Summary
The coordination of gene expression is critical for cell differentiation and the subsequent establishment of tissue function. We show here that a multiple zinc-finger transcription factor, zfp423/OAZ, is transiently expressed in newly differentiating olfactory receptor neurons (ORNs) and has a key role in coordinating the expression of immature and mature stage-specific genes. OAZ deletion in mice impairs aspects of ORN differentiation, particularly the patterns of axonal projection to the olfactory bulb. OAZ gain-of-function experiments show that sustained OAZ expression throughout ORN maturation arrests ORN development at an immature stage and alters OR gene expression. Importantly, reintroducing OAZ expression in mature ORNs suppresses mature marker expression and reactivates immature-specific markers. Together, theses experiments suggest that OAZ participates in a developmental switch regulating the transition from differentiation to maturation in ORNs.
Introduction
The olfactory receptor neurons (ORNs) are continuously generated from a population of neuroblast-like stem/progenitor cells located at the basal olfactory epithelium (OE). This lifelong olfactory neurogenesis is under the control of a temporal series of intrinsic transcriptional cascades (Nicolay et al., 2006), some of which are regulated by extrinsic cues (Kawauchi et al., 2004; Kawauchi et al., 2005; Wu et al., 2003). During ORN development, olfactory stem cells undergo infrequent asymmetric cell divisions to generate transient amplifying progenitors (Mash1-positive) and subsequently, Ngn1-positive immediate neuronal precursors (Calof et al., 2002; Cau et al., 2002). The division of Ngn1-positive cells generates daughter cells that differentiate into ORNs and express characteristic markers including neural cell adhesion molecule (NCAM), olfactory maker protein (OMP) and olfactory-specific genes essential for odorant transduction (Bakalyar and Reed, 1990; Brunet et al., 1996; Calof and Chikaraishi, 1989; Jones and Reed, 1989; Kawauchi et al., 2004).
Extrinsic signaling molecules implicated in olfactory neurogenesis include the fibroblast growth factor (FGF) and transforming growth factor-β (TGF-β) superfamilies (Kawauchi et al., 2004; Kawauchi et al., 2005). These signaling molecules, particular the bone morphogenic proteins (BMPs) exert both positive and negative effects on ORN neurogenesis, depending on the type of the signaling molecule, the concentration and the stage at which they act (Kawauchi et al., 2004; Shou et al., 1999; Shou et al., 2000; Wu et al., 2003). Despite considerable progress in defining transcription factors and signaling molecules in OE neurogenesis, much less is known about the key transcriptional regulators mediating the interaction between these two aspects of regulation.
The Zfp423/OAZ protein, containing 30 kruppel-like C2H2 zinc-fingers is an attractive candidate to mediate extrinsic regulation of the transcriptional cascade. OAZ (O/E associated zinc-finger protein) was identified as an inhibitor of O/E proteins, a family of transcriptional factors involved in terminal ORN differentiation (Wang and Reed, 1993; Wang et al., 1993; Wang et al., 1997). OAZ binds to O/E proteins using a subset of zinc-finger domains and abolishes O/E binding to consensus binding sites present in olfactory-specific promoters (Tsai and Reed, 1997; Tsai and Reed, 1998). This negative regulation of O/E activity was proposed to prevent premature differentiation of immature ORNs. Independently, OAZ was also identified as a cofactor of Smad proteins in the BMP-signaling pathway (Hata et al., 2000; Ku et al., 2006; Shim et al., 2002). The zinc-finger domains mediating Smad interaction are distinct from those that engage in O/E proteins (Hata et al., 2000). The presence of BMPs and their receptors in the olfactory system and the proposed role of BMPs in regulation of ORN differentiation (Dewulf et al., 1995; Wu et al., 2003; Zhang et al., 1998) suggest that OAZ might have an important role in integrating BMP signaling and O/E activity at critical stages of ORN differentiation.
In this study, we provide in vivo evidence that OAZ regulates the transition from differentiation to maturation in ORNs. OAZ is transiently expressed in newly generated postmitotic ORNs. OAZ deletion in mice impairs ORN differentiation. Importantly, reintroducing OAZ into mature cells is sufficient to revert mature cells to an immature phenotype. These arrested, immature cells accumulate in OE and are perturbed in normal olfactory receptor (OR) gene selection and expression.
Results
OAZ expression is downstream of Mash1 and partly overlaps with Ngn1+ cells
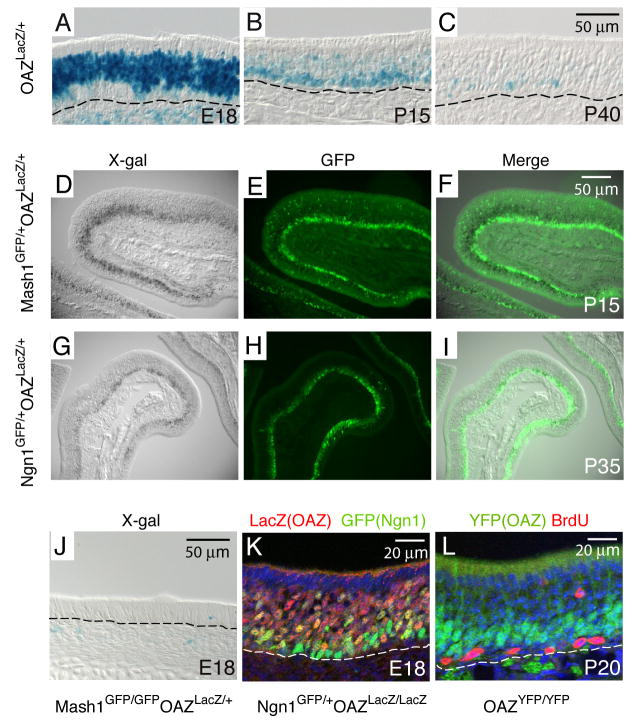
To elucidate the pattern of OAZ expression in OE, we initially utilized an OAZ-lacZ reporter mouse line (OAZlacZ, XB409), derived from a gene-trap screen in ES cells (Stryke et al., 2003). This line contains a β-geo gene-trap vector insertion into the last intron of OAZ gene, resulting in a deletion of nine amino acids from the OAZ C-terminus and expression of an OAZ-lacZ fusion protein. X-gal staining in OAZlacZ/+ mice at E18 revealed a broad OAZ expression in the developing OE (Figure 1A). The expression decreased postnatally and was restricted to the basal OE (Figure 1B, C), the site of continuous olfactory neurogenesis in adults.
Figure 1. OAZ is expressed in newly differentiating ORNs.
(A) In OAZlacZ/+ mice, X-gal positive cells are present in a broad region in the OE at E18. (B, C) At P15 and P40, X-gal positive cells become restricted to the basal OE. The basal lamina is indicated by dashed line. (D-F) OAZ is expressed downstream of Mash1. OAZ-expressing cells were identified by X-gal staining (D) and Mash1-expressing cells were identified with an anti-GFP antibody (E). In the merged picture (F), X-gal positive cells are located above GFP-positive cells. (G-I) In OAZlacZ/+Ngn1GFP/+ mice, X-gal positive OAZ-expressing cells are in the same layer as GFP-positive Ngn1-expressing cells. (J) OAZ-expressing cells are absent in Mash1-null mice. (K) Colocalization of OAZ and Ngn1 in the OE of OAZlacZ/lacZNgn1GFP/+ mice. (L) OAZ is expressed in postmitotic cells in the OE. OAZ-expressing cells were identified in homozygous OAZYFP/YFP mice (see figure 2) with an anti-GFP antibody (green) and proliferating cells were labeled with BrdU (red).
