Abstract
Background
Naphthalene is a volatile hydrocarbon that causes dose-, species-, and cell type–dependent cytotoxicity after acute exposure and hyperplasia/neoplasia after lifetime exposures in rodents. Toxicity depends on metabolic activation, and reactive metabolite binding correlates with tissue and site susceptibility.
Objectives
We compared proteins adducted in nasal epithelium from rats and rhesus macaques in vitro.
Methods
Adducted proteins recovered from incubations of nasal epithelium and 14C-naphthalene were separated by two-dimensional (2D) gel electrophoresis and imaged to register radioactive proteins. We identified proteins visualized by silver staining on complementary nonradioactive gels by peptide mass mapping.
Results
The levels of reactive metabolite binding in incubations of rhesus ethmoturbinates and maxilloturbinates are similar to those in incubations of target tissues, including rat septal/olfactory regions and murine dissected airway incubations. We identified 40 adducted spots from 2D gel separations of rat olfactory epithelial proteins; 22 of these were nonredundant. In monkeys, we identified 19 spots by mass spectrometry, yielding three nonredundant identifications. Structural proteins (actin/tubulin) were prominent targets in both species.
Conclusions
In this study we identified potential target proteins that may serve as markers closely associated with toxicity. The large differences in previously reported rates of naphthalene metabolism to water-soluble metabolites in dissected airways from mice and monkeys are not reflected in similar differences in covalent adduct formation in the nose. This raises concerns that downstream metabolic/biochemical events are very similar between the rat, a known target for naphthalene toxicity and tumorigenicity, and the rhesus macaque, a species similar to the human.
Keywords: monkey, naphthalene, nasal epithelium, protein adducts, rat, reactive metabolites, species comparisons
Lung disease is the fourth leading cause of morbidity and mortality in the U.S. population. Although death rates from heart disease, cancer, and cerebrovascular disease have decreased in the past 10 years, death rates from chronic lower respiratory disease have not. Although tobacco use and traffic-related pollutants account for a portion of disease statistics, the precise etiology of lung disease remains unknown. A significant problem in assessing the contribution of individual chemicals or mixtures of chemicals to lung disease is related to the uncertainty of the use of animal models for human lung disease. Numerous chemicals, including naphthalene (Plopper et al. 1992), trichloroethylene (Odum et al. 1992), and styrene (Green et al. 2001), cause differential toxicity in two common rodent models, mice and rats, raising the question of relevance of these rodent models for human risk assessment.
Naphthalene administration results in highly selective necrosis of nonciliated bronchiolar epithelial cells of mice regardless of the route of administration. Inhaled concentrations of 2–5 ppm for 4 hr, well below the current occupational standard, cause detectable toxicity (West et al. 2001). In contrast, injury to airway epithelium of the rat does not occur at concentrations ≤ 100 ppm (West et al. 2001). The nasal epithelium of both rats and mice is highly sensitive to naphthalene administered either parenterally or by inhalation (Lee et al. 2005; Plopper et al. 1992). Recent studies have demonstrated sensitivity of rat olfactory epithelium at concentrations similar to those in urban atmospheres (0.3 ppm) (Dodd et al. 2010).
The species-, tissue-, and site-selective injury caused by naphthalene has been used to identify mechanisms associated with toxicity (for review, see Buckpitt et al. 2002). Naphthalene undergoes metabolism by the cytochrome P450 monooxygenases to reactive metabolites that become bound covalently to tissue proteins. The primary P450 involved in metabolism in the mouse lung and rat nose appears to be cytochrome P450 2F (CYP2F); this protein has high affinity for naphthalene as a substrate (Km, ~ 4 μM) with high catalytic turnover (104/min) (Shultz et al. 1999). CYP2F orthologs found in the lungs of nonhuman primates (CYP2F5; Baldwin et al. 2005) and humans (CYP2F1; Nhamburo et al. 1990) had little or no catalytic activity when expressed as recombinant enzyme (Lanza et al. 1999). This was consistent with the finding that lung microsomes from monkeys and humans metabolized naphthalene to water-soluble metabolites at very low rates (Buckpitt and Bahnson 1986; Buckpitt et al. 1992) and that dissected monkey airways generated metabolites very slowly (Boland et al. 2004). Because the initial step in the turnover of naphthalene is critical to toxicity, these data would indicate that monkeys and humans would be considerably less susceptible to naphthalene exposure than rodents. However, more recent data have shown that, although the rates of water-soluble metabolite formation are low, reactive metabolite protein binding occurs at similar levels in sensitive rodent tissues (Cho et al. 1994) and in dissected airways of monkeys (Boland et al. 2004).
