Blackfan Anemia Foundation. This work was also supported by NIH grant PO1 HL 32262 and a grant from Amgen Corporation to HFL.
Footnotes
Conflict of interest statement: Colin A. Sieff, Jing Yang, Lilia B. Merida-Long, Harvey Lodish.
The authors have no competing financial interests
References
- Abkowitz JL, Sabo KM, Nakamoto B, Blau CA, Martin FH, Zsebo KM, Papayannopoulou T. Diamond-Blackfan anemia: in vitro response of erythroid progenitors to the ligand for c-kit. Blood. 1991;78:2198–2202. [PubMed] [Google Scholar]
- Ball SE, McGuckin CP, Jenkins G, Gordon-Smith EC. Diamond-Blackfan anaemia in the U.K.: analysis of 80 cases from a 20-year birth cohort. Br.J.Haematol. 1996;94:645–653. doi: 10.1046/j.1365-2141.1996.d01-1839.x. [DOI] [PubMed] [Google Scholar]
- Choesmel V, Bacqueville D, Rouquette J, Noaillac-Depeyre J, Fribourg S, Cretien A, Leblanc T, Tchernia G, Dacosta L, Gleizes PE. Impaired ribosome biogenesis in Diamond-Blackfan anemia. Blood. 2007;109:1275–1283. doi: 10.1182/blood-2006-07-038372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cmejla R, Cmejlova J, Handrkova H, Petrak J, Pospisilova D. Ribosomal protein S17 gene (RPS17) is mutated in Diamond-Blackfan anemia. Hum Mutat. 2007;28:1178–1182. doi: 10.1002/humu.20608. [DOI] [PubMed] [Google Scholar]
- Da Costa L, Narla G, Willig TN, Peters LL, Parra M, Fixler J, Tchernia G, Mohandas N. Ribosomal protein S19 expression during erythroid differentiation. Blood. 2003;101:318–324. doi: 10.1182/blood-2002-04-1131. [DOI] [PubMed] [Google Scholar]
- Dai MS, Lu H. Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. J Biol Chem. 2004;279:44475–44482. doi: 10.1074/jbc.M403722200. [DOI] [PubMed] [Google Scholar]
- Daniely Y, Dimitrova DD, Borowiec JA. Stress-dependent nucleolin mobilization mediated by p53-nucleolin complex formation. Mol Cell Biol. 2002;22:6014–6022. doi: 10.1128/MCB.22.16.6014-6022.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolznig H, Boulme F, Stangl K, Deiner EM, Mikulits W, Beug H, Mullner EW. Establishment of normal, terminally differentiating mouse erythroid progenitors: molecular characterization by cDNA arrays. Faseb J. 2001;15:1442–1444. doi: 10.1096/fj.00-0705fje. [DOI] [PubMed] [Google Scholar]
- Dolznig H, Kolbus A, Leberbauer C, Schmidt U, Deiner EM, Mullner EW, Beug H. Expansion and differentiation of immature mouse and human hematopoietic progenitors. Methods Mol Med. 2005;105:323–344. doi: 10.1385/1-59259-826-9:323. [DOI] [PubMed] [Google Scholar]
- Draptchinskaia N, Gustavsson P, Andersson B, Pettersson M, Willig TN, Dianzani I, Ball S, Tchernia G, Klar J, Matsson H, Tentler D, Mohandas N, Carlsson B, Dahl N. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat Genet. 1999;21:169–175. doi: 10.1038/5951. [DOI] [PubMed] [Google Scholar]
- Ebert BL, Lee MM, Pretz JL, Subramanian A, Mak R, Golub TR, Sieff CA. An RNA interference model of RPS19 deficiency in Diamond-Blackfan anemia recapitulates defective hematopoiesis and rescue by dexamethasone: identification of dexamethasone-responsive genes by microarray. Blood. 2005;105:4620–4626. doi: 10.1182/blood-2004-08-3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emambokus N, Vegiopoulos A, Harman B, Jenkinson E, Anderson G, Frampton J. Progression through key stages of haemopoiesis is dependent on distinct threshold levels of c-Myb. Embo J. 2003;22:4478–4488. doi: 10.1093/emboj/cdg434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrar JE, Nater M, Caywood E, McDevitt MA, Kowalski J, Takemoto CM, Talbot CC, Jr., Meltzer P, Esposito D, Beggs AH, Schneider HE, Grabowska A, Ball SE, Niewiadomska E, Sieff CA, Vlachos A, Atsidaftos E, Ellis SR, Lipton JM, Gazda HT, Arceci RJ. Abnormalities of the large ribosomal subunit protein, Rpl35a, in Diamond-Blackfan anemia. Blood. 2008;112:1582–1592. doi: 10.1182/blood-2008-02-140012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett DW. Hematopoiesis. In: Fawcett DW, editor. Bloom & Fawcett: Concise Histology. Hodder Arnold; 1997. [Google Scholar]