During ORN development, progenitors express the molecular markers Mash1 and Ngn1 at successive stages and migrate apically in the OE (Calof et al., 2002; Cau et al., 2002). To identify the stage at which OAZ is expressed, we introduced Mash1GFP/+ and Ngn1GFP/+ reporter knock-in mice, in which a nuclear GFP reporter precisely replaces the respective coding regions and faithfully mimic expression of the endogenous gene (Leung and Reed, unpublished). In OAZlacZ/+Mash1GFP/+ mice, OAZ-expressing cells were identified with X-gal staining (Figure 1D) and Mash1-expressing cells were labeled with an anti-GFP antibody (Figure 1E). OAZ-expressing cells were located apically to Mash1-expressing cells (Figure 1F), and were essentially absent in E18 Mash1-null mice (Figure 1J), suggesting that OAZ is downstream of Mash1. In OAZlacZ/+Ngn1GFP/+ mice, Ngn1 and OAZ-expressing cells were largely located in the same layer (Figures 1G–I). In cofocal images, Ngn1/OAZ double-labeled cells were predominantly located between the more basal Ngn1-expressing cells and more apical OAZ-expressing cells (Figure 1K), suggesting that OAZ expression may be initiated as cells ceased Ngn1 expression. Furthermore, OAZ was confined to postmitotic cells when examined by BrdU labeling (Figure 1L). Together, these observations indicate that OAZ is expressed in newly differentiating ORNs.
Mature cells are reduced in OAZ−/− mice
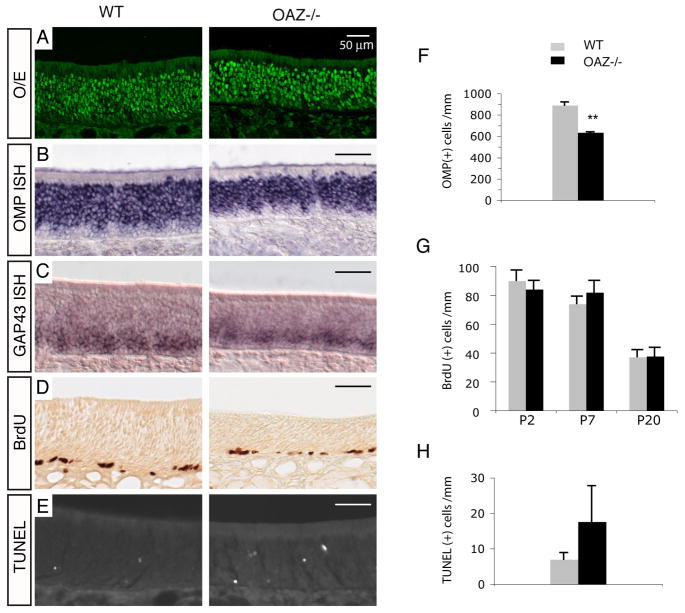
In parallel, we generated a reporter-tagged OAZ-null allele by targeted deletion of the OAZ large exon 4 and insertion of IRES-YFP-polyA. This presumptive functional null allele lacks 1180/1292 amino acids of the native OAZ protein including all the known DNA-binding and protein-binding domains and is hereafter referred to as OAZ−. Homozygous OAZ−/− mice exhibited profound cerebellar defects and died by approximately four weeks. The cerebellar phenotype of other OAZ alleles has been described elsewhere (Alcaraz et al., 2006; Cheng and Reed, 2007; Warming et al., 2006). The olfactory turbinates of OAZ−/− mice display normal organization with a slightly smaller nasal cavity and correspondingly reduced OE area. To examine subsequent stages of ORN development, OE sections (PD 20–25) were examined with the ORN lineage marker O/E, mature ORN marker OMP, and differentiation markers GAP43 and NeuroD1. The pattern of O/E immunostaining in the epithelium from wild-type and OAZ−/− mice were indistinguishable (Figure 2A). However, the number of mature cells visualized by OMP immunostaining was fewer in P20 OAZ−/− mice (data not shown) and, when quantified by in situ hybridization (Figure 2B), was reduced 30% (wt, 890±30/mm; OAZ−/−, 635±15/mm, Figure 2F). In contrast, there were no changes in the expression of GAP43 (Figure 2C) and NeuroD1 (Figure S1C in supplemental data). It is noteworthy that Evi3, an OAZ homolog gene that also interacts with O/E protein (Hentges et al., 2005; Warming et al., 2003), is present at low levels in the OE and displays a pattern of expression similar to OAZ (Figure S2), which may obscure a complete analysis of loss-of-function of OAZ in the OE.
Figure 2. Characterization of OE in OAZ−/− mice.
(A–E) O/E immunostaining, OMP and GAP43 in situ hybridization, BrdU labeling and TUNEL staining in wild-type and OAZ−/− mice. There is a reduction of mature cells and a two-fold increase of apoptotic cells in OAZ−/− mice. (F) Quantification of OMP-positive cells reveals a 30% reduction in OAZ−/− mice (wt, 890±30/mm; OAZ−/−, 635±15/mm; n=3, p<0.001). (G) Quantification of BrdU-positive cells in the OE at P2, P7 and P20 (n=2 mice for each genotype at each age group). (H) Quantification of TUNEL-positive cells in the OE (wt, 7.0±1.9/mm, n=6; OAZ−/−, 17.6±10/mm, n=5, p<0.03). Mice examined were from P20-P25 except that BrdU labeling was conducted at three ages.
The reduction of mature cells could arise from decreased proliferation of progenitors, delayed maturation, and/or reduction of cell lifespan. No significant changes in the number of BrdU-positive cells were observed between wild-type and OAZ−/− mice examined at P2, P7 and P20 (Figure 2D, G). TUNEL staining, however, showed a twofold increase in the number of apoptotic cells in OAZ−/− mice (wt, 7.0±1.9/mm; OAZ−/−, 17.6±10.0/mm, Figure 2E, H) that could account for the decease in mature neurons, but additional effects of OAZ on progression of maturation are possible. The observed OE phenotype was consistent along the anterior to posterior axis and across different dorsal-ventral zones in the OE (Figures S1A, B).
Olfactory axonal projection is impaired in OAZ−/− mice
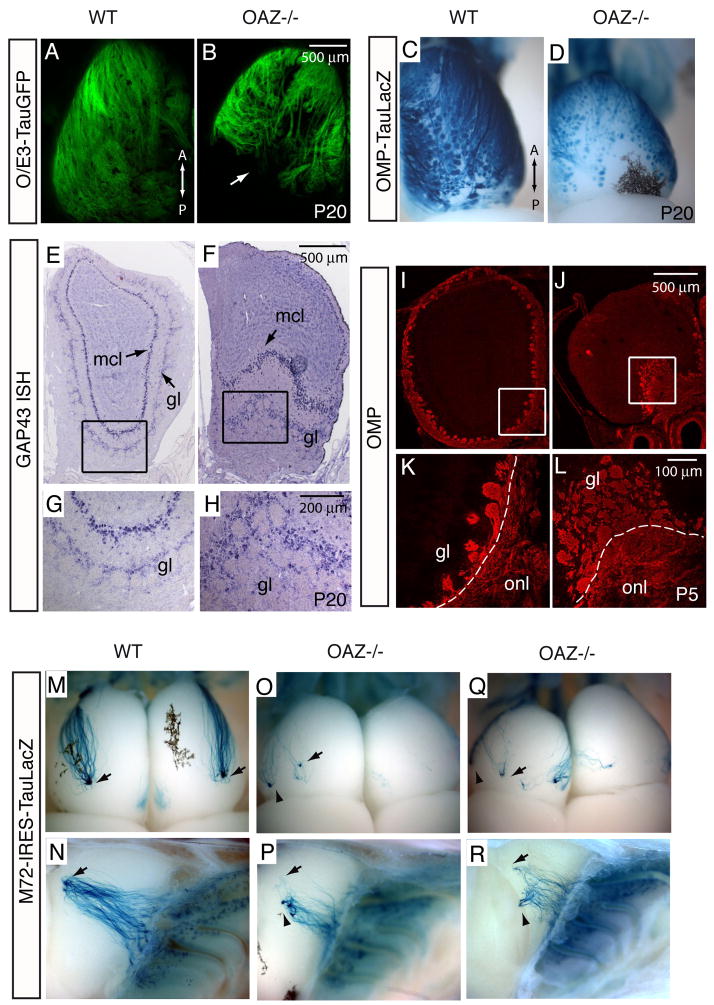
A hallmark of ORN organization is that cells expressing a particular OR gene project their axons to spatially defined glomeruli in the olfactory bulb (OB) (Vassar et al., 1994). The OB in OAZ−/− mice was smaller, a phenotype also observed in other genetic models where primary ORN afferents fail to project or lack odor-evoked activity (Baker et al., 1999; Hirata et al., 2006). We examined ORN projection by introducing the O/E3-tauGFP reporter (Wang et al., 2004). In whole-mount view, ORN axons failed to innervate the dorsal caudal region of the OB in OAZ−/− mice (Figure 3B and Figures S3B–D). In coronal sections, the caudal dorsal OB was essentially devoid of olfactory nerves and glomeruli; instead, additional glomeruli were concentrated on the ventral surface (Figures S3K–M). A similar OB phenotype was observed when projection were visualized with an OMP-taulacZ reporter (Mombaerts et al., 1996) (Figures 3C, D).