Human exposure to naphthalene is nearly universal; the recent National Health and Nutrition Examination Survey studies demonstrated detectable levels of 1-naphthol glucuronide in the urine of all 2,785 subjects tested (Li et al. 2008). However, epidemiologic studies have failed to establish a relationship between naphthalene exposure and any long-term health consequences in the respiratory tract. Either humans are not as susceptible to the compound as rodents are, or respiratory diseases are multifactorial in origin and epidemiologic studies have not been sufficiently powered to detect subtle contributions of naphthalene to the overall incidence of disease.
In the present study we focused on protein-bound metabolites in nasal epithelium of rats and rhesus macaques. In contrast to previous work showing 10- to 100-fold higher rates of naphthalene metabolism by dissected mouse airways compared with the rhesus airway (Boland et al. 2004; Plopper et al. 1991), the present study shows that the rates of formation of covalently bound metabolites in rhesus nasal epithelium are similar to those in rat nasal epithelium, a target for naphthalene toxicity. This study also demonstrates some concordance in the adducted proteins identified in rat and monkey nasal epithelial incubations. We identified proteins involved in the unfolded protein response in incubations with rat but not monkey nasal epithelium. Proteins identified in the present work could serve as potential biomarker targets for studies in exposed human populations.
Materials and Methods
Animals
All animal work was conducted under protocols approved by the University of California–Davis (UCD) Animal Use and Care Committee. Animals were treated humanely and with regard to alleviation of suffering. Male Sprague-Dawley rats (200–225 g body weight) were obtained from Harlan Laboratories (Indianapolis, IN) and housed in the UCD vivarium for at least 1 week before experimentation. Animals were euthanized with an overdose of pentobarbital, and olfactory and septal turbinates were removed. Ethmoturbinates and maxilloturbinates were obtained from eight male and seven female rhesus macaques through the California National Primate Research Center [animal characteristics are available in Supplemental Material, Table 1 (doi:10.1289/ehp.0901333)]. All rhesus macaque tissue was from animals euthanized for normal colony maintenance at the center.
Reagents
We obtained [1,4,5,8-14C]naphthalene (specific activity 58 mCi/mmol) from Moravek Radiochemicals (Brea, CA) and checked purity (99%) by high-performance liquid chromatography. Protease inhibitor cocktail III, nondetergent sulfobetaine-195 (NDSB-195), benzamidine, 1,10-phenanthroline, aprotinin, pepstatin, and leupeptin were purchased from Calbiochem (La Jolla, CA). Rhinohide acrylamide/bisacrylamide was from Invitrogen (Carlsbad, CA), and Bio-Rad Protein Assay solution was purchased from Bio-Rad (Hercules, CA). We obtained immobilized pH gradient (IPG) buffers and narrow-range IPG Immobiline dry strips (18 cm) from GE Health Care (Piscataway, NJ). Unless otherwise stated, all reagents were molecular biology grade or better.
Tissue incubation and preparation
Tissue was incubated in oxygenated Waymouth’s medium deficient in sulfur amino acids (glutathione, cysteine, methionine) containing 250 μM naphthalene or 250 μM 14C-naphthalene at a specific activity of 1.10 × 105 disintegrations per minute per nanomole. Incubations were performed for 2 hr at 37°C in a shaking water bath. For protein identification, we incubated tissue with unlabeled substrate.
After the incubation, tissues were rinsed with cold 5% dextrose and homogenized in lysis solution [2 M thiourea, 7 M urea, 4% wt/wt 3-(3-cholamidopropyl)-dimethylammoniopropane sulfonate (CHAPS), 0.5% Triton X-100, 1% dithiothreitol, and 2% protease inhibitor cocktail] using glass–glass homogenizers. Tissues were allowed to sit at room temperature for 1 hr after homogenization to allow further solubilization and denaturation of proteins. Insoluble proteins were removed by centrifugation (100,000 × g for 1 hr).
