Abstract
Guanine and adenine nucleotide triphosphatases, such as Ras proteins and protein kinases, undergo large conformational changes upon ligand binding in the course of their functions. New computer simulation methods have combined with experimental studies to deepen our understanding of these phenomena. In particular, a "conformational selection" picture is emerging, where alterations in the relative populations of pre-existing conformations can best describe the conformational switching activity of these important proteins.
1. Introduction
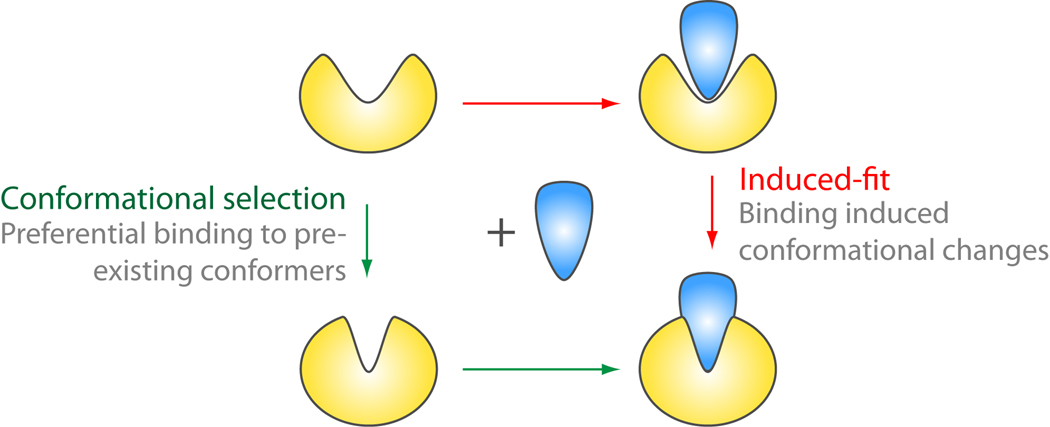
Structural fluctuations of proteins are often intimately coupled to biochemical function. Indeed, regulated conformational changes control diverse cellular processes, such as signal transduction, membrane trafficking, vesicular transport and polypeptide chain elongation. Key to the linkage of structural fluctuations and functional properties is the ability of alternate conformations to preferentially interact with different binding partners at different times or locations. This spatiotemporal modulation of function through dynamics allows eukaryotic cells to manage a large number of essential cellular processes with a limited number of distinct gene products. Perhaps the most prominent examples of proteins that undergo large-scale conformational changes to regulate cell processes are guanine and adenine nucleotide triphosphatases (NTPases). These ubiquitous enzymes function as conformational switches and regulators fueled by nucleotide binding and hydrolysis [1]. In this review we focus on two classes of such proteins: Ras-related G-proteins and kinases. In the following sections we outline current techniques for studying the dynamics of such systems, with special emphasis on advanced simulation methods. We then describe recent studies that highlight a shifting paradigm for how signaling enzymes operate, namely, via alterations in the relative populations of pre-existing conformations that are sampled through motions with a topologically preferred directionality (Figure 1).
Figure 1.
Conformational selection and induced-fit models. Conformational selection (green arrows) dictates that the unbound protein (left of figure) explores a range of conformations some of which are structurally similar to bound conformations. Interaction with binding partners (blue) leads to the preferential selection of favorable pre-existing conformations causing a corresponding shift in the population of microstates in the direction of bound conformations (right of figure). With the induced-fit model (red arrows) the bound like conformations form only after interaction with a binding partner due to specific induced structural changes rather than selection from the already present unbound ensemble.
2. Current techniques for probing conformational changes
Proteins exhibit a rich hierarchy of internal motions, from individual atomic displacements to collective large-scale motions, over a wide range of time scales [2,3]. The complex topography of the energy surface that underlies this dynamic behavior can give rise to multiple, significantly populated, conformational states that are separated by a distribution of energy barriers. Changes to the environment, such as temperature and pH, as well as binding events and enzymatic modifications can alter the extent of the accessible energy surface and thus both the equilibrium distribution of conformational states and their interconversion dynamics. Further, the internal motions and intrinsic dynamics of proteins have increasingly been recognized as essential for activities including ligand binding, enzymatic catalysis and bi-molecular recognition [4–7].