Figure 3. OAZ−/− mice exhibit ORN projection defects.
(A–D) The ORN projection to the OB is visualized by O/E3-tauGFP reporter (A, B) and OMP-taulacZ reporter (C, D). In whole-mount dorsal view, ORN axons fail to innervate the caudal OB in OAZ−/− mice (n>6). (E, H) GAP43 in situ hybridization labels mitral cells and periglomerular cells in the OB. In OAZ−/− mice, the dorsal OB surface is devoid of glomeruli. The mitral cell layer (mcl) at the ventral OB is pushed inwardly by the abnormally accumulated glomeruli. (I–L) Labeling of olfactory axons with OMP antibody reveals tangled axons and disorganized glomeruli on the ventral surface in a P5 OAZ−/− mouse. (M–R) Poor convergence and a ventral shift of the M72 glomeruli in OAZ−/− mice. Projection of the M72-expressing axons is visualized by X-gal staining in M72-IRES-taulacZ reporter mice. As M72-expressing neurons converge to one lateral and one medial glomerulus in each bulb in control mice (arrows in M, N), the M72 glomerulus in OAZ−/− mice is shifted ventrally (arrowheads in O-R, n=6) and a subset of axons miss their target. All reporter mice examined were from P20-P25.
We examined the laminar organization of the OB using makers for OB neurons and olfactory axons. In neonatal and adult mice, GAP43 mRNA is detected in mitral cells and periglomerular cells (Verhaagen et al., 1990) (Figure 3E–H). In OAZ−/− mice, the dorsal OB surface was devoid of periglomerular cells and the mitral cell layer was pushed inwardly by the ventrally accumulated glomeruli (Figure 3F, H). Consistently, when olfactory axons were labeled with OMP (Figures 3I–L), the dorsal OB surface lacked an olfactory nerve layer (ONL), but a fibrocellular mass-like structure composed of tangled axons and disorganized glomeruli was observed at the ventral surface (Figure 3L).
We next examined the projection of individual ORNs by introducing reporter-tagged OR alleles (Mombaerts et al., 1996; Zheng et al., 2000). In contrast to two topographically fixed M72 glomeruli in wild-type mice (arrows in Figure 3M, N), multiple M72 projection sites were detected (10/12 OBs), or only wandering axons could be observed (2/12 OBs) in OAZ−/− mice. Interestingly, a subset of M72 neurons projected to a glomerulus located in a similar position as in OAZ wild-type mice (arrows in Figure 3O–R), whereas the remaining majority axons projected to multiple, more ventral glomeruli (arrowheads). The disrupted projection pattern was also observed for the M71-expressing cells using the M71-IRES-TauGFP reporter (control, n=8 mice, OAZ−/−, n=12 mice). ORNs expressing the P2 OR extend much shorter axons and project to a ventral region of the bulb. In OAZ−/− mice, the P2 axons innervate glomeruli that were anteriorly shifted and wandering axons were observed at higher frequency (data not shown).
Gain-of-function: targeted-OAZ expression throughout ORN development
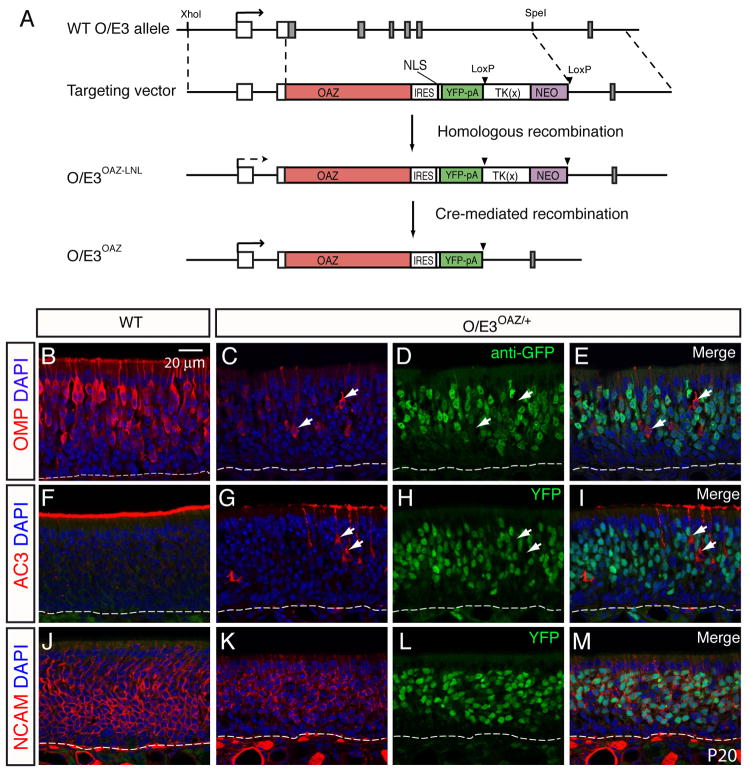
The loss-of-function experiments described above suggest that OAZ is important for the normal program of ORN differentiation, particularly the projection of olfactory axons. The proposed role of OAZ as an inhibitor of O/E transcription factor function (but not O/E protein expression) led us to ask whether sustained OAZ expression was sufficient to arrest ORN development. We designed a dominant gain-of-function knock-in allele that expresses OAZ from the O/E3 locus (Figure 4A). The O/E3 promoter would drive sustained OAZ expression throughout ORN development. An IRES-driven YFP reporter was included to facilitate identification of the O/E3-OAZ expressing cells.
Figure 4. Sustained OAZ expression arrests ORN maturation.
(A) Schematic representation of the OAZ knock-in strategy. Full length OAZ cDNA, followed by IRES-3NLS-YFP-pA and LTNL cassette was inserted in the 5′UTR of the O/E3 gene, replacing the first five exons of O/E3. Transcription initiation sites are indicated by arrow. The LTNL cassette can be deleted by CRE-mediated recombination. (B-M) OMP, AC3 and NCAM staining in wild-type (B, F, J) and O/E3OAZ/+ mice (C, G, K) at P20 revealed a dramatic reduction of mature cells in O/E3OAZ/+ mice. The distribution of OAZ-expressing cells was visualized by either GFP antibody (D) or direct YFP fluorescence (H, L). The merged images (E, I, M) show that OAZ expression does not overlap with mature markers but overlaps with neuronal marker NCAM.
