Abstract
Purpose
DNA methyltransferase-3B (DNMT3B) plays an important role in de novo CpG island methylation. Dnmt3b can induce colon tumor in mice with methylation in specific CpG islands. We hypothesized that cellular DNMT3B level might influence the occurrence of widespread CpG island methylation (i.e., the CpG island methylator phenotype, CIMP) in colon cancer.
Experimental Design
Utilizing 765 colorectal cancers in two cohort studies, we detected DNMT3B expression in 116 (15%) tumors by immunohistochemistry. We assessed microsatellite instability, quantified DNA methylation in repetitive long interspersed nucleotide element-1 (LINE-1) by Pyrosequencing, eight CIMP-specific promoters [CACNA1G, CDKN2A (p16), CRABP1, IGF2, MLH1, NEUROG1, RUNX3, and SOCS1], and eight other CpG islands (CHFR, HIC1, IGFBP3, MGMT, MINT1, MINT31, p14, and WRN) by real-time PCR (MethyLight).
Results
Tumoral DNMT3B overexpression was significantly associated with CIMP-high [≥6/8 methylated CIMP-specific promoters; odds ratio (OR), 3.34; 95% confidence interval, 2.11-5.29; P < 0.0001]. The relations between DNMT3B and methylation in 16 individual CpG islands varied substantially (OR, 0.80-2.96), suggesting variable locus-to-locus specificities of DNMT3B activity. DNMT3B expression was not significantly related with LINE-1 hypomethylation. In multivariate logistic regression, the significant relation between DNMT3B and CIMP-high persisted (OR, 2.39; 95% confidence interval, 1.11-5.14; P = 0.026) after adjusting for clinical and other molecular features, including p53, β-catenin, LINE-1, microsatellite instability, KRAS, PIK3CA,and BRAF. DNMT3B expression was unrelated with patient outcome, survival, or prognosis.
Conclusions
Tumoral DNMT3B overexpression is associated with CIMP-high in colorectal cancer. Our data support a possible role of DNMT3B in nonrandom de novo CpG island methylation leading to colorectal cancer.
Epigenetic aberrations are important mechanisms in carcinogenesis, and many tumor suppressor genes can be silenced by promoter CpG island methylation (1). Aberrant de novo CpG island methylation has been linked to DNA methyltransferase-3B (DNMT3B; refs. 2–7). DNMT3B plays an important role in the establishment and maintenance of genomic methylation patterns (2–5, 8, 9). Dnmt3b overexpression in mice can induce colon tumor with a nonrandom CpG island methylation pattern, suggesting some degree of locus specificity of Dnmt3b activity (7). DNMT3B is overexpressed in various human cancers (6, 10–12), including colorectal cancer (13), and inhibition of DNMT3B causes demethylation of CpG islands in cancer cells (10, 14–16). Certain DNMT3B polymorphisms have been associated with decreased CpG island methylation in ovarian cancer (17) and normal colonic mucosa (18), and increased risks of lung and colorectal cancers (19, 20). These data collectively suggest a pathogenetic link between DNMT3B, CpG island methylation, and cancer. DNMT3B is a potential target of chemotherapy and chemoprevention by DNMT inhibitors (1, 2).
Translational Relevance.
Causes of DNA methylation at specific CpG islands in cancer are enigmatic and probably multifactorial. In this study, we provide compelling evidence for the association between DNMT3B expression and methylation in specific promoter CpG islands in colorectal cancer. Together with previous experimental data, our findings suggest that DNMT3B may contribute to the occurrence of widespread CpG island methylation, which has been known as the CpG island methylator phenotype (CIMP). CIMP has been established as a unique phenotype in colorectal cancer, associated with distinct clinical, pathologic, and molecular features. Our findings are likely clinically important because DNMT inhibitors are promising chemopreventive and chemotherapeutic agents, and a clinical indication of DNMT inhibitors may depend on CIMP status of colorectal neoplasia.
The CpG island methylator phenotype (CIMP) in colorectal cancer is characterized by a widespread CpG island methylation (21–23), and is associated with older age, female gender, proximal tumor location, BRAF mutation, microsatellite instability (MSI), wild-type TP53, and high global DNA methylation level (24–28). However, the cause of widespread promoter CpG island methylation is currently unknown. Considering the role of DNMT3B in de novo methylation, we hypothesized that DNMT3B expression may influence the occurrence of CIMP in colorectal cancer.
In this study, we examined the relation between tumoral DNMT3B expression and CIMP in a large number of colorectal cancers (n = 765). Our findings of the significant relation between DNMT3B and CIMP support a potential role of DNMT3B in de novo CpG island methylation in carcinogenic process.
Materials and Methods
Study group
We used the databases of two large prospective cohort studies; the Nurses’ Health Study (n = 121,700 women followed since 1976; ref. 29), and the Health Professional Follow-up Study (n = 51,500 men followed since 1986; ref. 29). A subset of the cohort participants developed colorectal cancers during prospective follow-up. Thus, these colorectal cancers represented population-based, relatively unbiased samples (compared with retrospective or single hospital–based samples). Previous studies on the Nurses’ Health Study and Health Professionals Follow-up Study have described baseline characteristics of cohort participants and incident colorectal cancer cases, and confirmed that our colorectal cancers were well representative as a population-based sample (29). We collected paraffin-embedded tissue blocks from hospitals in which cohort participants with colorectal cancers had undergone resections of primary tumors. Based on the availability of adequate tissue specimens, a total of 765 colorectal cancers (diagnosed up to June 30, 2002) were included. Among our cohort studies, there was no significant difference in demographic features between cases with tissue available and those without available tissue (29). Many of the cases have been previously characterized for CIMP, MSI, p53, KRAS, and BRAF (28, 30). However, we have not examined DNMT3B expression in our tumors or the relationship between DNMT3B and other molecular events. Informed consent was obtained from all study subjects. Tissue collection and analyses were approved by the Harvard School of Public Health and Brigham and Women's Hospital Institutional Review Boards.
Measurement of mortality
Patients were observed until death or June 30, 2006, whichever came first. Ascertainment of deaths included reporting by the family or postal authorities. In addition, the names of persistent nonresponders were searched in the National Death Index. In rare patients who died as a result of colorectal cancer not previously reported, we obtained medical records with permission from next of kin. More than 98% of deaths in the cohorts were identified by these methods.