- Flygare J, Aspesi A, Bailey JC, Miyake K, Caffrey JM, Karlsson S, Ellis S. Human RPS19, the gene mutated in Diamond Blackfan anemia, encodes a ribosomal protein required for the maturation of 40S ribosomal subunits. Blood. 2007;109:980–986. doi: 10.1182/blood-2006-07-038232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franze-Fernandez MT, Pogo AO. Regulation of the nucleolar DNA-dependent RNA polymerase by amino acids in Ehrlich ascites tumor cells. Proc Natl Acad Sci U S A. 1971;68:3040–3044. doi: 10.1073/pnas.68.12.3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman MH, Amato D, Saunders EF. Erythroid colony growth in congenital hypoplastic anemia. J.Clin.Invest. 1976;57:673–677. doi: 10.1172/JCI108323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguli G, Back J, Sengupta S, Wasylyk B. The p53 tumour suppressor inhibits glucocorticoid-induced proliferation of erythroid progenitors. EMBO Rep. 2002;3:569–574. doi: 10.1093/embo-reports/kvf114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazda HT, Grabowska A, Merida-Long LB, Latawiec E, Schneider HE, Lipton JM, Vlachos A, Atsidaftos E, Ball SE, Orfali KA, Niewiadomska E, Da Costa L, Tchernia G, Niemeyer C, Meerpohl JJ, Stahl J, Schratt G, Glader B, Backer K, Wong C, Nathan DG, Beggs AH, Sieff CA. Ribosomal Protein S24 Gene is Mutated in Diamond-Blackfan Anemia. Am J Hum Genet. 2006a;79:1110–1118. doi: 10.1086/510020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazda HT, Kho AT, Sanoudou D, Zaucha JM, Kohane IS, Sieff CA, Beggs AH. Defective ribosomal protein gene expression alters transcription, translation, apoptosis, and oncogenic pathways in Diamond-Blackfan anemia. Stem Cells. 2006b;24:2034–2044. doi: 10.1634/stemcells.2005-0554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazda HT, Sheen MR, Vlachos A, Choesmel V, O'Donohue MF, Schneider H, Darras N, Hasman C, Sieff CA, Newburger PE, Ball SE, Niewiadomska E, Matysiak M, Zaucha JM, Glader B, Niemeyer C, Meerpohl JJ, Atsidaftos E, Lipton JM, Gleizes PE, Beggs AH. Ribosomal protein L5 and L11 mutations are associated with cleft palate and abnormal thumbs in Diamond-Blackfan anemia patients. Am J Hum Genet. 2008;83:769–780. doi: 10.1016/j.ajhg.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazda HT, Zhong R, Long L, Niewiadomska E, Lipton JM, Ploszynska A, Zaucha JM, Vlachos A, Atsidaftos E, Viskochil DH, Niemeyer CM, Meerpohl JJ, Rokicka-Milewska R, Pospisilova D, Wiktor-Jedrzejczak W, Nathan DG, Beggs AH, Sieff CA. RNA and protein evidence for haplo-insufficiency in Diamond-Blackfan anaemia patients with RPS19 mutations. Br J Haematol. 2004;127:105–113. doi: 10.1111/j.1365-2141.2004.05152.x. [DOI] [PubMed] [Google Scholar]
- Gregory CJ, Tepperman AD, McCulloch EA, Till JE. Erythropoietic progenitors capable of colony formation in culture: response of normal and genetically anemic W-W-V mice to manipulations of the erythron. J Cell Physiol. 1974;84:1–12. doi: 10.1002/jcp.1040840102. [DOI] [PubMed] [Google Scholar]
- Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- Idol RA, Robledo S, Du HY, Crimmins DL, Wilson DB, Ladenson JH, Bessler M, Mason PJ. Cells depleted for RPS19, a protein associated with Diamond Blackfan Anemia, show defects in 18S ribosomal RNA synthesis and small ribosomal subunit production. Blood Cells Mol Dis. 2007;39:35–43. doi: 10.1016/j.bcmd.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Ikuta K, Kina T, MacNeil I, Uchida N, Peault B, Chien YH, Weissman IL. A developmental switch in thymic lymphocyte maturation potential occurs at the level of hematopoietic stem cells. Cell. 1990;62:863–874. doi: 10.1016/0092-8674(90)90262-d. [DOI] [PubMed] [Google Scholar]