Crystallographic structures determined under different crystallization conditions or oligomerization states often yield valuable insights into conformational rearrangements. However, crystal structures represent only the average conformation for a particular condition. Important information on protein mobility has also been obtained from other experimental methods, such as nuclear magnetic resonance (NMR) spectroscopy, hydrogen-deuterium exchange, time-resolved X-ray crystallography, fluorescence spectroscopy and inelastic neutron scattering [8,9]. For example NMR measurements coupled with modeling and simulation, have been instrumental in elucidating the dynamic properties of many small proteins (under 25 kDa) [10,11]. Computational approaches based on empirically derived potential energy functions, such as molecular dynamics (MD) and normal mode analysis (NMA), have enabled an understanding of the relationship between protein structure and dynamics at the atomic level [12]. For instance, MD simulation of the trypsin inhibitor protein provided the first insight into the rather “fluid”-like nature of protein structures, when the “rigid-body” view was still dominant [13]. Subsequent computational efforts, most prominently via MD, helped uncover many biological functions of protein flexibility (for recent reviews, see [12,14– 17]). Here we focus on some of the most recent developments that are expanding the scope of conventional MD.
3. Simulation approaches
A number of diverse computational methodologies have been developed for modeling protein dynamics with varying degrees of resolution. Among these, atomistic classical MD (cMD) and replica exchange MD (reMD) are the most widely used, while cutting-edge methods such as accelerated MD (aMD) are beginning to provide exciting results. cMD remains the method of choice for sampling nanosecond and Angstrom scale fluctuations [12,14]. However, many biological processes involve motions that span microseconds to seconds and tens of nanometers or more. Such motions can in principle be sampled by cMD given sufficient computational resources. Indeed, microsecond length cMD simulations have been achieved for medium sized proteins [16], though this required specialized hardware and software combinations.
A number of recently developed cMD adaptations employ elevated temperatures, an external force, or a biasing boost potential to facilitate the more rapid crossing of energy barriers separating distinct conformations. For example the reMD method attempts to avoid kinetic traps by simulating frequently exchanging non-interacting replicas at different temperatures [18,19]. Another popular approach is the so-called steered MD (SMD), in which external forces are applied on the system of study to explore its mechanical responses (reviewed in [20,21]). Decreasing the depth of low-energy wells can also promote the sampling of high-energy conformations [22]. Along this line, the aMD method developed by Hamelberg et al. adds a bias potential (or boost energy) when the systems potential energy is below a certain threshold thus facilitating an accelerated rate of state-to-state evolution [23].
An alternative approach for probing large scale motions involves coarse-grained (CG) models that typically utilize Cα atom or reaction center representations and simplified potential energy functions. Methods in this category include those that step atoms through time with MD or Brownian dynamics (BD) and those that employ collective motions estimated through normal mode approaches [24–26]. There are multiple flavors of the latter, including the elastic and anisotropic network models (ENM & ANM). Their fundamental assumption is that functionally significant fluctuations can be captured by a few low-frequency collective modes that behave much like an elastic material. Despite their simplifying assumptions these methods are often capable of capturing long time and length scale protein dynamics (see reviews [27–30]).
4. NTP dependent cellular regulators
Proteins that hydrolyze guanosine or adenosine triphosphate (GTPases and ATPases) play a crucial role in many diverse cellular processes essential for growth and development. GTPase families, such as Ras, regulate processes including intracellular transport, sensory perception and protein synthesis [1,31]. ATPases including protein kinases control cell cycle regulation, cytoskeletal rearrangements and motility [1,31]. Remarkably, these structurally and functionally diverse families utilize the same underlying strategy: the addition or removal of inorganic phosphate to switch from active to inactive conformations.