Histopathologic evaluations
H&E-stained tissue sections were examined by a pathologist (S. Ogino) unaware of other data. The tumor grade was categorized as low (≥50% gland formation) versus high (<50% gland formation). The presence and extent of extracellular mucin were categorized as 0% (no mucin), 1% to 49%, or ≥50% of the tumor volume. The presence and extent of signet ring cells were categorized as 0% (no signet ring cells) or ≥1% of the tumor volume.
Sequencing of KRAS, BRAF and PIK3CA, and MSI analysis
Genomic DNA was extracted from dissected tumor tissue sections, and whole genome amplification was done by PCR using random 15-mer primers (31). PCR and Pyrosequencing targeted for KRAS (codons 12 and 13; ref. 31), BRAF (codon 600; ref. 32), and PIK3CA (exons 9 and 20; ref. 33) were done. MSI analysis was done, using 10 microsatellite markers (D2S123, D5S346, D17S250, BAT25, BAT26, BAT40, D18S55, D18S56, D18S67, and D18S487; ref. 30). MSI-high was defined as the presence of instability in ≥30% of the markers. MSI-low was defined as instability in <30% of the markers, and “microsatellite stable” tumors were defined as tumors without an unstable marker.
Real-time PCR to measure DNA methylation in CIMP-specific and other CpG islands
Sodium bisulfite treatment on genomic DNA and subsequent real-time PCR (MethyLight) were validated and done as previously described (34). We quantified DNA methylation in eight CIMP-specific promoters [CACNA1G, CDKN2A (p16), CRABP1, IGF2, MLH1, NEUROG1, RUNX3, and SOCS1; refs. 27, 30], all of which were selected from screening of 195 CpG islands (26, 27). Primers and probes were previously described (27). Methylation positivity at each locus was defined as the percentage of methylated reference (≥4), as previously validated (34). CIMP-high was defined as the presence of ≥6 of 8 methylated promoters, CIMP-low as the presence of 1/8 to 5/8 methylated promoters, and CIMP-0 as the absence (0/8) of methylated promoters, according to the previously established criteria (30).
In addition, we quantified DNA methylation in eight other CpG islands (not in the CIMP panel), including CHFR, HIC1, IGFBP3, MGMT, MINT1, MINT31, p14, and WRN. Primers and probes were previously described (HIC1, IGFBP3, p14, and WRN; refs. 27, 35). The PCR condition for all markers was initial denaturation at 95°C for 10 min followed by 45 cycles of 95°C for 15 s, and 60°C for 1 min.
Pyrosequencing to measure LINE-1 methylation
In order to accurately quantify relatively high methylation levels in long interspersed nucleotide element-1 (LINE-1) repetitive elements, we used Pyrosequencing as previously described (28). LINE-1 methylation level measured by Pyrosequencing has been shown to be a good indicator of 5-methylcytosine level (i.e., genome-wide DNA methylation level; ref. 36). LINE-1 methylation levels were distributed approximately normally (mean, 60.9%; median, 61.8%; SD, 9.8%).
Immunohistochemistry for p53, β-catenin, and DNMT3B
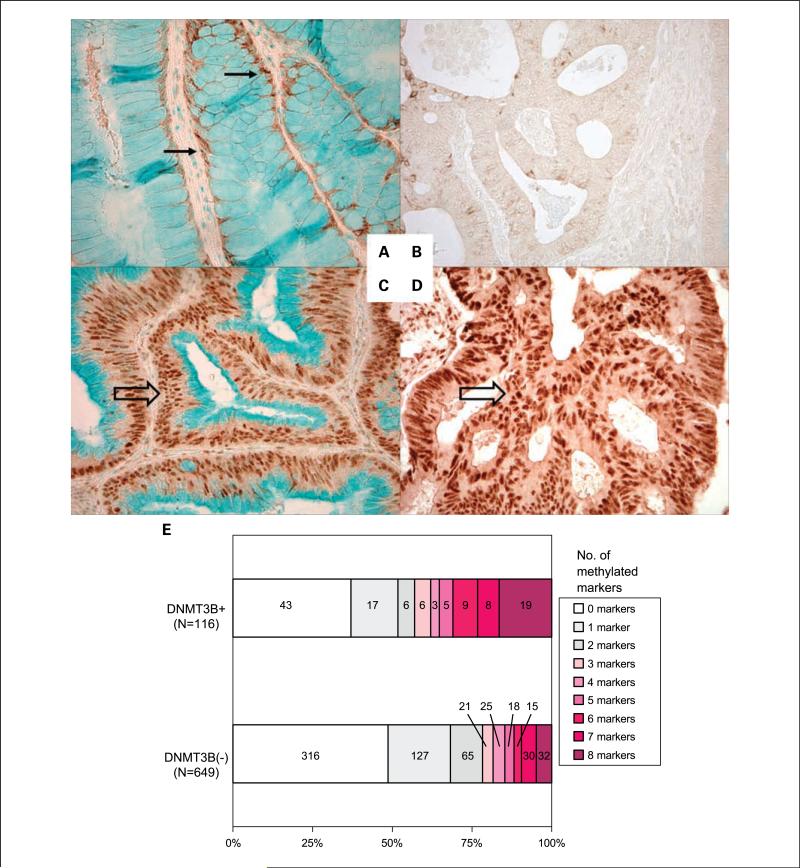
Tissue microarrays were constructed as previously described (29). Methods of immunohistochemical procedures and interpretations were previously described for p53 and β-catenin (30, 37). For DNMT3B, antigen retrieval was done, and deparaffinized tissue sections in Antigen Retrieval Citra Solution (Biogenex Laboratories) were microwaved for 15 min. Tissue sections were incubated with 10% normal goat serum (Vector Laboratories) in PBS (30 min). We applied primary monoclonal antibody against DNMT3B (1:150 dilution, clone 52A1018; Imgenex), and the slides were incubated overnight at room temperature. This antibody was generated using full-length mouse recombinant Dnmt3b, and has been shown to recognize human DNMT3B.8 Next, we applied an anti-mouse IgG antibody (Vector Laboratories) for 30 min, followed by an avidin-biotin complex conjugate (Vector Laboratories) for 30 min, diaminobenzidine (5 min) and methyl green counterstain. Nuclear DNMT3B expression was recorded as no expression, weak expression, or moderate/strong expression (Fig. 1). DNMT3B positivity (i.e., over-expression) was defined as ≥30% of tumor cells with at least weak nuclear staining. For DNMT3B expression analysis in normal mucosa, we used normal mucosa adjacent to colorectal cancer on the same tissue section for each case. In normal colonic mucosa, there were no cases with nuclear expression, and we recorded the intensity of cytoplasmic expression as no expression, weak expression, or moderate/strong expression. Appropriate positive and negative controls were included in each run of immunohistochemistry. All immunohistochemically stained slides were interpreted by one of the investigators (K. Nosho) unaware of other data. A random selection of 141 cases was examined by a second observer (K. Shima) unaware of other data, and concordance between the two observers was 0.91 (κ = 0.60; P < 0.0001), indicating substantial agreement.