The LTNL cassette used for ES cell selection affects gene expression from the O/E3 promoter. In the initial O/E3OAZ-LTNL mouse line that still carries LTNL, no YFP expression was detected in the OE. However, after CRE-mediated removal of LTNL, heterozygous O/E3OAZ/+ mice could be readily distinguished by their small size and YFP expression (mean weight at P21: wt, 11.1±0.6g, n=7; O/E3OAZ/+, 7.5±0.8g, n=7, P<0.001). We speculate that LTNL blocks a downstream enhancer of the O/E3 promoter, resulting in the absence of OAZ-YFP expression in cassette-containing mice. A similar phenomenon is seen with other insertions at this locus (unpublished data). Since both the O/E3OAZ-LTNL mice and O/E3OAZ/+ mice contain one deleted copy of the O/E3 gene, analysis of O/E3OAZ-LTNL mice (which display a wild-type phenotype) provides strong evidence that the phenotype we observed in O/E3OAZ/+ mice results from expression of OAZ, rather than simply the loss of one copy of O/E3. This special feature also provides for conditional control of OAZ-YFP expression.
Sustained OAZ expression arrests ORN maturation
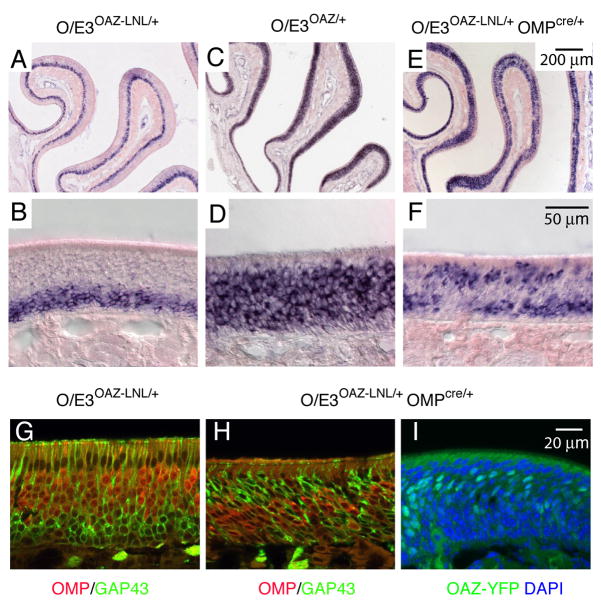
We characterized ORN development in O/E3OAZ/+ mice using mature markers OMP and Adenylyl cyclase 3 (AC3), the general neuronal marker NCAM, and the immature marker GAP43. In O/E3OAZ/+ mice, there was a dramatic reduction in the number of mature ORNs visualized by OMP (Figures 4B, C) and AC3 immunostaining (Figures 4F, G). Intriguingly, there was no overlap between expression of OAZ and the mature cell markers; cells that displayed high levels of OMP or AC3 expression were those that failed to express the OAZ-linked YFP reporter (arrows in Figures 4C–E and G–I). Furthermore, the YFP-positive cells in O/E3OAZ/+ mice expressed NCAM, indicative of a normal neuronal lineage (Figures 4J–M). In situ hybridization revealed that the majority of cells in the epithelium of O/E3OAZ/+ mice were positive for GAP43 (Figures 5C, D, n=6 mice), indicating that the cells were arrested at an immature stage. In contrast, GAP43 expression was restricted to the basal immature cell layer in cassette-containing, phenotypically wild-type mice (Figures 5A, B). Importantly, the number of GAP43 positive cells in O/E3OAZ/+ mice was markedly increased in comparison to wild-type OE.
Figure 5. OAZ expression is sufficient for an immature phenotype.
(A–D) GAP43 in situ hybridization demonstrates that the majority of cells in the epithelium are arrested at an immature stage in O/E3OAZ/+ mice and the absolute number of GAP43-positive cells in the epithelium is increased. (E–F) In O/E3OAZ-LNL/+OMPCRE/+ mice, in which OAZ-YFP is selectively expressed in mature cells, a second population of GAP43-expressing cell is identified at the apical epithelium. (G, H) OMP/GAP43 double labeling in O/E3OAZ-LNL/+OMPCRE/+ mice shows that OMP and GAP43 are regulated in a reciprocal fashion by OAZ; in the apical GAP43-expressing cells, OMP expression is repressed. (I) OAZ-YFP reporter shows the reintroduction of OAZ expression (green) in mature cells in O/E3OAZ-LNL/+OMPCRE/+ mice. Mice examined were from P20-P25.
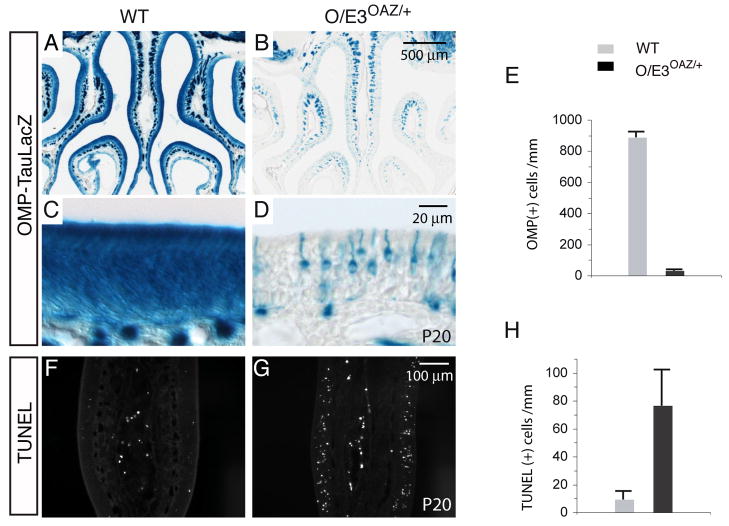
The reduction of mature cells was also confirmed by using an OMP-taulacZ reporter (Figures 6A–D). There was a 95% reduction of OMP-positive cells in O/E3OAZ/+ mice compared with OMP in situ hybridization of wild-type mice (wt, 865±66/mm; O/E3OAZ/+, 47±28/mm, Figure 6E). In parallel, in situ hybridization with several OR gene probes revealed that OR genes were not expressed at high levels in these immature cells (Figure S4). If these cells are blocked from completing the normal differentiation program by the persistent expression of OAZ, one might expect an increase in apoptosis. We observed an 8-fold increase in TUNEL-positive cells in O/E3OAZ/+ mice (Figures 6F, G and H, wt, 9.4±5.4/mm; O/E3OAZ/+, 76.5±25.2/mm). However, these apoptotic cells still represent only a small fraction of the total immature population in the epithelium. Taken together, these experiments indicate that ORN maturation is arrested at an immature neuronal stage and accumulate in greater numbers inO/E3OAZ/+ mice.
Figure 6. Defects of ORN maturation and increased apoptosis in O/E3OAZ/+ mice.
(A–D) X-gal staining of the OMP-taulacZ reporter confirms the reduction of mature cells in O/E3OAZ/+ mice. (E) Quantification of OMP-positive cells in wild-type and O/E3OAZ/+ mice (wt, 865±66/mm; O/E3OAZ/+, 47±28/mm, n=3, P<0.001). (F, G) TUNEL staining shows a dramatic increase of apoptotic cells in the OE of O/E3OAZ/+ mice. (H) Quantification of TUNEL-positive cells (wt, 9.4±5.4/mm, n=3; O/E3OAZ/+, 76.5±25.2/mm, n=3, P<0.01).