The tough, bony structure of primate turbinates dictated the use of modified procedures for protein recovery. Briefly, after the incubation, the incubation medium was transferred to a clean centrifuge tube. The tissue was washed with 5% dextrose to remove buffer salts, and fractions of the aqueous phase were combined. These samples were centrifuged to recover any tissue/cells removed, and the pellets were combined with the remaining cartilage containing the epithelium. Lysis buffer containing protease inhibitors was added, and the tube was agitated on a rotating shaker to dislodge cells.
Comparison of covalent binding levels
A portion (~ 1 mg protein) of each sample (from rat or monkey nasal epithelium) incubated with 14C-naphthalene was dialyzed in 0.1% sodium dodecyl sulfate, 1 mM disodium EDTA at 4°C [3,500 molecular weight (MW) cutoff Slide-A-Lyzers; Pierce, Rockford, IL]. The dialysis solution was replaced with fresh solution until the counts per minute was less than twice the background level. Finally, we determined the specific activity of adducted protein for each sample by scintillation counting and a protein assay (Bradford 1976).
Separation and identification of adducted protein
Procedures for the separation, localization, and identification of adducted proteins have been described in detail previously (Lin et al. 2005). Briefly, samples were not pooled, and separate gels were run for each incubation with labeled (for imaging) and unlabeled (for identification) naphthalene. Proteins were resolved by two-dimensional (2D) gel electrophoresis on 18-cm narrow-range pI (isoelectric point) strips for 4.5–5.5 and 5.5–6.7 pI. Strips were rehydrated with 300 μg protein for rat samples or 600 μg protein for monkey samples. Gels containing nonradioactive samples were silver stained and imaged on a 16-bit gray-scale scanner. 14C-Labeled sample gels were electroblotted to Sequi-Blot polyvinylidene fluoride membrane (0.2 μm; Bio-Rad), and these were placed against storage phosphor screens for 60 days. The patterns of 14C-labeled adducted proteins on the 2D gels were visualized using the GE Typhoon imager (GE Health Care). After excising protein spots of interest from nonradioactive gels, the gel plugs were destained, washed, and incubated in 1:1 (vol/vol) acetonitrile:50 mM ammonium bicarbonate followed by 100% acetonitrile. Tryptic digestion and matrix-assisted laser desorption ionization (MALDI) tandem mass spectroscopy (MS/MS) identification was carried out as described by Lin et al. (2006).
We used GPS Explorer Workstation software (version 3.5; Applied Biosystems, Foster City, CA) and MASCOT (Matrix Sciences, Boston, MA) to search protein databases in the International Protein Index (IPI) rat database (http://www.ebi.ac.uk/IPI/), the National Center for Biotechnology Information primate database (NCBI; http://www.ncbi.nlm.nih.gov/), and the UniProt database (http://www.ebi.ac.uk/uniprot/). Databases were searched for a precursor tolerance of 150 ppm, MS/MS fragment tolerance of 0.2 Da, a maximum of two missed cleavages, and variable modifications for carbamidomethylation and methionine oxidation. An identification with a probability-based MOWSE score > 60, obtained through MASCOT, was considered statistically significant (p < 0.05).
Results
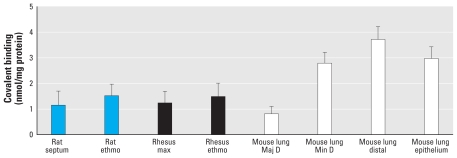
The specific activities of adducted proteins (nanomoles bound per milligram protein) generated in in vitro incubations of nasal epithelium from rats or rhesus macaques varied from 1.0 to 1.5 nmol/mg protein; we noted no statistically significant difference in the data obtained from rat olfactory and septal epithelium compared with rhesus ethmoturbinates and maxilloturbinates (Figure 1). Compared with incubations of target airway subcompartments (minor daughter and distal airway) from the mouse (Cho et al. 1994), the overall levels of adduct generated in vitro are slightly lower in rat and monkey nasal epithelial mucosal incubations.
Figure 1.
Covalent reactive metabolite binding to protein in rat, rhesus, and mouse tissue. Abbreviations: ethmo, ethmoturbinate; Maj D, major daughter; max, maxilloturbinate; Min D, minor daughter. Fresh samples of rat septum (n = 3), rat olfactory ethmoturbinate (n = 3), rhesus maxilloturbinate (n = 8), and rhesus ethmoturbinate (n = 8) were incubated with 14C-naphthalene (250 μM) for 2 hr at 37°C for determination of the levels of covalent reactive metabolite binding to protein. Values are mean ± SD. Reference values for tissue subcompartments of mouse lung are from Cho et al. (1994).