Fig. 1.
DNMT3B expression in colorectal cancer and normal mucosa. A, cytoplasmic expression of DNMT3B in normal colonic mucosa (arrows). B, no nuclear DNMT3B expression in colorectal cancer. C and D, strong nuclear DNMT3B expression in colorectal cancers (empty arrows). E, number of methylated CIMP markers according to DNMT3B status in colorectal cancer.
Statistical analysis
All statistical analyses used SAS program (version 9.1, SAS Institute). All P values were two-sided, and statistical significance was set at P ≤ 0.05. P values were conservatively interpreted when performing multiple hypotheses testing. For categorical data, the χ2 test (or Fisher's exact test when any expected cell count was <5) was done. To compare mean LINE-1 methylation levels, the t test assuming unequal variances was done. The κ coefficients were calculated to assess an agreement between the two interpreters in immunohistochemistry, and that between the two panels (8 markers versus 16 markers) for the diagnosis of CIMP-high. A correlation between the degree of tumoral DNMT3B expression and the degree of DNMT3B expression in adjacent normal colonic mucosa was assessed by the Spearman correlation test. To assess independent effect of tumoral DNMT3B expression on tumoral CIMP status, multivariate logistic regression analysis was done (with DNMT3B as an exposure of interest and CIMP as an outcome variable) and odds ratio (OR) was adjusted for age (as a continuous variable), sex, tumor location (proximal versus distal), stage (I-II versus III-IV), grade (low versus high), mucin (present versus absent), signet ring cells (present versus absent), MSI (high versus low/microsatellite stable), p53, nuclear β-catenin (positive versus negative), LINE-1 methylation (as a continuous variable), BRAF, KRAS,and PIK3CA (mutant versus wild-type). Likewise, to assess independent effect of tumoral DNMT3B on methylation in each of the 16 individual CpG islands, multivariate logistic regression analysis was done (with DNMT3B as an exposure variable and a given methylation marker as an outcome variable) with adjustment done as above.
For survival analysis, we did not take recurrence or relapse into account. For analyses of colorectal cancer–specific mortality, death as a result of colorectal cancer was the primary end point, and deaths as a result of other causes were censored. For analyses of all-cause mortality as the end point, participants were censored only at the end of followup. The Kaplan-Meier method was used to describe the distribution of colon cancer–specific and overall survival time, and a log rank test was done. We also used stage-matched, conditional Cox proportional hazard models to calculate hazard ratios of death according to tumoral DNMT3B status adjusted for age, sex, year of diagnosis, tumor location, stage, grade, MSI, CIMP, LINE-1, KRAS, BRAF, p53, and β-catenin. To adjust for potential confounding, tumor stage was used as a matching variable, and age, year of diagnosis, and LINE-1 methylation were used as continuous variables. All of the other covariates were used as dichotomous categorical variables. For missing information in covariates [including tumor grade (0.3% missing), MSI (1.0%), KRAS (0.7%), BRAF (3.2%), p53 (0.7%), and β-catenin (1.7%)], we included those cases in a majority category in the particular variable in order to eliminate “missing” indicator variables and maximize the efficiency of multivariate Cox regression analyses. We confirmed that excluding cases with missing information in any of the covariates did not substantially alter results (data not shown). An interaction with CIMP (or LINE-1) was assessed by including the cross product term of the DNMT3B variable and the CIMP (or LINE-1) variable in a multivariate Cox model, and the likelihood ratio test was done.
Results
DNMT3B expression in colorectal cancers
Among the 765 colorectal cancers assessed by immunohistochemistry, 116 (15%) tumors overexpressed DNMT3B. Table 1 shows clinical, pathologic and molecular features of colorectal cancer according to tumoral DNMT3B status. DNMT3B overexpression was significantly associated with proximal location (P < 0.0001), high tumor grade (P = 0.007), mucinous component (P = 0.0001), signet ring cells (P = 0.0007), CIMP-high (P < 0.0001), MSI-high (P < 0.0001), and BRAF mutation (P < 0.0001; Table 1).
Table 1.