OAZ expression is sufficient for an immature ORN phenotype
The OAZ-mediated arrest of ORN maturation is consistent with its ability to block O/E-mediated expression from reporter constructs in heterologous cell lines (Tsai and Reed, 1997). We next asked whether OAZ expression is sufficient for an immature phenotype by utilizing the gain-of-function O/E3OAZ-LNL/+ allele to reintroduce OAZ expression in mature ORNs. Specifically, O/E3OAZ-LNL/+ mice were crossed with mice in which the CRE recombinase is under the control of the OMP promoter (OMP-CRE) (Li et al., 2004). As expected, LTNL cassette removal led to appearance of OAZ-YFP in the mature neuronal layer (Figure 5I). In these mice, GAP43 in situ hybridization revealed a second population of GAP43-expressing cells present in the apical region (Figures 5E, F, n=4) in addition to the basal layer of the endogenous GAP43-expressing cells. This pattern contrasts with the basally-restricted GAP43-positive cells observed in the O/E3OAZ-LNL/+ mice (Figure 5B). Subsequent OMP/GAP43 double-labeling revealed that OMP expression was suppressed in these reactivated GAP43-positive cells (Figures 5G, H). Messenger RNA for ORs and other mature ORN markers examined by in situ hybridization were also reduced (data not shown). Together, these experiments indicate that OAZ expression is sufficient to establish an immature ORN phenotype.
Altered axonal projection and OR gene expression in O/E3OAZ/+ mice
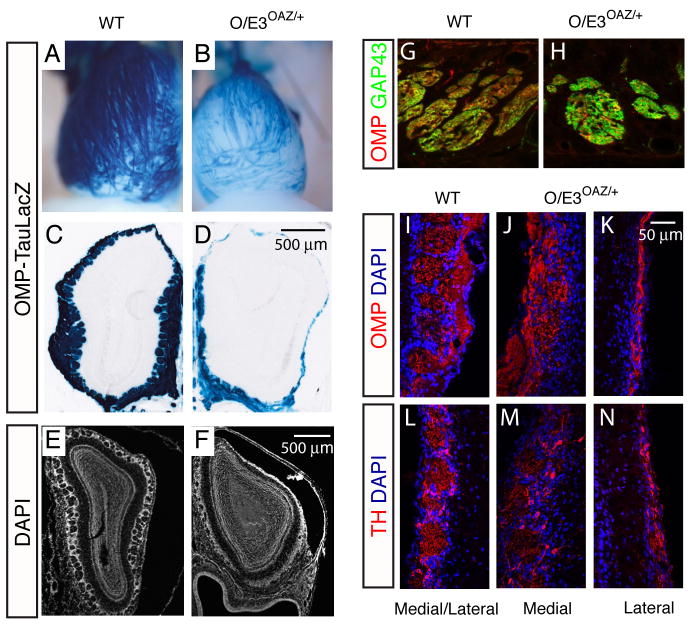
We further characterized the OAZ-mediated arrest of ORN maturation and examined ORN projection using the OMP-tauLacZ reporter. In O/E3OAZ/+ mice, the dorsal OB surface was largely devoid of glomeruli; the few projecting axons reaching the dorsal surface migrated along aberrant paths instead of converging (Figures 7A, B). Coronal sections confirmed the lack of an extensive ONL and glomerular layer at the dorsal and lateral OB surface, while some glomeruli remained at the medial surface (Figures 7C–F). Further analysis showed that even though GAP43-expressing immature cells could initiate their axonal growth in O/E3OAZ/+ mice (Figures 7G, H), the majority failed to innervate the bulb (Figure S5). The medial OB surface contained OMP-positive axons and disorganized glomerular structures, but the lateral surface contained only a thin ONL (Figures 7I–K) that presumable derive from the “escaper” cells. The expression of tyrosine hydroxylase, a marker for periglomerular interneurons, was also reduced at the lateral OB surface consistent with the reduced innervation by ORN axons (Figures 7L–N).
Figure 7. Defects in the ORN projection in O/E3OAZ/+ mice.
(A, B) Whole-mount X-gal staining reveals that the dorsal OB is largely devoid of glomeruli. (C–F) Coronal sections through the OB confirm the reduced ONL and absence of dorsal glomeruli in O/E3OAZ/+ mice. (G, H) OMP/GAP43 double-labeling of axon bundles. In O/E3OAZ/+ mice, axon bundles are GAP43-positive and a small fraction of axons are OMP-positive, suggesting that GAP43-expressing immature cells initiate their axonal growth in O/E3OAZ/+ mice. (I–K) OMP staining demonstrates that the medial OB surface contains OMP-positive olfactory axons and disorganized glomerular structures, while the lateral surface is covered by thin ONL. (L–N) The expression of tyrosine hydroxylase in periglomerular interneurons is greatly reduced at the lateral OB in O/E3OAZ/+ mice. Mice examined were from P20-P25.
We next examined OR-dependent axonal convergence. In O/E3OAZ/+ mice, the projections from both P2 and M72-expressing cells to the OB were greatly reduced. The remaining P2 and M72 projecting axons were unable to converge but spread along the medial surface of the bulb (Figures 8A, B and Figure S6).
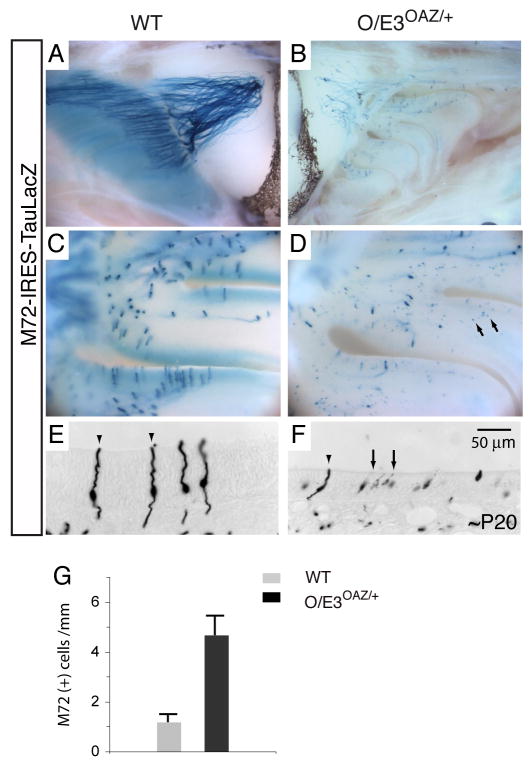
Figure 8. Sustained OAZ expression alters OR gene expression.
(A, B) Whole-mount X-gal staining of M72-IRES-taulacZ reporter. The axons from M72-expressing cells in O/E3OAZ/+ mice do not converge, but project along the dorsal to ventral surface of the OB. (C, D) In the O/E3OAZ/+ epithelium, the strongly stained axon-projecting M72-expressing cells are accompanied by a large number of weakly stained non-projecting cells (arrows). (E, F) In coronal sections, the weakly stained cells (arrows) display short dendrites and no obvious axon extensions. The signal below the basal lamina reflects individual axons stained in cross sections. (G) Quantification of M72-expressing cells in control and O/E3OAZ/+ mice (wt, 1.2±0.3/mm, n=3; O/E3OAZ/+, 4.7±0.7/mm, n=3, P<0.002). Mice examined were from P20-P25.