Representative images from the phosphorimaging screens and complementary silver-stained gels/phosphor images indicating the spot locations chosen for MALDI dual time of flight (TOF/TOF) MS analysis are available in Supplemental Material, Figures 1 and 2 (doi:10.1289/ehp.0901333). In general, more adducts were apparent on the images from incubations of rat nasal epithelium than on those from rhesus macaques, but the patterns of major adducts were similar. Spots were matched using the alignment function of Progenesis SameSpots and Progenesis PG220 (version 2006; Nonlinear Dynamics, Durham, NC) matches were manually verified. For identification, we used only those features that could be aligned with the storage phosphor images and selected from the gel with high confidence.
We identified a large number of adducted proteins after incubation of rat nasal olfactory epithelium with naphthalene (Table 1). These included a number of redundant identifications of structural proteins (actin and tubulin), catalytic proteins (some of which are involved in energy metabolism such as ATP synthase), and a number of proteins involved in the unfolded protein response, including one of the master regulators of that response, glucose regulated protein (heat shock 70 kDa protein 5). In contrast, fewer features were identified from the gels of monkey ethmoturbinates, representing only three nonredundant proteins (Table 2). Two of these were structural proteins (actin and tubulin), and the third was a protease inhibitor with a myriad of functions.
Table 1.
Adducted proteins identified from incubation of rat olfactory ethmoturbinate with 14C-naphthalene.
| Gel spot | Protein name (abbreviated) | MOWSE scorea | Peptide countb | Protein score CI%c | Accession no.d | Protein MW (Da) | Protein pI | Summary of function |
|---|---|---|---|---|---|---|---|---|
| N1 | Sorcin (P30626) | 133 | 5 | 100 | SORCN CRILOe | 22017.4 | 5.32 | Calcium-binding protein |
| N4 | Golgi autoantigen | 90 | 24 | 99.951 | GI: 808869f | 372014.8 | 4.96 | May participate in forming intercisternal cross bridges of the Golgi complex |
| N7 | pyruvate dehydrogenase | 69 | 7 | 99.596 | ODPB_RATe | 39,336 | 5.94 | Glycolysis |
| N8 | mu-crystallin | 135 | 4 | 100 | IPI00214448 | 33704.1 | 5.34 | Ammonia elimination |
| N9 | pyrophosphatase | 67 | 7 | 99.283 | IPI00371957 | 33206.3 | 5.26 | Signal transduction |
| N12 | creatinine kinase | 543 | 20 | 100 | IPI00470288 | 42983.4 | 5.39 | Energy transduction |
| N13 | Sec 1413 protein | 131 | 6 | 100 | IPI00208939 | 46453.5 | 5.64 | Hydrophobic ligand binding |
| N19 | creatinine kinase | 299 | 10 | 100 | IPI00470288 | 42983.4 | 5.39 | Energy transduction |
| N23 | actin | 259 | 10 | 100 | IPI00369618 | 59162.6 | 5.67 | Structural |
| N24 | eukaryotic translation initiation factor 4A1 | 129 | 14 | 100 | IPI00369618 | 46352.6 | 5.32 | Protein synthesis initiation |
| N25 | beta actin | 447 | 10 | 100 | GI: 62897625f | 41737.8 | 5.37 | Structural |
| N26 | actin, cytoplasmic | 420 | 15 | 100 | IPI00767505 | 42108.9 | 5.31 | Structural |
| N27 | actin | 753 | 17 | 100 | IPI00194087 | 42,334 | 5.23 | Structural |