Ras GTPases
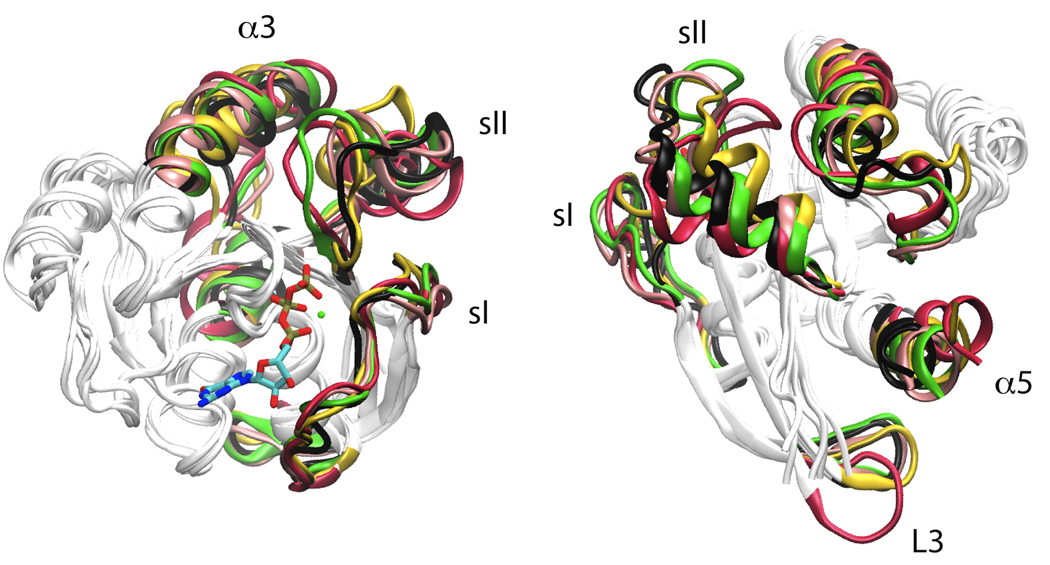
Ras GTPases are ubiquitously expressed conformational switches that cycle between GTP bound on and GDP bound off states to mediate signaling pathways that control cell proliferation and differentiation [32]. Recent analysis confirmed that the majority of H-ras crystal structures represent one of two major conformations, either GDP or GTP like [33]. Interestingly, a number of intermediate conformations for structures with oncogenic point mutations in the vicinity of the nucleotide binding site were also noted [33]. cMD simulations initiated from one such intermediate, G12V with a docked GTP, resulted in a spontaneous transition toward the main GTP conformation [33,34]. The activating transition involved a multiphase process in which L4-α2 (known as sII) reorganization was followed by that of L2 (known as sI, see Figure 2) [33]. The work also predicted flexibility at L3 and the C-terminus of α5 [33,34]. This was recapitulated by subsequent aMD simulations [34] on wild-type H-ras that revealed concerted motions and unique conformations for these regions during the step-wise transition of sI and sII [34]. The combined application of cMD, aMD and NMA [33–35] indicated that conformational changes in Ras are best described by a population-shift mechanism rather than by the popular “loaded spring” [31] or induced-fit on/off switch. First, nucleotide free aMD simulations sampled the regions populated by the major GTP, GDP and intermediate mutant conformers [34]. Second, NMA qualitatively captured the differences between GDP and GTP conformations and had high overlap with the eigenvectors obtained from aMD trajectories. Together, these results suggest that nucleotide dependent dynamics is facilitated by low frequency global motions that are intrinsic to the structure. Other recent simulation [36] and 31P NMR studies [37] have also highlighted the complex and global nature of the Ras dynamics. Furthermore, these results are consistent with earlier cMD simulations [35] and subsequent experiments [38,39] of membrane-bound H-ras. For instance, cMD predicted that a pair of basic residues in helix α4 stabilizes the membrane orientation of GTP-H-ras while that of GDP-H-ras is stabilized by a different pair of basic residues at the flexible C-terminal region [35]. Experimentally, the ERK-phosphorylating ability of H-ras variants in which these residues were replaced by alanine was respectively weaker- and significantly higher than the oncogenic G12V H-ras [38,39]. This can be best understood in terms of a population-shift mechanism wherein mutations “re-distribute” the population of conformers centered around the two predicted conformations with distinct GDP and GTP membrane orientations [38,39].
Figure 2.
Major conformational clusters in a single aMD simulation of GTP bound H-ras include GDP, GTP and intermediate like conformations. See [37] for further details.