Clinical, pathologic, and molecular features according to tumoral DNMT3B status in colorectal cancer
| Clinical or molecular feature | Total N | DNMT3B+ | DNMT3B– | P |
|---|---|---|---|---|
| All cases | 765 | 116 | 649 | |
| Gender | 0.15 | |||
| Men | 337 (44%) | 44 (38%) | 293 (45%) | |
| Women | 428 (56%) | 72 (62%) | 356 (55%) | |
| Age | 0.11 | |||
| ≤59 | 191 (25%) | 20 (17%) | 171 (26%) | |
| 60-69 | 317 (41%) | 52 (45%) | 265 (41%) | |
| ≥70 | 257 (34%) | 44 (38%) | 213 (33%) | |
| Tumor location | <0.0001 | |||
| Distal (splenic flexure to rectum) | 408 (56%) | 42 (38%) | 366 (59%) | |
| Proximal (cecum to transverse) | 326 (44%) | 70 (63%) | 256 (41%) | |
| Stage | 0.36 | |||
| I | 159 (23%) | 21 (20%) | 138 (24%) | |
| II | 225 (33%) | 37 (35%) | 188 (33%) | |
| III | 206 (30%) | 28 (27%) | 178 (31%) | |
| IV | 92 (13%) | 19 (18%) | 73 (13%) | |
| Tumor grade | 0.007 | |||
| Low | 686 (91%) | 98 (84%) | 588 (92%) | |
| High | 67 (8.9%) | 18 (16%) | 49 (7.7%) | |
| Mucinous component (%) | 0.0006 | |||
| 0 | 403 (61%) | 54 (50%) | 349 (63%) | |
| 1-49 | 161 (24%) | 26 (24%) | 135 (25%) | |
| ≥50 | 94 (14%) | 28 (26%) | 66 (12%) | |
| Signet ring cell component (%) | 0.0007 | |||
| 0 | 569 (92%) | 83 (84%) | 486 (94%) | |
| ≥1 | 48 (7.8%) | 16 (16%) | 32 (6.2%) | |
| p53 expression | 0.79 | |||
| (–) | 431 (57%) | 67 (58%) | 364 (56%) | |
| (+) | 330 (43%) | 49 (42%) | 281 (44%) | |
| CIMP status | <0.0001 | |||
| CIMP-0 | 359 (47%) | 43 (37%) | 316 (49%) | |
| CIMP-low | 293 (38%) | 37 (32%) | 256 (39%) | |
| CIMP-high | 113 (15%) | 36 (31%) | 77 (12%) | |
| MSI status | <0.0001 | |||
| MSS | 585 (77%) | 79 (69%) | 506 (79%) | |
| MSI-low | 64 (8.4%) | 4 (3.5%) | 60 (9.3%) | |
| MSI-high | 109 (14%) | 32 (28%) | 77 (12%) | |
| BRAF mutation | <0.0001 | |||
| (–) | 641 (86%) | 86 (74%) | 555 (89%) | |
| (+) | 102 (14%) | 30 (26%) | 72 (11%) | |
| KRAS mutation | 0.49 | |||
| (–) | 484 (64%) | 77 (66%) | 407 (63%) | |
| (+) | 278 (36%) | 39 (34%) | 239 (37%) | |
| PIK3CA mutation | 0.69 | |||
| (–) | 600 (85%) | 91 (83%) | 509 (85%) | |
| (+) | 108 (15%) | 18 (17%) | 90 (15%) | |
| Nuclear β-catenin expression | 0.098 | |||
| (–) | 343 (48%) | 59 (56%) | 284 (47%) | |
| (+) | 368 (52%) | 47 (44%) | 321 (53%) | |
| LINE-1 methylation level (%) | 0.58 | |||
| ≥70 | 131 (18%) | 23 (20%) | 108 (17%) | |
| 50-70 | 518 (70%) | 79 (69%) | 439 (70%) | |
| <50 | 96 (13%) | 12 (11%) | 84 (13%) | |
| DNMT3B in adjacent normal colon | <0.0001 | |||
| (–) | 365 (51%) | 29 (26%) | 336 (55%) | |
| Weak+ | 214 (30%) | 39 (35%) | 175 (29%) | |
| (+) | 139 (19%) | 43 (39%) | 96 (16%) |
NOTE: Percentages indicate the proportion of cases with a specific clinical, pathologic, or molecularfeature among DNMT3B+ cases (or DNMT3B – cases).
Abbreviation: MSS, microsatellite stable.
Because methylation in LINE-1 repetitive DNA elements is a good indicator of global DNA methylation level (36), we examined the relation between DNMT3B expression and LINE-1 methylation. The mean LINE-1 methylation level was higher in DNMT3B-positive tumors [62.6%; 95% confidence interval (CI), 60.9-64.4%] than in DNMT3B-negative tumors (60.6%; 95% CI, 59.9-61.4%; P = 0.041), but the difference was rather subtle.
DNMT3B expression in cancer and adjacent normal colon mucosa
In 718 cases, we were able to compare DNMT3B expression in colorectal cancer to DNMT3B expression in adjacent normal mucosa. Tumoral DNMT3B expression seemed to be significantly associated with DNMT3B expression in adjacent normal colonic mucosa (Table 1). The intensity of DNMT3B expression in normal colonic mucosa (absent, 1+, 2+, and 3 +) was significantly correlated with the intensity of tumoral DNMT3B expression (absent, 1+, 2+,and 3+; Spearman r = 0.27; P < 0.0001) and with the percentage of tumor cells with DNMT3B nuclear staining (Spearman r = 0.26; P < 0.0001). This phenomenon could be explained by field effect or field cancerization by DNMT3B expression. Another possibility was that this phenomenon could be, at least in part, due to the presence of tumors with poor antigenicity, driving an overall relation towards a concordant pattern.
Association between tumoral DNMT3B and CIMP-high
Using real-time quantitative PCR (MethyLight) assays, we have extensively validated the use of the eight markers (CACNA1G, CDKN2A, CRABP1, IGF2, MLH1, NEUROG1, RUNX3, and SOCS1) as the CIMP diagnostic panel (30), among which CACNA1G, IGF2, NEUROG1, RUNX3, and SOCS1 were selected from screening of 195 CpG islands (27). We also examined methylation in eight other CpG islands (CHFR, HIC1, IGFBP3, MGMT, MINT1, MINT31, p14, and WRN). In the current analysis, CIMP-high tumors determined by this eight-marker panel were nearly identical to CIMP-high tumors determined by the panel using all of the 16 markers (with 97.2% concordance, κ = 0.89; P <0.0001). Because of our previous extensive validation, we defined CIMP-high as ≥6/8 methylated CIMP-specific markers (CACNA1G, CDKN2A, CRABP1, IGF2, MLH1, NEUROG1, RUNX3, and SOCS1).
We examined the frequencies of CIMP marker methylation in DNMT3B-positive and DNMT3B-negative tumors (Fig. 1E). It was evident that the DNMT3B-positive tumor group included more heavily methylated tumors than the DNMT3B-negative group.
Tumoral DNMT3B is independently associated with CIMP-high
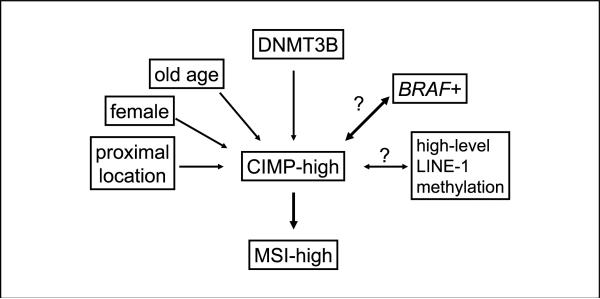
To confirm an independent relation between DNMT3B and CIMP-high, we did a multivariate logistic regression analysis (Table 2). DNMT3B was associated with CIMP-high (multivariate OR, 2.39; 95% CI, 1.11-5.14) after adjusting for age, sex, tumor location, stage, grade, mucinous component, signet ring cells, MSI, LINE-1, p53, nuclear β-catenin, KRAS, PIK3CA, and BRAF. Based on these results, Fig. 2 summarizes hypothetical relations with CIMP-high in colorectal cancer.
Table 2.
