Abstract
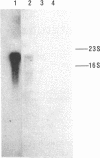
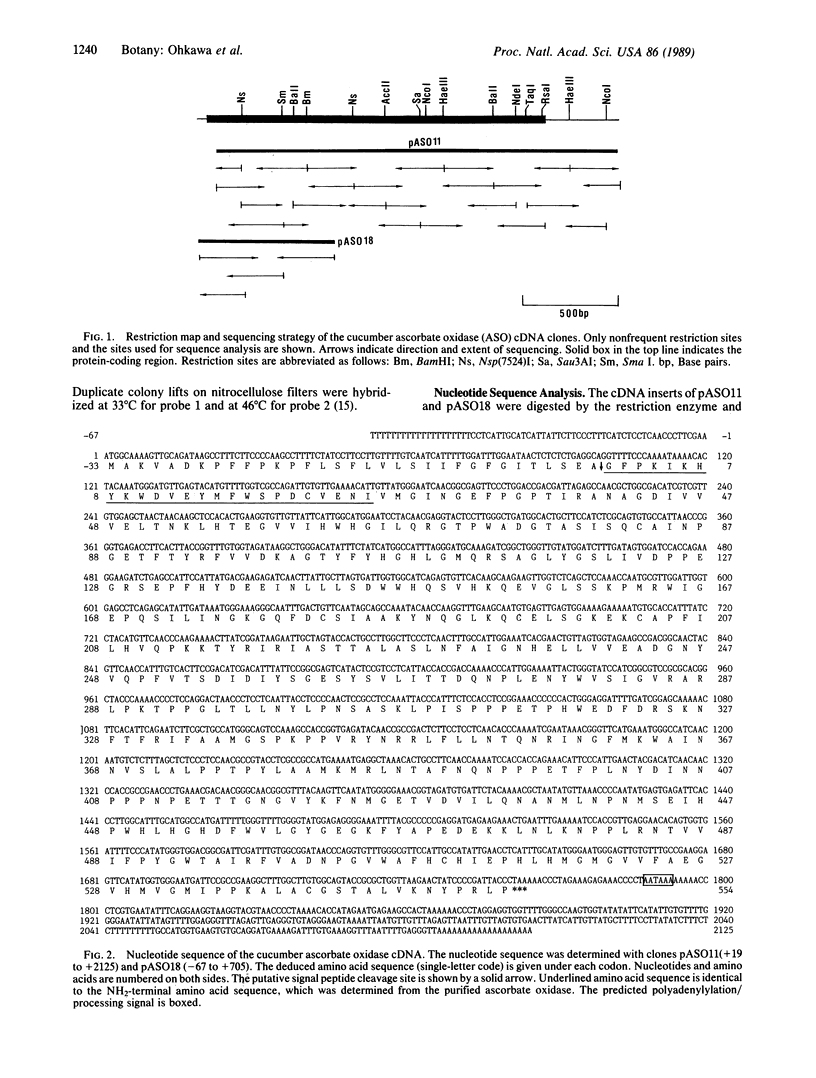
cDNA clones for ascorbate oxidase were isolated from a cDNA library made from cucumber (Cucumis sativus) fruit mRNA. The library was screened with synthetic oligonucleotides that encode the NH2-terminal sequence of this enzyme. Nucleotide sequence analysis of the cloned cDNA inserts revealed a 1761-base-pair open reading frame that encoded an NH2-terminal signal peptide of 33 amino acids and a mature enzyme of 554 amino acids (Mr, 62,258). The amino acid sequence deduced from nucleotide sequence analysis agrees with the NH2-terminal amino acid sequence of the purified ascorbate oxidase, as determined by microsequencing methods. Cucumber ascorbate oxidase contained four histidine-rich regions with striking sequence homology to the corresponding parts of the other multicopper oxidases such as Neurospora crassa laccase and human ceruloplasmin and, to some extent, to a low molecular weight copper protein such as plastocyanin. Moreover, these data further support the hypothesis that the small blue copper proteins and the multicopper oxidases have evolved from the same ancestral gene. By RNA blot hybridization analysis, the mRNA for the ascorbate oxidase was found to be abundant in cucumber fruit tissue while expressed at very low levels in leaf and root tissues.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Fujiyama K., Takemura H., Shibayama S., Kobayashi K., Choi J. K., Shinmyo A., Takano M., Yamada Y., Okada H. Structure of the horseradish peroxidase isozyme C genes. Eur J Biochem. 1988 May 2;173(3):681–687. doi: 10.1111/j.1432-1033.1988.tb14052.x. [DOI] [PubMed] [Google Scholar]
- Germann U. A., Lerch K. Isolation and partial nucleotide sequence of the laccase gene from Neurospora crassa: amino acid sequence homology of the protein to human ceruloplasmin. Proc Natl Acad Sci U S A. 1986 Dec;83(23):8854–8858. doi: 10.1073/pnas.83.23.8854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germann U. A., Müller G., Hunziker P. E., Lerch K. Characterization of two allelic forms of Neurospora crassa laccase. Amino- and carboxyl-terminal processing of a precursor. J Biol Chem. 1988 Jan 15;263(2):885–896. [PubMed] [Google Scholar]
- Gubler U., Hoffman B. J. A simple and very efficient method for generating cDNA libraries. Gene. 1983 Nov;25(2-3):263–269. doi: 10.1016/0378-1119(83)90230-5. [DOI] [PubMed] [Google Scholar]