Protein Kinases
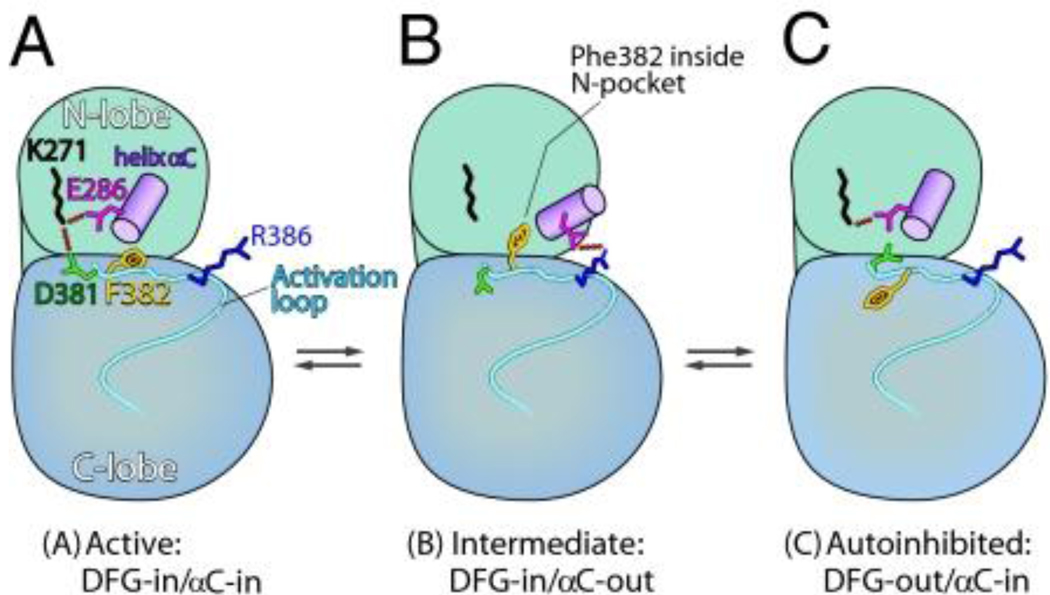
Protein kinases are a large superfamily involved in the regulation of diverse cell signaling processes. They operate by transferring a phosphate group from ATP to specific serine, threonine, or tyrosine residues. The added phosphate alters the conformation and function of the target protein, with different kinases recognizing different target sequences. Structural studies have shown that major rearrangements occur during the inactive to active transition of kinases, including closure of the active-site cleft, packing of the activation loop, and rotation of the C-helix [40]. These rearrangements were found to involve set of conserved motifs that are anchored at the F-helix [41]. A number of computational studies have probed the plasticity and allosteric nature of these rearrangements [42–46]. In some kinases structural changes during activation are linked to rearrangements of a conserved DFG motif that props open the active site cleft between kinase domains [40]. Recently, Shaw and collaborators formulated a mechanism for DFG “flipping” based on microsecond cMD simulations together with crystallographic and kinetic experiments [47]. The proposed mechanism involves a pH dependent DFG rearrangement driven by electrostatic changes inherent to the catalytic cycle. This motion is correlated with a corresponding in-out displacement of the C-helix during activation (Figure 3). An intermediate structure that exhibits a DFG-in and C-helix-out configuration has also been identified [47].
Figure 3.
Key features of the proposed DFG flipping mechanism in Abl kinases include rearrangements of helix αC and the salt bridge partner of Glu-286. Schematics are based on the kinase domain of active (A) and autoinhibited crystal structures (C), along with a cMD simulated intermediate (B). See [45] for further details © 2008 by The National Academy of Sciences of the USA
In another recent study Roux and colleagues examined the catalytic domain of Src tyrosine kinase using a CG model [48] and an atomistic “string method with swarms-of-trajectories” [49]. By analyzing “structural networks” among clusters of conformations from their CG simulations, the authors found two major ensembles of pathways for activation: one involving interactions of the C-helix, the activation-loop, and the beta strands in the N-lobe, and another involving partial unfolding of the N-lobe [48]. Free energy profiles from the “string method” predicted a two step activation pathway with opening of the activation loop preceding rotation of the C-helix [49]. A very similar pathway has been found in other kinases, such as in Lyn [50] and CDK5 [51]. A multiphase activation mechanism has also been proposed for Abl and EGFR kinases [52]. An intermediate structure that lies between the active and inactive structures of Src tyrosine kinase has also been identified [49]. Interestingly, both the two-step transition and the presence of an intermediate mirror the observations in Ras discussed in the previous section. Thus, conformational selection rather than induction might also operate in kinases.