Univariate and multivariate logistic regression analysis of the relation of tumoral DNMT3B with CIMP-high in colorectal cancer
| Variable independently associated with CIMP-high | Univariate OR (95% CI) | P | Multivariate OR (95% CI) | P |
|---|---|---|---|---|
| DNMT3B expression | 3.34 (2.11-5.29) | <0.0001 | 2.39 (1.11-5.14) | 0.026 |
| Other significant variables | ||||
| MSI-high (vs MSI-low/MSS) | 33.1 (19.7-55.4) | <0.0001 | 18.6 (8.27-41.9) | <0.0001 |
| BRAF mutation | 26.6 (15.9-44.5) | <0.0001 | 13.6 (5.71-32.5) | <0.0001 |
| Age (for 10 year increment) | 1.79 (1.38-2.33) | <0.0001 | 3.39 (2.00-5.76) | <0.0001 |
| Proximal location | 11.8 (6.49-21.5) | <0.0001 | 3.46 (1.57-7.64) | 0.002 |
| Nuclear β-catenin expression | 0.17 (0.10-0.28) | <0.0001 | 0.30 (0.13-0.68) | 0.004 |
| LINE-1 methylation (for 10% increment) | 1.68 (1.33-2.11) | <0.0001 | 1.57 (1.08-2.29) | 0.018 |
| High tumor grade | 5.43 (3.28-9.00) | <0.0001 | 2.86 (1.14-7.22) | 0.026 |
| Sex (female vs male) | 1.83 (1.20-2.80) | 0.005 | 2.44 (1.10-5.43) | 0.028 |
| p53 expression | 0.28 (0.17-0.46) | <0.0001 | 0.42 (0.19-0.92) | 0.031 |
NOTE: Multivariate logistic regression analysis assessed the relation of DNMT3B (an exposure variable of interest) with CIMP (an outcome variable), adjusting for tumor stage, mucin, signet ring cells, KRAS, PIK3CA, and the variables listed in the table. Only significant variables in the multivariate analysis are listed.
Abbreviation: MSS, microsatellite stable.
Fig. 2.
Hypothetical relations with CIMP-high in colorectal cancer. Question marks indicate uncertainty in the direction of the relation. The thick lines with BRAF and MSI-high imply the particularly tight relations with CIMP-high.
Relationship between tumoral DNMT3B and methylation in individual CpG islands
Because we found the association between tumoral DNMT3B expression and CIMP-high, we examined if there was any differential association between tumoral DNMT3B and methylation in individual CpG islands. For each CpG island locus (CACNA1G, CDKN2A, CRABP1, IGF2, MLH1, NEUROG1, RUNX3, SOCS1, CHFR, HIC1, IGFBP3, MGMT, MINT1, MINT31, p14, or WRN), we did univariate and multivariate logistic regression analyses of the relation of tumoral DNMT3B (as an exposure variable of interest) with each methylation marker (as an outcome variable; Table 3). Tumoral DNMT3B expression was significantly associated with CpG island methylation at each locus, except for HIC1, MGMT, and MINT1 in univariate analyses. Interestingly, the strength of the relationship with DNMT3B varied from marker to marker [OR ranging from 0.80 (for MGMT) to 2.96 (for CACNA1G)], suggesting that DNMT3B might have varying specificities against different CpG islands. In multivariate analysis, tumoral DNMT3B expression remained significantly associated with CACNA1G, IGF2, and SOCS1. The strength of the relationship with tumoral DNMT3B varied from marker to marker [multivariate OR ranging from 0.75 (for MGMT) to 1.96 (for CACNA1G)].
Table 3.
Univariate and multivariate logistic regression analysis for the relation of tumoral DNMT3B with each methylation marker
| Outcome variable | Univariate OR, 95% CI (DNMT3B and methylation marker) | P | Multivariate OR, 95% CI (DNMT3B and methylation marker) | P |
|---|---|---|---|---|
| CIMP panel markers | ||||
| CACNA1G | 2.96 (1.94-4.51) | <0.0001 | 1.96 (1.15-3.34) | 0.014 |
| CDKN2A (pl6) | 2.10 (1.40-3.16) | 0.0003 | 1.24 (0.76-2.02) | 0.39 |
| CRABP1 | 2.06 (1.38-3.09) | 0.0004 | 1.31 (0.79-2.16) | 0.29 |
| IGF2 | 2.84 (1.87-4.30) | <0.0001 | 1.87 (1.09-3.21) | 0.023 |
| MLH1 | 2.31 (1.39-3.84) | 0.001 | 1.31 (0.63-2.71) | 0.48 |
| NEUROG1 | 1.69(1.12-2.54) | 0.012 | 0.85 (0.50-1.44) | 0.55 |
| RUNX3 | 2.52 (1.64-3.89) | <0.0001 | 1.26 (0.68-2.34) | 0.47 |
| SOCS1 | 2.84 (1.81-4.44) | <0.0001 | 1.93 (1.13-3.30) | 0.016 |
| Other CpG islands | ||||
| CHFR | 1.66 (1.12-2.47) | 0.012 | 1.02 (0.65-1.60) | 0.93 |
| HIC1 | 1.46 (0.98-2.18) | 0.064 | 1.04 (0.67-1.62) | 0.87 |
| IGFBP3 | 1.61 (1.06-2.46) | 0.025 | 1.24 (0.79-1.97) | 0.35 |
| MGMT | 0.80 (0.53-1.21) | 0.29 | 0.75 (0.48-1.17) | 0.21 |
| MINT1 | 1.18 (0.78-1.77) | 0.43 | 0.77 (0.49-1.21) | 0.26 |
| MINT31 | 2.36 (1.58-3.53) | <0.0001 | 1.58 (0.99-2.54) | 0.057 |
| p14 (CDKN2A/ARF) | 1.75 (1.12-2.74) | 0.014 | 1.09 (0.64-1.85) | 0.76 |
| WRN | 1.75 (1.18-2.61) | 0.005 | 1.08 (0.67-1.75) | 0.75 |
NOTE: Multivariate logistic regression analysis assessed the relation of tumoral DNMT3B (an exposure variable) with each methylation marker (an outcome variable), adjusted for age, sex, tumor location, stage, grade, mucin, signet ring cells, LINE-1, KRAS, BRAF, PIK3CA, β-catenin, and p53.