- Koschinsky M. L., Funk W. D., van Oost B. A., MacGillivray R. T. Complete cDNA sequence of human preceruloplasmin. Proc Natl Acad Sci U S A. 1986 Jul;83(14):5086–5090. doi: 10.1073/pnas.83.14.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res. 1984 Jan 25;12(2):857–872. doi: 10.1093/nar/12.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagrimini L. M., Burkhart W., Moyer M., Rothstein S. Molecular cloning of complementary DNA encoding the lignin-forming peroxidase from tobacco: Molecular analysis and tissue-specific expression. Proc Natl Acad Sci U S A. 1987 Nov;84(21):7542–7546. doi: 10.1073/pnas.84.21.7542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkin R., Malmström B. G. The state and function of copper in biological systems. Adv Enzymol Relat Areas Mol Biol. 1970;33:177–244. doi: 10.1002/9780470122785.ch4. [DOI] [PubMed] [Google Scholar]
- Marchesini A., Kroneck P. M. Ascorbate oxidase from Cucurbita pepo medullosa. New method of purification and reinvestigation of properties. Eur J Biochem. 1979 Nov 1;101(1):65–76. doi: 10.1111/j.1432-1033.1979.tb04217.x. [DOI] [PubMed] [Google Scholar]
- Messerschmidt A., Rossi A., Ladenstein R., Huber R., Bolognesi M., Marchesini A., Petruzelli R., Finazzi-Agro A. Preliminary X-ray crystal structure and partial cDNA-sequence of ascorbate oxidase from zucchini. Prog Clin Biol Res. 1988;274:285–288. [PubMed] [Google Scholar]
- Nakamura T., Makino N., Ogura Y. Purification and properties of ascorbate oxidase from cucumber. J Biochem. 1968 Aug;64(2):189–195. doi: 10.1093/oxfordjournals.jbchem.a128879. [DOI] [PubMed] [Google Scholar]
- Nawa H., Hirose T., Takashima H., Inayama S., Nakanishi S. Nucleotide sequences of cloned cDNAs for two types of bovine brain substance P precursor. Nature. 1983 Nov 3;306(5938):32–36. doi: 10.1038/306032a0. [DOI] [PubMed] [Google Scholar]
- STARK G. R., DAWSON C. R. On the accessibility of sulfhydryl groups in ascorbic acid oxidase. J Biol Chem. 1962 Mar;237:712–716. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N., Ortel T. L., Putnam F. W. Single-chain structure of human ceruloplasmin: the complete amino acid sequence of the whole molecule. Proc Natl Acad Sci U S A. 1984 Jan;81(2):390–394. doi: 10.1073/pnas.81.2.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson M. E. Compilation of published signal sequences. Nucleic Acids Res. 1984 Jul 11;12(13):5145–5164. doi: 10.1093/nar/12.13.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. Patterns of amino acids near signal-sequence cleavage sites. Eur J Biochem. 1983 Jun 1;133(1):17–21. doi: 10.1111/j.1432-1033.1983.tb07424.x. [DOI] [PubMed] [Google Scholar]
- von Heijne G. Signal sequences are not uniformly hydrophobic. J Mol Biol. 1982 Aug 15;159(3):537–541. doi: 10.1016/0022-2836(82)90300-x. [DOI] [PubMed] [Google Scholar]