5. Conclusions
Our current view of conformational transitions owes much to X-ray crystallography. Examination of different static structures of a protein or its complexes has led to a predominance of Koshland-Nemethy-Filmer like models that depict conformational inter-conversion as arising from induction by ligand binding or mutation [53]. As a consequence, signaling proteins such Ras were believed to operate via a loaded-spring mechanism wherein the GTP gamma-phosphate forces rearrangement of distal sites [31]. However, the simulation results discussed above, and other earlier studies [54], indicate that these proteins harbor an intrinsic susceptibility to sample multiple conformational states regardless of the bound nucleotide, i.e., perturbations do not directly induce conformational change but rather shift or bias pre-existing conformations. It is increasingly apparent that the conformational plasticity of signaling proteins can be exploited for therapeutic benefit. For example the cancer drug Gleevic/imatinib selectively binds and stabilizes an inactive form of Abl kinases [55,56]. Selectivity for Abl with respect to related kinases such as Src can be traced to distinct conformational preferences of the different families [57,58]. Clearly knowledge of the complete conformational repertoire of signaling proteins such as Ras and kinases has applications to drug development and may facilitate the selective conformational targeting of distinct inactive conformations. The examples highlighted in this review demonstrate the power of modeling and simulation to decipher the dynamic features of signaling molecules and their increasing ability to provide mechanistic insight into large-scale conformational changes.
Acknowledgement
We thank Dr Ana Rodrigues for critical reading of the manuscript, members of the McCammon and Gorfe laboratories for helpful discussions, and the NSF Supercomputer Centers and the Center for Theoretical Biological Physics (CTBP) for computational resources. This work was in part supported by grants from National Institutes of Health, National Science Foundation, Howard Hughes Medical Institute and CTBP. AAG acknowledges support from the University of Texas Medical School at Houston.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Vetter IR, Wittinghofer A. Nucleoside triphosphate-binding proteins: different scaffolds to achieve phosphoryl transfer. Q Rev Biophys. 1999;32:1–56. doi: 10.1017/s0033583599003480. [DOI] [PubMed] [Google Scholar]
- 2.Frauenfelder H, Sligar SG, Wolynes PG. The energy landscapes and motions of proteins. Science. 1991;254:1598–1603. doi: 10.1126/science.1749933. [DOI] [PubMed] [Google Scholar]
- 3.McCammon JA. Computer-aided molecular design. Science. 1987;238:486–491. doi: 10.1126/science.3310236. [DOI] [PubMed] [Google Scholar]
- 4.Karplus M, McCammon JA. Dynamics of proteins: elements and function. Annu Rev Biochem. 1983;52:263–300. doi: 10.1146/annurev.bi.52.070183.001403. [DOI] [PubMed] [Google Scholar]
- 5.Berendsen HJ, Hayward S. Collective protein dynamics in relation to function. Curr Opin Struct Biol. 2000;10:165–169. doi: 10.1016/s0959-440x(00)00061-0. [DOI] [PubMed] [Google Scholar]
- 6.Sinha N, Smith-Gill SJ. Protein structure to function via dynamics. Protein Pept Lett. 2002;9:367–377. doi: 10.2174/0929866023408508. [DOI] [PubMed] [Google Scholar]
- 7.Frauenfelder H, Fenimore PW, Young RD. Protein dynamics and function: insights from the energy landscape and solvent slaving. IUBMB Life. 2007;59:506–512. doi: 10.1080/15216540701194113. [DOI] [PubMed] [Google Scholar]
- 8.Forsen S, Kordel J. Biomolecular structure and dynamics--experiment and theory. J Pharm Biomed Anal. 1996;14:233–236. doi: 10.1016/0731-7085(95)01640-6. [DOI] [PubMed] [Google Scholar]
- 9.Vendruscolo M, Paci E. Protein folding: bringing theory and experiment closer together. Curr Opin Struct Biol. 2003;13:82–87. doi: 10.1016/s0959-440x(03)00007-1. [DOI] [PubMed] [Google Scholar]
- 10.Bouguet-Bonnet S, Buck M. Compensatory and long-range changes in picosecond-nanosecond main-chain dynamics upon complex formation: 15N relaxation analysis of the free and bound states of the ubiquitinlike domain of human plexin-B1 and the small GTPase Rac1. J Mol Biol. 2008;377:1474–1487. doi: 10.1016/j.jmb.2008.01.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Billeter M, Wagner G, Wuthrich K. Solution NMR structure determination of proteins revisited. J Biomol NMR. 2008;42:155–158. doi: 10.1007/s10858-008-9277-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karplus M, McCammon JA. Molecular dynamics simulations of biomolecules. Nat Struct Biol. 2002;9:646–652. doi: 10.1038/nsb0902-646. [DOI] [PubMed] [Google Scholar]
- 13.McCammon JA, Gelin BR, Karplus M. Dynamics of folded proteins. Nature. 1977;267:585–590. doi: 10.1038/267585a0. [DOI] [PubMed] [Google Scholar]
- 14. Adcock SA, McCammon JA. Molecular dynamics: survey of methods for simulating the activity of proteins. Chem Rev. 2006;106:1589–1615. doi: 10.1021/cr040426m.. This paper surveys various flavors of molecular dynamics simulation methods used to study protein function.