- Jin A, Itahana K, O'Keefe K, Zhang Y. Inhibition of HDM2 and activation of p53 by ribosomal protein L23. Mol Cell Biol. 2004;24:7669–7680. doi: 10.1128/MCB.24.17.7669-7680.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kina T, Ikuta K, Takayama E, Wada K, Majumdar AS, Weissman IL, Katsura Y. The monoclonal antibody TER-119 recognizes a molecule associated with glycophorin A and specifically marks the late stages of murine erythroid lineage. Br J Haematol. 2000;109:280–287. doi: 10.1046/j.1365-2141.2000.02037.x. [DOI] [PubMed] [Google Scholar]
- Kuramitsu M, Hamaguchi I, Takuo M, Masumi A, Momose H, Takizawa K, Mochizuki M, Naito S, Yamaguchi K. Deficient RPS19 protein production induces cell cycle arrest in erythroid progenitor cells. Br J Haematol. 2008;140:348–359. doi: 10.1111/j.1365-2141.2007.06930.x. [DOI] [PubMed] [Google Scholar]
- Levine EM, Becker Y, Boone CW, Eagle H. Contact Inhibition, Macromolecular Synthesis, and Polyribosomes in Cultured Human Diploid Fibroblasts. Proc Natl Acad Sci U S A. 1965;53:350–356. doi: 10.1073/pnas.53.2.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohrum MA, Ludwig RL, Kubbutat MH, Hanlon M, Vousden KH. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell. 2003;3:577–587. doi: 10.1016/s1535-6108(03)00134-x. [DOI] [PubMed] [Google Scholar]
- Marechal V, Elenbaas B, Piette J, Nicolas JC, Levine AJ. The ribosomal L5 protein is associated with mdm-2 and mdm-2-p53 complexes. Mol Cell Biol. 1994;14:7414–7420. doi: 10.1128/mcb.14.11.7414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan KA, Li JZ, Park CY, Beaudry V, Tabor HK, Sabnis AJ, Zhang W, Fuchs H, de Angelis MH, Myers RM, Attardi LD, Barsh GS. Ribosomal mutations cause p53-mediated dark skin and pleiotropic effects. Nat Genet. 2008;40:963–970. doi: 10.1038/ng.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake K, Utsugisawa T, Flygare J, Kiefer T, Hamaguchi I, Richter J, Karlsson S. Ribosomal protein S19 deficiency leads to reduced proliferation and increased apoptosis but does not affect terminal erythroid differentiation in a cell line model of Diamond-Blackfan anemia. Stem Cells. 2008;26:323–329. doi: 10.1634/stemcells.2007-0569. [DOI] [PubMed] [Google Scholar]
- Mucenski ML, McLain K, Kier AB, Swerdlow SH, Schreiner CM, Miller TA, Pietryga DW, Scott WJ, Jr., Potter SS. A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell. 1991;65:677–689. doi: 10.1016/0092-8674(91)90099-k. [DOI] [PubMed] [Google Scholar]
- Nathan DG, Clarke BJ, Hillman DG, Alter BP, Housman DE. Erythroid precursors in congenital hypoplastic (Diamond-Blackfan) anemia. J.Clin.Invest. 1978;61:489–498. doi: 10.1172/JCI108960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohene-Abuakwa Y, Orfali KA, Marius C, Ball SE. Two-phase culture in Diamond Blackfan Anemia: localization of erythroid defect. Blood. 2005;105:838–846. doi: 10.1182/blood-2004-03-1016. [DOI] [PubMed] [Google Scholar]
- Oliner JD, Pietenpol JA, Thiagalingam S, Gyuris J, Kinzler KW, Vogelstein B. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature. 1993;362:857–860. doi: 10.1038/362857a0. [DOI] [PubMed] [Google Scholar]
- Olivieri NF, Grunberger T, Ben-David Y, Ng J, Williams DE, Lyman S, Anderson DM, Axelrad AA, Correa P, Bernstein A, Freedman MH. Diamond-Blackfan anemia: heterogenous response of hematopoietic progenitor cells in vitro to the protein product of the Steel locus. Blood. 1991;78:2211–2215. [PubMed] [Google Scholar]
- Orfali KA, Ohene-Abuakwa Y, Ball SE. Diamond Blackfan anaemia in the UK: clinical and genetic heterogeneity. Br J Haematol. 2004;125:243–252. doi: 10.1111/j.1365-2141.2004.04890.x. [DOI] [PubMed] [Google Scholar]
- Perdahl EB, Naprstek BL, Wallace WC, Lipton JM. Erythroid failure in Diamon-Blackfan anemia is characterized by apoptosis. Blood. 1994;83:645–650. [PubMed] [Google Scholar]