Interestingly, whole-mount X-gal staining of taulacZ-tagged M72 receptor revealed a large number of weakly stained cells in the epithelium (arrows in Figure 8D). In coronal sections, these weakly stained cells displayed short dendrites and no obvious axon extensions although the faint signal might prevent visualization of these structures. (Figure 8F). Confocal imaging revealed that most of these cells (94%, n=89) were also expressing OAZ-YFP. The level of M72 gene expression in these immature cells, inferred by the intensity of X-gal staining, was much lower than that observed for axon-projecting M72 cells in wild-type mice (arrowheads in Figure 8E) and could not be detected by in situ hybridization (Figure S4). Remarkably, the total number of M72-taulacZ expressing cells was three-fold greater in the epithelium of O/E3OAZ/+ mice than in control littermates (wt, 1.2±0.3/mm, n=3; O/E3OAZ/+, 4.7±0.7/mm, n=3, P<0.002, Figure 8G). The OAZ-mediated arrest of ORN maturation appears to perturb OR expression and alters the number of cells that have chosen a particular receptor for expression.
Discussion
OAZ mediates the transition from differentiation to maturation in ORNs
First identified as a transcriptional partner of O/E proteins, OAZ was also found to mediate BMP-signaling pathways through independent domains of the protein. The loss-of-function and gain-of function experiments described here suggest that in ORN development, OAZ may coordinately regulate the duration of immature gene expression and the onset of mature gene expression (Figure S7). These two developmental processes may arise through distinct O/E and BMP-mediated pathways.
In the OE, OAZ is transiently expressed in newly generated differentiating ORNs. Consistent with its postmitotic expression, OAZ deletion does not affect proliferation of olfactory progenitors. Instead, we observed decreased mature cells, impaired axonal targeting and increased apoptosis in OAZ−/− mice. Similar but less severe axonal targeting defects were also observed in O/E2 and O/E3-null mice (Wang et al., 2004). Therefore, the O/E-OAZ interaction may regulate (directly or indirectly) ORN axonal growth and/or targeting. The overlapping expression of Evi3 and OAZ in the OE may moderate the phenotype in the loss-of-function OAZ-null mice and obscure a critical role for OAZ in the transition from immature to mature ORNs. However, the series of OAZ knock-in experiments described here provide strong evidence that the presence of OAZ can perturb normal developmental timing and alter immature and mature ORN gene expression patterns. In this scenario, the transient OAZ expression during early differentiation contributes to a critical developmental switch during OE neurogenesis.
Consistent with the ability of OAZ to inhibit O/E-mediated transcription in vitro, ORNs that maintain OAZ expression in vivo are arrested at a differentiating stage precisely when the known targets of O/E (OMP, ORs and other olfactory transduction components) are first expressed. However, the re-expression of GAP43 when OAZ is reintroduced into mature cells (O/E3OAZ-LNL/+OMPCRE/+ mice) suggests that OAZ has additional functions beyond its action as an O/E inhibitor. A potential pathway involved in this transcriptional activation is the BMP-signaling pathway. OAZ is normally expressed at a stage when low-doses of BMP4 promotes survival of newly generated NCAM-positive immature cells in olfactory cultures (Shou et al., 2000). It would be interesting to determine whether BMP4 regulates GAP43 expression. Taken together, our experiments support a model in which OAZ may coordinately regulate immature and mature gene expression by integrating extrinsic microenvironmental cues (BMP-pathway) with an intrinsic transcriptional cascade (O/E pathway).
Involvement of OAZ in OR gene selection
The molecular mechanisms controlling OR gene choice and expression remains largely unresolved. While a specific genomic element, the “H region”, is suggested to function as a global enhancer promoting OR expression (Lomvardas et al., 2006; Serizawa et al., 2003), the molecular interactions and proteins that participate in this complex process are unknown. These studies and related experiments on feedback mechanisms in OR gene choice suggest that the OR selection takes place within a defined time window during ORN development (Lewcock and Reed, 2003; Lewcock and Reed, 2004; Mombaerts, 2004). The expression of OAZ during this window and the phenotype observed in O/E3OAZ/+ mice suggest that it may participate in OR selection. The increased number of immature M72-expressing cells in O/E3OAZ/+ mice may derive from neuronal precursors that are arrested in development by sustained OAZ expression and accumulate multiple, activated OR genes in each cell. Unfortunately, the low level of reporter expression in these immature cells has precluded direct examination of this interesting question. Alternatively, maintained OAZ expression could promote differential selection of particular OR gene family members. The O/E3OAZ/+ mice may also facilitate the study of OR selection by providing large numbers of genetically-tagged differentiating cells arrested in development.
ORNs in O/E3OAZ/+ mice escape the gain-of-function phenotype at low frequency
The dramatic reduction of mature ORNs in O/E3OAZ/+ mice is accompanied by a fraction of cells (~5% - “escapers”) that mature and express OMP and AC3. Double-label experiments reveal that the escaper cells do not express the reporter-tagged OE3-OAZ allele. The arrest phenotype associated with the OE3-OAZ allele is dominant and individual cells that silence this allele epigenetically or by loss-of-heterozygosity (LOH) during mitotic recombination will have a distinct survival advantage. The inability of these escapers to converge to discrete glomeruli may reflect their abnormal developmental history, altered OR expression or the reduced number of cells of each OR type in the epithelium.
OAZ knock-in functions a dominant O/E suppressor
The O/E transcription factors are involved in many developmental processes (Corradi et al., 2003; Garel et al., 1999; Garel et al., 2002; Kieslinger et al., 2005; Lin and Grosschedl, 1995). The redundant expression of different O/E family members has limited our ability to study these processes by conventional gene-targeted inactivation. For example, disruption of EBF1, the founding member of this family, leads to profound developmental defects in B cells and striatum tissues that express only this member of the O/E family (Garel et al., 1999; Lin and Grosschedl, 1995), but its loss results in modest, if any, phenotype in the OE where it is highly expressed. Similarly, O/E2 and O/E3 knockout mice as well as O/E2O/E3 double heterozygous mice displayed modest defects of ORN projection suggesting complex interactions and considerable functional redundancy with the additional O/E family members expressed in the OE (Wang et al., 2004). The ability of the OAZ protein to inhibit the function of all O/E family members in vitro (Tsai and Reed, 1997) and the expression of the O/E3-OAZ allele in all olfactory neuronal progenitors provides powerful tools to functionally disrupt all O/E function in this tissue. The abundant, persistent expression of the O/E3 locus in mature olfactory neurons may make these cells particular susceptible to this dominant suppression.
Experimental Procedures
Mouse Genetics
The generation of OAZlacZ and OAZ-null mice, and their phenotype outside of the olfactory system are described elsewhere (Cheng and Reed, 2007; see supplemental methods). The OAZ knock-in was created from the O/E3 knock-out targeting vector (Wang et al., 2004) and modified by inserting the full-length OAZ cDNA 261 base pairs before the initiation codon for O/E3. The OAZ cDNA was followed by an IRES-3NLS-YFP-pA cassette and LoxP-TK(Δ)-Neo-LoxP (LTNL) cassette. The OR reporter mice, OMP-taulacZ and OMP-CRE mice were obtained from Dr. P. Mombaerts. The sequences of primers for genotyping each of the lines are available from the authors.
X-gal Staining, Immunolocalization and in situ Hybridization
The X-gal staining (Mombaerts et al., 1996) and immunohistochemistry was performed on PFA-fixed cryostat sections. The primary antibodies were: AC3 (rabbit, 1:1000; Santa Cruz Biotechnology), GAP43 (rabbit, 1:200, Chemicon), GFP (rabbit, 1:1000, Molecular Probes), LacZ (mouse, 1:500, Promega), O/E (rabbit, 1:1000), OMP (goat, 1: 3000; provided by Dr. F. Margolis), Tyrosine hydroxylase (rabbit, 1:500; Chemicon), NCAM (mouse, 1:1000; Sigma). Alexa 488- or Alexa 546-labled secondary antibodies were used at 1:1000 dilution. For double staining of X-gal and GFP, sections were stained with X-gal staining solution overnight, fixed with 4% PFA and then followed by normal antibody staining procedure using the anti-GFP antibody.