- 15.van Gunsteren WF, Bakowies D, Baron R, Chandrasekhar I, Christen M, Daura X, Gee P, Geerke DP, Glattli A, Hunenberger PH, et al. Biomolecular modeling: Goals, problems, perspectives. Angew Chem Int Ed Engl. 2006;45:4064–4092. doi: 10.1002/anie.200502655. [DOI] [PubMed] [Google Scholar]
- 16.Klepeis JL, Lindorff-Larsen K, Dror RO, Shaw DE. Long-timescale molecular dynamics simulations of protein structure and function. Curr Opin Struct Biol. 2009;19:120–127. doi: 10.1016/j.sbi.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 17.Khalili-Araghi F, Gumbart J, Wen PC, Sotomayor M, Tajkhorshid E, Schulten K. Molecular dynamics simulations of membrane channels and transporters. Curr Opin Struct Biol. 2009;19:128–137. doi: 10.1016/j.sbi.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sugita Y. Free-energy landscapes of proteins in solution by generalized-ensemble simulations. Front Biosci. 2009;14:1292–1303. doi: 10.2741/3309. [DOI] [PubMed] [Google Scholar]
- 19.Mitsutake A, Sugita Y, Okamoto Y. Generalized-ensemble algorithms for molecular simulations of biopolymers. Biopolymers. 2001;60:96–123. doi: 10.1002/1097-0282(2001)60:2<96::AID-BIP1007>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 20.Genchev GZ, Kallberg M, Gursoy G, Mittal A, Dubey L, Perisic O, Feng G, Langlois R, Lu H. Mechanical Signaling on the Single Protein Level Studied Using Steered Molecular Dynamics. Cell Biochem Biophys. 2009 doi: 10.1007/s12013-009-9064-5. [DOI] [PubMed] [Google Scholar]
- 21.Sotomayor M, Schulten K. Single-molecule experiments in vitro and in silico. Science. 2007;316:1144–1148. doi: 10.1126/science.1137591. [DOI] [PubMed] [Google Scholar]
- 22.Voter AF. A method for accelerating the molecular dynamics simulation of infrequent events. Journal of Chemical Physics. 1997;106:4665–4677. [Google Scholar]
- 23.Hamelberg D, Mongan J, McCammon JA. Accelerated molecular dynamics: a promising and efficient simulation method for biomolecules. J Chem Phys. 2004;120:11919–11929. doi: 10.1063/1.1755656. [DOI] [PubMed] [Google Scholar]
- 24.Chu JW, Ayton GS, Izvekov S, Voth GA. Emerging methods for multiscale simulation of biomolecular systems. Molecular Physics. 2007;105:167–175. [Google Scholar]
- 25.Ayton GS, Noid WG, Voth GA. Multiscale modeling of biomolecular systems: in serial and in parallel. Curr Opin Struct Biol. 2007;17:192–198. doi: 10.1016/j.sbi.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 26.Chang CE, Trylska J, Tozzini V, McCammon JA. Binding pathways of ligands to HIV-1 protease: coarse-grained and atomistic simulations. Chem Biol Drug Des. 2007;69:5–13. doi: 10.1111/j.1747-0285.2007.00464.x. [DOI] [PubMed] [Google Scholar]
- 27. Bahar I, Rader AJ. Coarse-grained normal mode analysis in structural biology. Curr Opin Struct Biol. 2005;15:586–592. doi: 10.1016/j.sbi.2005.08.007.. This paper provides an overview of the application of normal mode analysis to study functionally relevant protein motions.
- 28. Ma J. Usefulness and limitations of normal mode analysis in modeling dynamics of biomolecular complexes. Structure. 2005;13:373–380. doi: 10.1016/j.str.2005.02.002.. This paper reviews the kinds of questions that can be addressed by normal mode analysis approaches.