Patient survival, tumoral DNMT3B, and interaction with CIMP or LINE-1
Finally, we examined the effect of DNMT3B expression on patient outcome and potential modifying effect of CIMP or LINE-1. Both CIMP and LINE-1 methylation level have been shown to be significantly associated with patient survival in our cohort studies (38, 39). Among 733 eligible cases with a total of 5,922 person-years of follow-up, there were 313 deaths, including 191 colorectal cancer–specific deaths. In the Kaplan-Meier analysis, there was no significant difference in survival time distributions according to tumoral DNMT3B status (log rank P > 0.20). In stage-matched Cox regression analysis or multivariate Cox regression analysis, DNMT3B expression in tumor was not significantly related with patient outcome (Supplemental Table). In addition, there was no evidence for a significant modifying effect of CIMP or LINE-1 methylation on the relation between DNMT3B and patient outcome (P for interaction > 0.19).
Discussion
We conducted this study to examine the relationship between DNMT3B and the CIMP in colorectal cancer, and the relation between DNMT3B expression in cancer and matched normal colonic mucosa. Molecular correlates with DNMT3B activation may be important for better understanding of epigenetic and epigenomic aberrations during the carcinogenic process. We have shown that DNMT3B overexpression is associated with CIMP-high independent of other clinical and molecular variables, including age, sex, tumor location, stage, pathologic features, MSI, and BRAF mutation. Our data support the hypothesis that DNMT3B may contribute to CpG island methylation, which may eventually lead to the development of CIMP-high colorectal cancer. Considering that DNMT3B is a potential target of chemotherapy and chemoprevention, our findings may have considerable clinical implications.
To measure DNA methylation, we used real-time PCR (MethyLight technology) for DNA methylation at the eight CIMP-specific loci and in eight other CpG islands. We also used Pyrosequencing to measure LINE-1 methylation, which is a good indicator of cellular 5-methylcytosine level (i.e., genome-wide DNA methylation level; ref. 36). Our resource of a large number of colorectal cancers derived from the two prospective cohort studies has enabled us to precisely estimate the frequency of colorectal cancers with a specific molecular feature (such as DNMT3B overexpression, CIMP-high, etc.). The large number of cases has also provided a sufficient power in our multivariate logistic regression analysis.
DNMT3B activation has been implicated in aberrant de novo methylation of CpG islands in colorectal cancer (2, 4, 5, 7). Dnmt3b can induce colon tumor in mice, and those tumors exhibit DNA methylation in specific CpG islands (7). In addition, a recent study using the MDA-MB231 breast cancer cell line has shown that Dnmt3b plays an important role in both methylation and demethylation (9). DNMT3B mRNA is overexpressed in human colorectal cancers compared with matched normal colonic mucosa (11, 13). In the current study, we have shown DNMT3B overexpression in 15% of colorectal cancers and the positive association between DNMT3B expression and CIMP-high, independent of other variables. DNMT3B overexpression has also been linked with the methylator phenotype in ovarian cancer cell lines (6). Our data are likely important because no study has comprehensively examined the relationship between DNMT3B expression and CIMP in human colorectal cancer.
The molecular features of colorectal cancer have been extensively studied (40–43). Although BRAF mutation has been tightly linked to CIMP-high in colorectal cancer (25, 27, 30), the exact mechanism of high propensity for widespread CpG island methylation leading to CIMP-high has remained uncertain. In the current study, we have shown the relation between DNMT3B and CIMP-high, independent of BRAF mutation and other factors associated with CIMP-high. On the other hand, our data do not support a direct link between DNMT3B and high-level methylation in LINE-1, a surrogate of genome-wide DNA methylation level. A recent study using a murine colon cancer model has shown that Dnmt3b overexpression induces DNA methylation in some, but not all, CpG islands, suggesting that Dnmt3b may facilitate CpG island methylation in a non-random fashion (7). DNMT3B depletion in cancer cells causes gene-specific demethylation, rather than genome-wide demethylation (14). In addition, different splicing variants of DNMT3B seem to regulate gene-specific CpG island methylation patterns (10, 44). These experimental data are consistent with our data of the significant relation between DNMT3B and CIMP-high, but no significant relation between DNMT3B and genome-wide DNA methylation level. Based on our current results and data in the literature, Fig. 2 represents hypothetical relations with CIMP-high in colorectal cancer. Further studies are necessary to elucidate the exact mechanism of DNMT3B activity on specific CpG islands without substantially affecting genome-wide DNA methylation level.
Regarding the different subtypes of CIMP in colorectal cancer, accumulating evidence suggests that CIMP-low is associated with KRAS mutation and is molecularly different from CIMP-high and CIMP-negative (CIMP-0; refs. 32, 35, 45–50). It has been suggested that KRAS mutation is associated with a random CpG island methylation pattern that leads to CIMP-low tumors (35). Together with the previous study suggesting nonrandom targeting of DNMT3B activity (7), our data further support a molecular difference between CIMP-high (associated with DNMT3B up-regulation, a nonrandom CpG island methylation pattern and BRAF mutation) and CIMP-low (associated with a random methylation pattern and KRAS mutation). Further studies are necessary to elucidate the pathogenic mechanisms of these different epigenomic aberrations.
In conclusion, DNMT3B overexpression is associated with CIMP-high in colorectal cancer, independent of other clinical and molecular characteristics. Our data support the hypothesis that DNMT3B may play an important role in widespread promoter CpG island methylation during the carcinogenic process. Considering that DNMT3B is a potential target of chemotherapy and chemoprevention, our findings may have considerable clinical implications.
Acknowledgments
We thank the Nurses’ Health Study and Health Professionals Follow-up Study cohort participants who have generously agreed to provide us with biological specimens and information through responses to questionnaires. We thank Frank Speizer, Walter Willett, Susan Hankinson, Meir Stampfer, and many other staff members who implemented and maintained the cohort studies.