In situ hybridization was performed on PFA-fixed sections following a standard protocol (Wang et al., 2004). The probes were: I7 (NM_010983), M4 (NM_146937) and M72 (NM_030553) coding sequence, GAP43 (nucleotides 147 to 860 of NM_008083), NeuroD1 (nucleotides 640 to 1192 of BC018241), Evi3 (nucleotides 1 to 1172 of BC021376) and OMP (NM_011010) partial cDNA. Images were obtained with a Zeiss Axioplan microscope or a Zeiss LSM 510 confocal laser-scanning microscope.
BrdU Labeling and TUNEL Staining
Adult mice or pregnant females were injected intraperitoneally with BrdU (Sigma) 50μg/g body weight 30min before they were sacrificed. PFA-fixed sections were incubated with 3N HCl for 30min before immunostaining with anti-BrdU antibody (rat, 1:100; Abcam). To visualize, either an Alexa 546-labled secondary antibody or ABC kit (vector) and FAST DAB tablet sets (Sigma) were used. For double labeling of BrdU with anti-GFP antibody, sections were incubated with anti-GFP antibody first, then washed and fixed with 4% PFA before proceeding with the acid treatment. The secondary antibody incubations were performed simultaneously. TUNEL staining was performed using Apoptag® in situ apoptosis detection kits (Chemicon) according to the manufacturer’s instruction.
Cell Counting in Epithelium and Statistics
OMP counting was performed on images taken at 20× magnification (4 images at matched locations for each animal, the total length of OE was approximately 5mm). The total number of BrdU, TUNEL and specific OR cells were counted under the microscope from overlapping 20× fields for the entire OE region on matched sections (4–5 sections for each animal, the total length of OE sampled was from 85mm to 160mm). OE length was determined by tracing the outline of the epithelium basal lamina using Axiovision® software. The data is represented as mean±s.d. and two-tailed Student’s t-test was used for statistical analysis.
Supplementary Material
Acknowledgments
We thank Dr. P. Mombaerts for the OR-reporter mice, OMP-lacZ and OMP-CRE mice (Rockefeller University); Dr. F. Margolis for the OMP antibody (University of Maryland); Dr. S. Warming for OAZ cDNA (National Cancer Institute). We thank members of the Reed lab and Dr. J. Nathans for helpful discussions. This work was supported by the NIDCD and the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alcaraz WA, Gold DA, Raponi E, Gent PM, Concepcion D, Hamilton BA. Zfp423 controls proliferation and differentiation of neural precursors in cerebellar vermis formation. Proc Natl Acad Sci U S A. 2006 doi: 10.1073/pnas.0609184103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakalyar HA, Reed RR. Identification of a specialized adenylyl cyclase that may mediate odorant detection. Science. 1990;250:1403–1406. doi: 10.1126/science.2255909. [DOI] [PubMed] [Google Scholar]
- Baker H, Cummings DM, Munger SD, Margolis JW, Franzen L, Reed RR, Margolis FL. Targeted deletion of a cyclic nucleotide-gated channel subunit (OCNC1): biochemical and morphological consequences in adult mice. J Neurosci. 1999;19:9313–9321. doi: 10.1523/JNEUROSCI.19-21-09313.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet LJ, Gold GH, Ngai J. General anosmia caused by a targeted disruption of the mouse olfactory cyclic nucleotide-gated cation channel. Neuron. 1996;17:681–693. doi: 10.1016/s0896-6273(00)80200-7. [DOI] [PubMed] [Google Scholar]
- Calof AL, Chikaraishi DM. Analysis of neurogenesis in a mammalian neuroepithelium: proliferation and differentiation of an olfactory neuron precursor in vitro. Neuron. 1989;3:115–127. doi: 10.1016/0896-6273(89)90120-7. [DOI] [PubMed] [Google Scholar]
- Calof AL, Bonnin A, Crocker C, Kawauchi S, Murray RC, Shou J, Wu HH. Progenitor cells of the olfactory receptor neuron lineage. Microsc Res Tech. 2002;58:176–188. doi: 10.1002/jemt.10147. [DOI] [PubMed] [Google Scholar]
- Cau E, Casarosa S, Guillemot F. Mash1 and Ngn1 control distinct steps of determination and differentiation in the olfactory sensory neuron lineage. Development. 2002;129:1871–1880. doi: 10.1242/dev.129.8.1871. [DOI] [PubMed] [Google Scholar]
- Cheng LE, Reed RR. The transcription factor Zfp423/OAZ is required for cerebellar dvelopment and CNS midline patterning. Dev Biol. 2007 doi: 10.1016/j.ydbio.2007.04.005. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corradi A, Croci L, Broccoli V, Zecchini S, Previtali S, Wurst W, Amadio S, Maggi R, Quattrini A, Consalez GG. Hypogonadotropic hypogonadism and peripheral neuropathy in Ebf2-null mice. Development. 2003;130:401–410. doi: 10.1242/dev.00215. [DOI] [PubMed] [Google Scholar]
- Dewulf N, Verschueren K, Lonnoy O, Moren A, Grimsby S, Vande Spiegle K, Miyazono K, Huylebroeck D, Ten Dijke P. Distinct spatial and temporal expression patterns of two type I receptors for bone morphogenetic proteins during mouse embryogenesis. Endocrinology. 1995;136:2652–2663. doi: 10.1210/endo.136.6.7750489. [DOI] [PubMed] [Google Scholar]
- Garel S, Marin F, Grosschedl R, Charnay P. Ebf1 controls early cell differentiation in the embryonic striatum. Development. 1999;126:5285–5294. doi: 10.1242/dev.126.23.5285. [DOI] [PubMed] [Google Scholar]
- Garel S, Yun K, Grosschedl R, Rubenstein JL. The early topography of thalamocortical projections is shifted in Ebf1 and Dlx1/2 mutant mice. Development. 2002;129:5621–5634. doi: 10.1242/dev.00166. [DOI] [PubMed] [Google Scholar]
- Hata A, Seoane J, Lagna G, Montalvo E, Hemmati-Brivanlou A, Massague J. OAZ uses distinct DNA- and protein-binding zinc fingers in separate BMP-Smad and Olf signaling pathways. Cell. 2000;100:229–240. doi: 10.1016/s0092-8674(00)81561-5. [DOI] [PubMed] [Google Scholar]
- Hentges KE, Weiser KC, Schountz T, Woodward LS, Morse HC, Justice MJ. Evi3, a zinc-finger protein related to EBFAZ, regulates EBF activity in B-cell leukemia. Oncogene. 2005;24:1220–1230. doi: 10.1038/sj.onc.1208243. [DOI] [PubMed] [Google Scholar]
- Hirata T, Nakazawa M, Yoshihara S, Miyachi H, Kitamura K, Yoshihara Y, Hibi M. Zinc-finger gene Fez in the olfactory sensory neurons regulates development of the olfactory bulb non-cell-autonomously. Development. 2006;133:1433–1443. doi: 10.1242/dev.02329. [DOI] [PubMed] [Google Scholar]
- Jones DT, Reed RR. Golf: an olfactory neuron specific-G protein involved in odorant signal transduction. Science. 1989;244:790–795. doi: 10.1126/science.2499043. [DOI] [PubMed] [Google Scholar]
- Kawauchi S, Beites CL, Crocker CE, Wu HH, Bonnin A, Murray R, Calof AL. Molecular signals regulating proliferation of stem and progenitor cells in mouse olfactory epithelium. Dev Neurosci. 2004;26:166–180. doi: 10.1159/000082135. [DOI] [PubMed] [Google Scholar]