- 29.Ma J. Usefulness and limitations of normal mode analysis in modeling dynamics of biomolecular complexes. Structure. 2005;13:373–380. doi: 10.1016/j.str.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 30.Tama F. Normal mode analysis with simplified models to investigate the global dynamics of biological systems. Protein Pept Lett. 2003;10:119–132. doi: 10.2174/0929866033479077. [DOI] [PubMed] [Google Scholar]
- 31. Vetter IR, Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science. 2001;294:1299–1304. doi: 10.1126/science.1062023.. A highly informative account of available x-ray structures of guanine nucleotide binding proteins; discusses the mechanisms of guanine nucleotide exchange protein-assisted GDP release and GTPAse activating protein-catalyzed GTP hydrolysis. The loaded-spring mechanism of conformational change is also outlined.
- 32. Karnoub AE, Weinberg RA. Ras oncogenes: split personalities. Nat Rev Mol Cell Biol. 2008;9:517–531. doi: 10.1038/nrm2438.. This review article contains a wealth of data on Ras biology, including a timeline of the major milestones made over the 40 year period since the discovery of the RAS gene. It discusses associated diseases and the potential for cancer therapeutics.
- 33.Gorfe AA, Grant BJ, McCammon JA. Mapping the nucleotide and isoform-dependent structural and dynamical features of Ras proteins. Structure. 2008;16:885–896. doi: 10.1016/j.str.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Grant BJ, Gorfe AA, McCammon JA. Ras conformational switching: simulating nucleotide-dependent conformational transitions with accelerated molecular dynamics. PLoS Comput Biol. 2009;5:e1000325. doi: 10.1371/journal.pcbi.1000325.. This paper discusses the application of accelerated molecular dynamics for sampling large-scale conformational changes. It also highlights the discovery of previously uncharacterized highly populated intermediate conformations sampled during the inter-conversion of a Ras protein between its active and inactive states.
- 35.Gorfe AA, Hanzal-Bayer M, Abankwa D, Hancock JF, McCammon JA. Structure and dynamics of the full-length lipid-modified H-Ras protein in a 1,2- dimyristoylglycero-3-phosphocholine bilayer. J Med Chem. 2007;50:674–684. doi: 10.1021/jm061053f. [DOI] [PubMed] [Google Scholar]
- 36.Fuentes G, Valencia A. Ras classical effectors: new tales from in silico complexes. Trends Biochem Sci. 2009;34:533–539. doi: 10.1016/j.tibs.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 37.O'Connor C, Kovrigin EL. Global conformational dynamics in ras. Biochemistry. 2008;47:10244–10246. doi: 10.1021/bi801076c. [DOI] [PubMed] [Google Scholar]
- 38. Abankwa D, Hanzal-Bayer M, Ariotti N, Plowman SJ, Gorfe AA, Parton RG, McCammon JA, Hancock JF. A novel switch region regulates H-ras membrane orientation and signal output. Embo J. 2008;27:727–735. doi: 10.1038/emboj.2008.10.. This paper discusses the discovery of a novel switch region that mediates nucleotide-dependent membrane re-orientation of Ras. It represents an example of how simulations can drive new experiments.
- 39.Abankwa D, Gorfe AA, Hancock JF. Mechanisms of Ras membrane organization and signalling: Ras on a rocker. Cell Cycle. 2008;7:2667–2673. doi: 10.4161/cc.7.17.6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huse M, Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- 41. Kornev AP, Taylor SS, Ten Eyck LF. A helix scaffold for the assembly of active protein kinases. Proc Natl Acad Sci U S A. 2008;105:14377–14382. doi: 10.1073/pnas.0807988105.. Using a spatial pattern recognition algorithm, the authors identified set of functionally relevant motifs that were not observed by other techniques Application of similar methods will likely yield fresh insights into the activation process of other signaling protein families.
- 42.Banavali NK, Roux B. Anatomy of a structural pathway for activation of the catalytic domain of Src kinase Hck. Proteins. 2007;67:1096–1112. doi: 10.1002/prot.21334. [DOI] [PubMed] [Google Scholar]
- 43.Banavali NK, Roux B. The N-terminal end of the catalytic domain of SRC kinase Hck is a conformational switch implicated in long-range allosteric regulation. Structure. 2005;13:1715–1723. doi: 10.1016/j.str.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 44. Yang S, Banavali NK, Roux B. Mapping the conformational transition in Src activation by cumulating the information from multiple molecular dynamics trajectories. Proc Natl Acad Sci U S A. 2009;106:3776–3781. doi: 10.1073/pnas.0808261106.. The authors combine 78 independent classical molecular dynamics simulations to build a “connectivity map” of the conformational transition between inactive and active states of Src kinases.