| N29 | actin, cytoplasmic | 198 | 6 | 100 | IPI00765011 | 59162.6 | 5.67 | Structural |
| N32 | actin, cytoplasmic | 167 | 4 | 100 | IPI00765011 | 59162.6 | 5.67 | Structural |
| N33 | actin, cytoplasmic | 186 | 5 | 100 | IPI00765011 | 59162.6 | 5.67 | Structural |
| N41 | ATP synthase | 919 | 21 | 100 | IPI00551812 | 56318.5 | 5.19 | ATP catabolism |
| N43 | ATP synthase | 160 | 17 | 100 | IPI00551812 | 56318.5 | 5.19 | ATP catabolism |
| N47 | heat shock protein 1A (chaperonin) (Hsp 60) | 220 | 27 | 100 | IPI00339148 | 61088.4 | 5.91 | Stress-induced chaperone; protein folding |
| N51 | heat shock 70 kDa 9 A (predicted) | 419 | 9 | 100 | IPI00363265 | 74,097 | 5.97 | Protein folding; protein export; binds ATP, nucleotides, and proteins |
| N53 | heat shock 70 kDa 9 A (predicted) | 64 | 5 | 98.428 | IPI00858386 | 74098.9 | 5.97 | Protein folding; protein export; binds ATP, nucleotides, and proteins |
| N55 | Vomeromodulin | 87 | 8 | 99.992 | IPI00209782 | 79527.3 | 5.92 | Putative pheromone and odorant transporter |
| N56 | heat shock 70 kDa, protein 5 | 621 | 15 | 100 | IPI00206624 | 72473.5 | 5.07 | Assembly of multimeric protein complexes |
| N57 | heat shock 70 kDa, protein 5 | 896 | 27 | 100 | IPI00206624 | 72473.5 | 5.07 | Assembly of multimeric protein complexes |
| N61 | tyrosine 3-mono-oxygenase Tryptophan 5 | 243 | 12 | 100 | IPI00230835 | 28,456 | 4.8 | Adapter protein; signal transduction |
| N63 | tubulin | 485 | 12 | 100 | IPI00189795 | 50787.9 | 4.94 | Structural |
| N64 | tubulin | 174 | 16 | 100 | IPI00339167 | 50119.6 | 4.94 | Structural |
| N65 | tubulin | 148 | 14 | 100 | IPI00339167 | 50119.6 | 4.94 | Structural |
| N66 | tubulin | 258 | 23 | 100 | IPI00400573 | 49,769 | 4.79 | Structural |
| N67 | tubulin | 464 | 24 | 100 | IPI00197579 | 49,639 | 4.78 | Structural |
| N68 | protein disulfide isomerase | 272 | 17 | 100 | IPI00198887 | 56915.7 | 4.82 | Cell redox homeostasis |
| N69 | protein disulfide isomerase | 139 | 15 | 100 | IPI00198887 | 56915.7 | 4.82 | Cell redox homeostasis |
| N70 | protein disulfide isomerase | 227 | 25 | 100 | IPI00198887 | 56915.7 | 4.82 | Cell redox homeostasis |
| N73 | ubiquinol-cytochrome c reductase core protein 1 | 162 | 16 | 100 | IPI00471577 | 57815.4 | 5.57 | Mitochondrial electron transport |
| N74 | ubiquinol-cytochrome c reductase core protein 1 | 78 | 19 | 99.939 | IPI00471577 | 52815.4 | 5.57 | Mitochondrial electron transport |
| N75 | chloride intracellular channel 1 | 212 | 14 | 100 | IPI00421995 | 26963.8 | 5.09 | Macrophage activation |
| N76 | tubulin | 166 | 12 | 100 | IPI0018975 | 5103.6 | 4.94 | Structural |
| N77 | tubulin | 126 | 19 | 100 | IPI00400573 | 49,769 | 4.79 | Structural |
The probability-based MOWSE score from MASCOT.
Peptides used for matching within the database.
Reflects the probability that this same match would be made in a different database.
Accession numbers are from the IPI rat database except where noted.
From the Swiss-Prot database searching mammals.
From the NCBI database searching mammals.
Table 2.
Adducted proteins identified from incubations of monkey ethmoturbinate with 14C-naphthalene.