- Pestov DG, Strezoska Z, Lau LF. Evidence of p53-dependent cross-talk between ribosome biogenesis and the cell cycle: effects of nucleolar protein Bop1 on G(1)/S transition. Mol Cell Biol. 2001;21:4246–4255. doi: 10.1128/MCB.21.13.4246-4255.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramenghi U, Garelli E, Valtolina S, Campagnoli MF, Timeus F, Crescenzio N, Mair M, Varotto S, D'Avanzo M, Nobili B, Massolo F, Mori PG, Locatelli F, Gustavsson P, Dahl N, Dianzani I. Diamond-Blackfan anaemia in the Italian population. Br J Haematol. 1999 Mar;104:841–848. doi: 10.1046/j.1365-2141.1999.01267.x. [DOI] [PubMed] [Google Scholar]
- Rubbi CP, Milner J. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. Embo J. 2003;22:6068–6077. doi: 10.1093/emboj/cdg579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieff CA, Yokoyama CT, Zsebo KM, Nathan DG, Williams DA. The enhancement of erythropoiesis by Steel factor (SF) and the production of SF mRNA is normal in Diamond Blackfan anemia (DBA) Blood. 1991;78:95a. abstr. 369. [Google Scholar]
- Socolovsky M, Nam H, Fleming MD, Haase VH, Brugnara C, Lodish HF. Ineffective erythropoiesis in Stat5a(−/−)5b(−/−) mice due to decreased survival of early erythroblasts. Blood. 2001;98:3261–3273. doi: 10.1182/blood.v98.12.3261. [DOI] [PubMed] [Google Scholar]
- Stephenson JR, Axelrad AA, McLeod DL, Shreeve MM. Induction of colonies of hemoglobin-synthesizing cells by erythropoietin in vitro. Proc Natl Acad Sci U S A. 1971;68:1542–1546. doi: 10.1073/pnas.68.7.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todaro GJ, Lazar GK, Green H. The initiation of cell division in a contact-inhibited mammalian cell line. J Cell Physiol. 1965;66:325–333. doi: 10.1002/jcp.1030660310. [DOI] [PubMed] [Google Scholar]
- Vegiopoulos A, Garcia P, Emambokus N, Frampton J. Coordination of erythropoiesis by the transcription factor c-Myb. Blood. 2006;107:4703–4710. doi: 10.1182/blood-2005-07-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volarevic S, Stewart MJ, Ledermann B, Zilberman F, Terracciano L, Montini E, Grompe M, Kozma SC, Thomas G. Proliferation, but not growth, blocked by conditional deletion of 40S ribosomal protein S6. Science. 2000;288:2045–2047. doi: 10.1126/science.288.5473.2045. [DOI] [PubMed] [Google Scholar]
- Ward GA, Plagemann PG. Fluctuations of DNA-dependent RNA polymerase and synthesis of macromolecules during the growth cycle of Novikoff rat hepatoma cells in suspension culture. J Cell Physiol. 1969;73:213–231. doi: 10.1002/jcp.1040730307. [DOI] [PubMed] [Google Scholar]
- Wessely O, Deiner EM, Beug H, von Lindern M. The glucocorticoid receptor is a key regulator of the decision between self-renewal and differentiation in erythroid progenitors. Embo J. 1997;16:267–280. doi: 10.1093/emboj/16.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willig T-N, Niemeyer C, Leblanc T, Tiemann C, Robert A, Budde J, Lambiliotte A, Kohne E, Souillet G, Stephan JL, Girot R, Bordigoni P, Cornu G, Blanche S, Guillard JM, Tchernia G. Diamond Blackfan anemia: clinical and epidemiological studies with identification of new long-term prognosis factors from the analysis of a registry of 234 patients. Pediatric Research. 1999 doi: 10.1203/00006450-199911000-00011. [DOI] [PubMed] [Google Scholar]
- Ying GG, Proost P, van Damme J, Bruschi M, Introna M, Golay J. Nucleolin, a novel partner for the Myb transcription factor family that regulates their activity. J Biol Chem. 2000;275:4152–4158. doi: 10.1074/jbc.275.6.4152. [DOI] [PubMed] [Google Scholar]
- Zhang J, Socolovsky M, Gross AW, Lodish HF. Role of Ras signaling in erythroid differentiation of mouse fetal liver cells: functional analysis by a flow cytometry-based novel culture system. Blood. 2003a;102:3938–3946. doi: 10.1182/blood-2003-05-1479. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wolf GW, Bhat K, Jin A, Allio T, Burkhart WA, Xiong Y. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol Cell Biol. 2003b;23:8902–8912. doi: 10.1128/MCB.23.23.8902-8912.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]