- 45.Kastner J, Loeffler HH, Roberts SK, Martin-Fernandez ML, Winn MD. Ectodomain orientation, conformational plasticity and oligomerization of ErbB1 receptors investigated by molecular dynamics. J Struct Biol. 2009;167:117–128. doi: 10.1016/j.jsb.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 46.Lu T, Tan H, Lee D, Chen G, Jia Z. New insights into the activation of Escherichia coli tyrosine kinase revealed by molecular dynamics simulation and biochemical analysis. Biochemistry. 2009;48:7986–7995. doi: 10.1021/bi900811p. [DOI] [PubMed] [Google Scholar]
- 47. Shan Y, Seeliger MA, Eastwood MP, Frank F, Xu H, Jensen MO, Dror RO, Kuriyan J, Shaw DE. A conserved protonation-dependent switch controls drug binding in the Abl kinase. Proc Natl Acad Sci U S A. 2009;106:139–144. doi: 10.1073/pnas.0811223106.. The paper presents an appealing model for DFG flipping in Abl kinase based primarily on long classical molecular dynamics simulations.
- 48. Yang S, Roux B. Src kinase conformational activation: thermodynamics, pathways, and mechanisms. PLoS Comput Biol. 2008;4:e1000047. doi: 10.1371/journal.pcbi.1000047.. A coarse-grained model is employed to characterize possible pathways for activation of Src kinases. See also reference 44.
- 49.Gan W, Yang S, Roux B. Atomistic view of the conformational activation of Src kinase using the string method with swarms-of-trajectories. Biophys J. 2009;97:L8–L10. doi: 10.1016/j.bpj.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ozkirimli E, Post CB. Src kinase activation: A switched electrostatic network. Protein Sci. 2006;15:1051–1062. doi: 10.1110/ps.051999206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Berteotti A, Cavalli A, Branduardi D, Gervasio FL, Recanatini M, Parrinello M. Protein conformational transitions: the closure mechanism of a kinase explored by atomistic simulations. J Am Chem Soc. 2009;131:244–250. doi: 10.1021/ja806846q. [DOI] [PubMed] [Google Scholar]
- 52.Dixit A, Verkhivker GM. Hierarchical modeling of activation mechanisms in the ABL and EGFR kinase domains: thermodynamic and mechanistic catalysts of kinase activation by cancer mutations. PLoS Comput Biol. 2009;5:e1000487. doi: 10.1371/journal.pcbi.1000487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koshland DE, Jr, Nemethy G, Filmer D. Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry. 1966;5:365–385. doi: 10.1021/bi00865a047. [DOI] [PubMed] [Google Scholar]
- 54.Nevo R, Stroh C, Kienberger F, Kaftan D, Brumfeld V, Elbaum M, Reich Z, Hinterdorfer P. A molecular switch between alternative conformational states in the complex of Ran and importin beta1. Nat Struct Biol. 2003;10:553–557. doi: 10.1038/nsb940. [DOI] [PubMed] [Google Scholar]
- 55.Druker BJ, Talpaz M, Resta DJ, Peng B, Buchdunger E, Ford JM, Lydon NB, Kantarjian H, Capdeville R, Ohno-Jones S, et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med. 2001;344:1031–1037. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- 56.Schindler T, Bornmann W, Pellicena P, Miller WT, Clarkson B, Kuriyan J. Structural mechanism for STI-571 inhibition of abelson tyrosine kinase. Science. 2000;289:1938–1942. doi: 10.1126/science.289.5486.1938. [DOI] [PubMed] [Google Scholar]
- 57.Seeliger MA, Ranjitkar P, Kasap C, Shan Y, Shaw DE, Shah NP, Kuriyan J, Maly DJ. Equally potent inhibition of c-Src and Abl by compounds that recognize inactive kinase conformations. Cancer Res. 2009;69:2384–2392. doi: 10.1158/0008-5472.CAN-08-3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Johnson LN, Noble ME, Owen DJ. Active and inactive protein kinases: structural basis for regulation. Cell. 1996;85:149–158. doi: 10.1016/s0092-8674(00)81092-2. [DOI] [PubMed] [Google Scholar]