- Kawauchi S, Shou J, Santos R, Hebert JM, McConnell SK, Mason I, Calof AL. Fgf8 expression defines a morphogenetic center required for olfactory neurogenesis and nasal cavity development in the mouse. Development. 2005;132:5211–5223. doi: 10.1242/dev.02143. [DOI] [PubMed] [Google Scholar]
- Kieslinger M, Folberth S, Dobreva G, Dorn T, Croci L, Erben R, Consalez GG, Grosschedl R. EBF2 regulates osteoblast-dependent differentiation of osteoclasts. Dev Cell. 2005;9:757–767. doi: 10.1016/j.devcel.2005.10.009. [DOI] [PubMed] [Google Scholar]
- Ku M, Howard S, Ni W, Lagna G, Hata A. OAZ Regulates Bone Morphogenetic Protein Signaling through Smad6 Activation. J Biol Chem. 2006;281:5277–5287. doi: 10.1074/jbc.M510004200. [DOI] [PubMed] [Google Scholar]
- Lewcock JW, Reed RR. Neuroscience. ORs rule the roost in the olfactory system. Science. 2003;302:2078–2079. doi: 10.1126/science.1093397. [DOI] [PubMed] [Google Scholar]
- Lewcock JW, Reed RR. A feedback mechanism regulates monoallelic odorant receptor expression. Proc Natl Acad Sci U S A. 2004;101:1069–1074. doi: 10.1073/pnas.0307986100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Ishii T, Feinstein P, Mombaerts P. Odorant receptor gene choice is reset by nuclear transfer from mouse olfactory sensory neurons. Nature. 2004;428:393–399. doi: 10.1038/nature02433. [DOI] [PubMed] [Google Scholar]
- Lin H, Grosschedl R. Failure of B-cell differentiation in mice lacking the transcription factor EBF. Nature. 1995;376:263–267. doi: 10.1038/376263a0. [DOI] [PubMed] [Google Scholar]
- Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126:403–413. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- Mombaerts P, Wang F, Dulac C, Chao SK, Nemes A, Mendelsohn M, Edmondson J, Axel R. Visualizing an olfactory sensory map. Cell. 1996;87:675–686. doi: 10.1016/s0092-8674(00)81387-2. [DOI] [PubMed] [Google Scholar]
- Mombaerts P. Odorant receptor gene choice in olfactory sensory neurons: the one receptor-one neuron hypothesis revisited. Curr Opin Neurobiol. 2004;14:31–36. doi: 10.1016/j.conb.2004.01.014. [DOI] [PubMed] [Google Scholar]
- Nicolay DJ, Doucette JR, Nazarali AJ. Transcriptional regulation of neurogenesis in the olfactory epithelium. Cell Mol Neurobiol. 2006;26:801–819. doi: 10.1007/s10571-006-9058-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serizawa S, Miyamichi K, Nakatani H, Suzuki M, Saito M, Yoshihara Y, Sakano H. Negative feedback regulation ensures the one receptor-one olfactory neuron rule in mouse. Science. 2003;302:2088–2094. doi: 10.1126/science.1089122. [DOI] [PubMed] [Google Scholar]
- Shim S, Bae N, Han JK. Bone morphogenetic protein-4-induced activation of Xretpos is mediated by Smads and Olf-1/EBF associated zinc finger (OAZ) Nucleic Acids Res. 2002;30:3107–3117. doi: 10.1093/nar/gkf437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shou J, Rim PC, Calof AL. BMPs inhibit neurogenesis by a mechanism involving degradation of a transcription factor. Nat Neurosci. 1999;2:339–345. doi: 10.1038/7251. [DOI] [PubMed] [Google Scholar]
- Shou J, Murray RC, Rim PC, Calof AL. Opposing effects of bone morphogenetic proteins on neuron production and survival in the olfactory receptor neuron lineage. Development. 2000;127:5403–5413. doi: 10.1242/dev.127.24.5403. [DOI] [PubMed] [Google Scholar]
- Stryke D, Kawamoto M, Huang CC, Johns SJ, King LA, Harper CA, Meng EC, Lee RE, Yee A, L’Italien L, et al. BayGenomics: a resource of insertional mutations in mouse embryonic stem cells. Nucleic Acids Res. 2003;31:278–281. doi: 10.1093/nar/gkg064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai RY, Reed RR. Cloning and functional characterization of Roaz, a zinc finger protein that interacts with O/E-1 to regulate gene expression: implications for olfactory neuronal development. J Neurosci. 1997;17:4159–4169. doi: 10.1523/JNEUROSCI.17-11-04159.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai RY, Reed RR. Identification of DNA recognition sequences and protein interaction domains of the multiple-Zn-finger protein Roaz. Mol Cell Biol. 1998;18:6447–6456. doi: 10.1128/mcb.18.11.6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassar R, Chao SK, Sitcheran R, Nunez JM, Vosshall LB, Axel R. Topographic organization of sensory projections to the olfactory bulb. Cell. 1994;79:981–991. doi: 10.1016/0092-8674(94)90029-9. [DOI] [PubMed] [Google Scholar]
- Verhaagen J, Greer CA, Margolis FL. B-50/GAP43 Gene Expression in the Rat Olfactory System During Postnatal Development and Aging. Eur J Neurosci. 1990;2:397–407. doi: 10.1111/j.1460-9568.1990.tb00432.x. [DOI] [PubMed] [Google Scholar]
- Wang MM, Reed RR. Molecular cloning of the olfactory neuronal transcription factor Olf-1 by genetic selection in yeast. Nature. 1993;364:121–126. doi: 10.1038/364121a0. [DOI] [PubMed] [Google Scholar]
- Wang MM, Tsai RY, Schrader KA, Reed RR. Genes encoding components of the olfactory signal transduction cascade contain a DNA binding site that may direct neuronal expression. Mol Cell Biol. 1993;13:5805–5813. doi: 10.1128/mcb.13.9.5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SS, Tsai RY, Reed RR. The characterization of the Olf-1/EBF-like HLH transcription factor family: implications in olfactory gene regulation and neuronal development. J Neurosci. 1997;17:4149–4158. doi: 10.1523/JNEUROSCI.17-11-04149.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SS, Lewcock JW, Feinstein P, Mombaerts P, Reed RR. Genetic disruptions of O/E2 and O/E3 genes reveal involvement in olfactory receptor neuron projection. Development. 2004;131:1377–1388. doi: 10.1242/dev.01009. [DOI] [PubMed] [Google Scholar]
- Warming S, Liu P, Suzuki T, Akagi K, Lindtner S, Pavlakis GN, Jenkins NA, Copeland NG. Evi3, a common retroviral integration site in murine B-cell lymphoma, encodes an EBFAZ-related Kruppel-like zinc finger protein. Blood. 2003;101:1934–1940. doi: 10.1182/blood-2002-08-2652. [DOI] [PubMed] [Google Scholar]
- Warming S, Rachel RA, Jenkins NA, Copeland NG. Zfp423 is required for normal cerebellar development. Mol Cell Biol. 2006;26:6913–6922. doi: 10.1128/MCB.02255-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HH, Ivkovic S, Murray RC, Jaramillo S, Lyons KM, Johnson JE, Calof AL. Autoregulation of neurogenesis by GDF11. Neuron. 2003;37:197–207. doi: 10.1016/s0896-6273(02)01172-8. [DOI] [PubMed] [Google Scholar]
- Zhang D, Mehler MF, Song Q, Kessler JA. Development of bone morphogenetic protein receptors in the nervous system and possible roles in regulating trkC expression. J Neurosci. 1998;18:3314–3326. doi: 10.1523/JNEUROSCI.18-09-03314.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng C, Feinstein P, Bozza T, Rodriguez I, Mombaerts P. Peripheral olfactory projections are differentially affected in mice deficient in a cyclic nucleotide-gated channel subunit. Neuron. 2000;26:81–91. doi: 10.1016/s0896-6273(00)81140-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.