Grant support: U.S. NIH grants P01 CA87969 (S. Hankinson), P01 CA55075 (W. Willett), P50 CA127003 (C.S. Fuchs), and K07 CA122826 (S. Ogino), and in part by grants from the Bennett Family Fund and from the Entertainment Industry Foundation National Colorectal Cancer Research Alliance. K. Nosho was supported by a fellowship grant from the Japan Society for the Promotion of Science.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the NIH. Funding agencies did not have any role in the design of the study; the collection, analysis, or interpretation of the data; the decision to submit the manuscript for publication; or the writing of the manuscript.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
K. Nosho, K. Shima, and N. Irahara contributed equally.
The manuscript has not been published previously and is not being considered concurrently by any other publication. This manuscript acknowledges all sources of support for the work.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–92. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kanai Y, Hirohashi S. Alterations of DNA methylation associated with abnormalities of DNA methyltransferases in human cancers during transition from a precancerous to a malignant state. Carcinogenesis. 2007;28:2434–42. doi: 10.1093/carcin/bgm206. [DOI] [PubMed] [Google Scholar]
- 3.Rhee I, Bachman KE, Park BH, et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002;416:552–6. doi: 10.1038/416552a. [DOI] [PubMed] [Google Scholar]
- 4.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–57. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 5.Bachman KE, Rountree MR, Baylin SB. Dnmt3a and Dnmt3b are transcriptional repressors that exhibit unique localization properties to heterochromatin. J Biol Chem. 2001;276:32282–7. doi: 10.1074/jbc.M104661200. [DOI] [PubMed] [Google Scholar]
- 6.Roll JD, Rivenbark AG, Jones WD, Coleman WB. DNMT3b overexpression contributes to a hypermethylator phenotype in human breast cancer cell lines. Mol Cancer. 2008;7:15. doi: 10.1186/1476-4598-7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Linhart HG, Lin H, Yamada Y, et al. Dnmt3b promotes tumorigenesis in vivo by gene-specific de novo methylation and transcriptional silencing. Genes Dev. 2007;21:3110–22. doi: 10.1101/gad.1594007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–93. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 9.Metivier R, Gallais R, Tiffoche C, et al. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452:45–50. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- 10.Saito Y, Kanai Y, Sakamoto M, Saito H, Ishii H, Hirohashi S. Overexpression of a splice variant of DNA methyltransferase 3b, DNMT3b4, associated with DNA hypomethylation on pericentromeric satellite regions during human hepatocarcinogenesis. Proc NatlAcad Sci U S A. 2002;99:10060–5. doi: 10.1073/pnas.152121799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robertson KD, Uzvolgyi E, Liang G, et al. The human DNA methyltransferases (DNMTs) 1, 3a and 3b: coordinate mRNA expression in normal tissues and overexpression in tumors. Nucleic Acids Res. 1999;27:2291–8. doi: 10.1093/nar/27.11.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahluwalia A, Hurteau JA, Bigsby RM, Nephew KP. DNA methylation in ovarian cancer: I.I. Expression of DNA methyltransferases in ovarian cancer cell lines and normal ovarian epithelial cells. Gynecol Oncol. 2001;82:299–304. doi: 10.1006/gyno.2001.6284. [DOI] [PubMed] [Google Scholar]
- 13.Schmidt WM, Sedivy R, Forstner B, Steger GG, Zochbauer-Muller S, Mader RM. Progressive up-regulation of genes encoding DNA methyltransferases in the colorectal adenoma-carcinoma sequence. Mol Carcinog. 2007;46:766–72. doi: 10.1002/mc.20307. [DOI] [PubMed] [Google Scholar]
- 14.Beaulieu N, Morin S, Chute IC, Robert MF, Nguyen H, MacLeod AR. An essential role for DNA methyltransferase DNMT3B in cancer cell survival. J Biol Chem. 2002;277:28176–81. doi: 10.1074/jbc.M204734200. [DOI] [PubMed] [Google Scholar]
- 15.Fini L, Selgrad M, Fogliano V, et al. Annurca apple polyphenols have potent demethylating activity and can reactivate silenced tumor suppressor genes in colorectal cancer cells. J Nutr. 2007;137:2622–8. doi: 10.1093/jn/137.12.2622. [DOI] [PubMed] [Google Scholar]
- 16.Leu YW, Rahmatpanah F, Shi H, et al. Double RNA interference of DNMT3b and DNMT1 enhances DNA demethylation and gene reactivation. Cancer Res. 2003;63:6110–5. [PubMed] [Google Scholar]
- 17.Teodoridis JM, Hall J, Marsh S, et al. CpG island methylation of DNA damage response genes in advanced ovarian cancer. Cancer Res. 2005;65:8961–7. doi: 10.1158/0008-5472.CAN-05-1187. [DOI] [PubMed] [Google Scholar]
- 18.Kawakami K, Ruszkiewicz A, Bennett G, et al. DNA hypermethylation in the normal colonic mucosa of patients with colorectal cancer. Br J Cancer. 2006;94:593–8. doi: 10.1038/sj.bjc.6602940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen H, Wang L, Spitz MR, Hong WK, Mao L, Wei Q. A novel polymorphism in human cytosine DNA-methyltransferase-3B promoter is associated with an increased risk of lung cancer. Cancer Res. 2002;62:4992–5. [PubMed] [Google Scholar]
- 20.Jones JS, Amos CI, Pande M, et al. DNMT3b polymorphism and hereditary nonpolyposis colorectal cancer age of onset. Cancer Epidemiol Biomarkers Prev. 2006;15:886–91. doi: 10.1158/1055-9965.EPI-05-0644. [DOI] [PubMed] [Google Scholar]
- 21.Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A. 1999;96:8681–6. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grady WM. CIMP and colon cancer gets more complicated. Gut. 2007;56:1498–500. doi: 10.1136/gut.2007.125732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Teodoridis JM, Hardie C, Brown R. CpG island methylator phenotype (CIMP) in cancer: causes and implications. Cancer Lett. 2008;268:177–86. doi: 10.1016/j.canlet.2008.03.022. [DOI] [PubMed] [Google Scholar]
- 24.Kambara T, Simms LA, Whitehall VLJ, et al. BRAF mutation is associated with DNA methylation in serrated polyps and cancers of the colorectum. Gut. 2004;53:1137–44. doi: 10.1136/gut.2003.037671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Samowitz W, Albertsen H, Herrick J, et al. Evaluation of a large, population-based sample supports a CpG island methylator phenotype in colon cancer. Gastroenterology. 2005;129:837–45. doi: 10.1053/j.gastro.2005.06.020. [DOI] [PubMed] [Google Scholar]