| Gel spot | Protein name (abbreviated) | MOWSE scorea | Peptide countb | Protein score CIc | Accession no.d | Protein MW (Da) | Protein pI | Function |
|---|---|---|---|---|---|---|---|---|
| M1 | actin | 333 | 22 | 100 | P60709 | 40978.4 | 5.56 | Structural |
| M2 | actin | 628 | 25 | 100 | GI: 62897625 | 41737.8 | 5.37 | Structural |
| M3 | actin | 526 | 23 | 100 | P60709 | 40978.4 | 5.56 | Structural |
| M4 | tubulin | 95 | 17 | 99.992 | GI: 114627728 | 7398.9 | 4.72 | Structural |
| M5 | actin | 107 | 11 | 100 | P68133 | 37799.8 | 5.39 | Structural |
| M6 | actin | 469 | 18 | 100 | Q8WvW5 | 40477.2 | 5.78 | Structural |
| M7 | actin | 690 | 23 | 100 | GI: 4501883 | 41981.8 | 5.23 | Structural |
| M8 | actin | 479 | 20 | 100 | GI: 4501883 | 41981.8 | 5.23 | Structural |
| M9 | actin | 181 | 13 | 100 | GI: 4885049 | 41981.9 | 5.23 | Structural |
| M10 | actin | 147 | 10 | 100 | GI: 149758067 | 37799.8 | 5.39 | Structural |
| M11 | fibrinogen | 162 | 12 | 100 | GI: 109075971 | 36492.6 | 5.61 | Various |
| M12 | protease inhibitor | 514 | 22 | 100 | GI: 109084715 | 46669.9 | 5.55 | Various |
| M13 | actin | 528 | 23 | 100 | GI: 15277503 | 40194.1 | 5.55 | Structural |
| M14 | actin | 588 | 26 | 100 | GI: 62897671 | 41693.7 | 5.29 | Structural |
| M15 | tubulin | 455 | 25 | 100 | GI: 18088719 | 49639.9 | 4.75 | Structural |
| M16 | tubulin | 253 | 20 | 100 | GI: 23958133 | 49808 | 4.83 | Structural |
| M17 | actin | 226 | 15 | 100 | GI: 14250401 | 40978.9 | 5.56 | Structural |
| M18 | tubulin | 338 | 20 | 100 | GI: 114627728 | 47398.9 | 4.72 | Structural |
| M19 | actin | 491 | 19 | 100 | GI: 4885049 | 41991.9 | 5.23 | Structural |
| M20 | tubulin | 314 | 15 | 100 | GI: 109096498 | 42214.1 | 4.94 | Structural |
| M21 | protease inhibitor | 106 | 11 | 100 | GI: 109069435 | 42214.1 | 5.08 | Various |
The probability-based MOWSE score from MASCOT.
Peptides used for matching within the database.
Reflects the probability that this same match would be made in a different database.
From the NCBI database searching mammals.
Discussion
The chronic bioassays conducted by the National Toxicology Program on the potential carcinogenicity of naphthalene in mice (Abdo et al. 1992) and rats (Abdo et al. 2001) have raised concerns about the possible effects of this compound on human health. In mice, an increased incidence of bronchioloalveolar adenoma was observed in females at the highest concentration tested (30 ppm) (Abdo et al. 1992), and in rats, the nasal olfactory epithelium was a prominent target after 105 weeks of exposure (Abdo et al. 2001). Long et al. (2003) observed dose-dependent increases in adenomas of the respiratory epithelium and a significant increase in olfactory epithelial neuroblastomas in male and female rats, respectively. In the olfactory epithelium, the incidence of hyperplasia and chronic inflammation was nearly 100%, even at the lowest concentration tested (10 ppm). Overall, the high degree of hyperplastic and inflammatory effects and the relative susceptibility of areas of the nasal epithelium after long-term exposures correlate well with the susceptible sites for acute toxicity (Lee et al. 2005) and raise the possibility that the tumorigenic effects of naphthalene observed in animals are related to repeated cycles of injury and repair (North et al. 2008).
The published assessments by regulatory bodies on the potential carcinogenicity of naphthalene in humans differ (e.g., International Agency for Research on Cancer 2002; National Toxicology Program 2004). Although the toxicologic data in animals are very clear, human epidemiologic studies in populations of exposed workers are uninformative. In most short-term mutagenicity assays, naphthalene is negative (reviewed by Brusick 2008). Recent evidence for the formation of DNA adducts comes both from in vitro studies with naphthalene metabolites (Saeed et al. 2007) and from skin-painting studies using both naphthalene and naphthalene metabolites (Saeed et al. 2009). The skin-painting studies demonstrated the presence of both depurinating and stable adducts, suggesting that this may be a possible mechanism for tumorigenesis associated with naphthalene in rodent models.