- 26.Ogino S, Cantor M, Kawasaki T, et al. CpG island methylator phenotype (CIMP) of colorectal cancer is best characterised by quantitative DNA methylation analysis and prospective cohort studies. Gut. 2006;55:1000–6. doi: 10.1136/gut.2005.082933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weisenberger DJ, Siegmund KD, Campan M, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–93. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 28.Ogino S, Kawasaki T, Nosho K, et al. LINE-1 hypomethylation is inversely associated with microsatellite instability and CpG island methylator phenotype in colorectal cancer. Int J Cancer. 2008;122:2767–73. doi: 10.1002/ijc.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chan AT, Ogino S, Fuchs CS. Aspirin and the risk of colorectal cancer in relation to the expression of COX-2. New Engl J Med. 2007;356:2131–42. doi: 10.1056/NEJMoa067208. [DOI] [PubMed] [Google Scholar]
- 30.Ogino S, Kawasaki T, Kirkner GJ, Kraft P, Loda M, Fuchs CS. Evaluation of markers for CpG island methylator phenotype (CIMP) in colorectal cancer by a large population-based sample. J Mol Diagn. 2007;9:305–14. doi: 10.2353/jmoldx.2007.060170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ogino S, Kawasaki T, Brahmandam M, et al. Sensitive sequencing method for KRAS mutation detection by Pyrosequencing. J Mol Diagn. 2005;7:413–21. doi: 10.1016/S1525-1578(10)60571-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ogino S, Kawasaki T, Kirkner GJ, Loda M, Fuchs CS. CpG island methylator phenotype-low (CIMP-low) in colorectal cancer: possible associations with male sex and KRAS mutations. J Mol Diagn. 2006;8:582–8. doi: 10.2353/jmoldx.2006.060082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nosho K, Kawasaki T, Ohnishi M, et al. PIK3CA mutation in colorectal cancer: relationship with genetic and epigenetic alterations. Neoplasia. 2008;10:534–41. doi: 10.1593/neo.08336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ogino S, kawasaki T, Brahmandam M, et al. Precision and performance characteristics of bisulfite conversion and real-time PCR (MethyLight) for quantitative DNA methylation analysis. J Mol Diagn. 2006;8:209–17. doi: 10.2353/jmoldx.2006.050135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nosho K, Irahara N, Shima K, et al. Comprehensive biostatistical analysis of CpG island methylator phenotype in colorectal cancer using a large population-based sample. PLoS ONE. 2008;3:e3698. doi: 10.1371/journal.pone.0003698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang AS, Estecio MR, Doshi K, Kondo Y, Tajara EH, Issa JP. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res. 2004;32:e38. doi: 10.1093/nar/gnh032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kawasaki T, Nosho K, Ohnishi M, et al. Correlation of β-catenin localization with cyclooxygenase-2 expression and CpG island methylator phenotype (CIMP) in colorectal cancer. Neoplasia. 2007;9:569–77. doi: 10.1593/neo.07334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ogino S, Nosho K, Kirkner GJ, et al. A cohort study of tumoral LINE-1 hypomethylation and prognosis in colon cancer. J Natl Cancer Inst. 2008;100:1734–8. doi: 10.1093/jnci/djn359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ogino S, Nosho K, Kirkner GJ, et al. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. GUT. 2009;58:90–6. doi: 10.1136/gut.2008.155473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bettstetter M, Dechant S, Ruemmele P, et al. Distinction of hereditary nonpolyposis colorectal cancer and sporadic microsatellite-unstable colorectal cancer through quantification of MLH1 methylation by real-time PCR. Clin Cancer Res. 2007;13:3221–8. doi: 10.1158/1078-0432.CCR-06-3064. [DOI] [PubMed] [Google Scholar]
- 41.Derks S, Postma C, Carvalho B, et al. Integrated analysis of chromosomal, microsatellite and epigenetic instability in colorectal cancer identifies specific associations between promoter methylation of pivotal tumour suppressor and DNA repair genes and specific chromosomal alterations. Carcinogenesis. 2008;29:434–9. doi: 10.1093/carcin/bgm270. [DOI] [PubMed] [Google Scholar]
- 42.Ying J, Li H, Yu J, et al. WNT5A exhibits tumor- suppressive activity through antagonizing the Wnt/β-catenin signaling, and is frequently methylated in colorectal cancer. Clin Cancer Res. 2008;14:55–61. doi: 10.1158/1078-0432.CCR-07-1644. [DOI] [PubMed] [Google Scholar]
- 43.Weichert W, Roske A, Niesporek S, et al. Class I histone deacetylase expression has independent prognostic impact in human colorectal cancer: specific role of class I histone deacetylases in vitro and in vivo. Clin Cancer Res. 2008;14:1669–77. doi: 10.1158/1078-0432.CCR-07-0990. [DOI] [PubMed] [Google Scholar]
- 44.Wang J, Bhutani M, Pathak AK, et al. ΔDNMT3B variants regulate DNA methylation in a promoter-specific manner. Cancer Res. 2007;67:10647–52. doi: 10.1158/0008-5472.CAN-07-1337. [DOI] [PubMed] [Google Scholar]
- 45.Ogino S, Kawasaki T, Kirkner GJ, Ohnishi M, Fuchs CS. 18q loss of heterozygosity in microsatellite stable colorectal cancer is correlated with CpG island methylator phenotype-negative (CIMP-0) and inversely with CIMP-low and CIMP-high. BMC Cancer. 2007;7:72. doi: 10.1186/1471-2407-7-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ogino S, kawasaki T, Kirkner GJ, Suemoto Y, Meyerhardt JA, Fuchs CS. Molecular correlates with MGMT promoter methylation and silencing support CpG island methylator phenotype-low (CIMP-low) in colorectal cancer. Gut. 2007;56:1409–16. doi: 10.1136/gut.2007.119750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shen L, Toyota M, Kondo Y, et al. Integrated genetic and epigenetic analysis identifies three different subclasses of colon cancer. Proc Natl Acad Sci U S A. 2007;104:18654–9. doi: 10.1073/pnas.0704652104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ogino S, Goel A. Molecular classification and correlates in colorectal cancer. J Mol Diagn. 2008;10:13–27. doi: 10.2353/jmoldx.2008.070082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kawasaki T, Ohnishi M, Nosho K, et al. CpG island methylator phenotype-low (CIMP-low) colorectal cancer shows not only few methylated CIMP-high-specific CpG islands, but also low-level methylation at individual loci. Mod Pathol. 2008;21:245–55. doi: 10.1038/modpathol.3800982. [DOI] [PubMed] [Google Scholar]
- 50.Barault L, Charon-Barra C, Jooste V, et al. Hypermethylator phenotype in sporadic colon cancer: study on a population-based series of 582 cases. Cancer Res. 2008;68:8541–6. doi: 10.1158/0008-5472.CAN-08-1171. [DOI] [PubMed] [Google Scholar]