Previous studies demonstrating 10- to 100-fold differences in the rates of pulmonary microsomal formation of glutathione conjugates of naphthalene between rodents and human/nonhuman primates (Buckpitt et al. 1992; Buckpitt and Bahnson 1986) and in dissected airways of rodents and nonhuman primates (Boland et al. 2004; Buckpitt et al. 1995) suggested that humans would be considerably less susceptible to naphthalene than are rodents. Although the dramatic differences in susceptibility to naphthalene of airway epithelium in the mouse and rat appear to be related to metabolism of naphthalene by CYP2F, recent work showing that several human CYPs can metabolize naphthalene with high efficiency suggests that other CYP enzymes may catalyze the turnover of this substrate in humans (Cho et al. 2006; Fukami et al. 2008). In particular, CYP2A13, which has been reported in both lung and nasal epithelium of humans (for review, see Ding and Kaminsky 2003), shows a Km of 36 μM and high catalytic efficiency with naphthalene. In addition, our finding of similar levels of covalent protein binding in nasal epithelium of the rodent and monkey (Figure 1), as well as observations that some (but not all) of the proteins adducted by reactive metabolites of naphthalene are the same in dissected airways and nasal epithelium of rodents and nonhuman primates (Lin et al. 2005, 2006; see also Tables 1 and 2), supports the need for additional studies on the possible sensitivity of humans to naphthalene exposure.
The concept that posttranslational modification of proteins by electrophiles begins a series of events leading to cellular necrosis was proposed by Brodie et al. (1971) some 40 years ago, and was supported by studies showing correlations between overall electrophile protein adduct formation and toxicity (for a recent review, see Hanzlik et al. 2009). The advent of new MS techniques coupled with 2D gel electrophoresis has resulted in the identification of numerous protein targets for cytotoxic agents that require metabolic activation; a current database of these targets shows many commonalities (Fang et al. 2009). Despite these efforts, a final unifying hypothesis tying the adduction of specific proteins with toxicity has not emerged, and additional work is needed to answer several questions. In those cases where adduction occurs to an abundant protein, is this protein simply acting as a sink to trap electrophiles? Does the formation of an adduct with an individual protein alter the function of that protein directly, or is the deleterious effect mediated by protein-interacting partners? These will clearly be challenging questions but, if solved, could greatly enhance our ability to more quickly and precisely assess the human risk of exposure to agents that undergo metabolic activation to protein-reactive electrophiles.
As noted above, much of the progress in identifying proteins targeted by reactive metabolites has derived from the application of 2D gel electrophoresis and more rapid MS-based identifications. Although 2D gel electrophoresis arguably provides the greatest resolving power for whole proteins of any separations technique available, there is still the potential of selecting a spot that contains two unseparated proteins, one that is low abundance and adducted and one that is much higher abundance and identified by MS. Although we cannot exclude this as a possibility in the present study, we used IPG strips with very narrow ranges to lessen the potential for misidentification. In only one case that we are aware of—with thiobenzamide—were the investigators able to show that the features removed from the gel were adducted by using isotope ratios with equal quantities of hydrogen/deuterium-labeled substrate (Ikehata et al. 2008). This was likely possible only because the adduct levels generated from thiobenzamide are considerably greater than those for other metabolically activated toxicants.
In the present study, adducts with the structural proteins actin and tubulin were isolated from both rat and monkey nasal epithelial incubations. These are high-abundance proteins in the cell that have been identified in several other studies as common targets for other reactive metabolites, including butylated hydroxytoluene (BHT) (Meier et al. 2005), benzene (Williams et al. 2002), thiobenzamide (Ikehata et al. 2008), and the model thiol-reactive electrophiles biotinyl-iodoacetamidyl-3,6-dioxaoctanediamine and 1-biotinamido-4-(4′[maleidoethylcyclohexane]-carboxyamido)butane (Dennehy et al. 2006). The formation of covalent adducts with actin and tubulin is not generally considered to be a critical step in the pathway to cellular injury because the specific activity of these adducted proteins (nanomoles bound per nanomole of protein) is likely to be quite low. However, one of the earliest signs of cellular disruption in naphthalene-treated mice involves the formation of apical membrane blebs and apparent disruption of cytoplasmic filaments (Van Winkle et al. 1999). Similar signs of disruption of cytoskeletal elements have been noted in airway epithelial cells after administration of diethyl maleate, but these changes resolved and did not lead to frank necrosis (Phimister et al. 2005). This is consistent with previous work showing that S-glutathionylation of cysteine-374 of actin reduces the rate of polymerization of this structural molecule (Dalle-Donne et al. 2003) and that glutathionylation of this protein leads to decreased affinity of its partner protein tropomyosin (Chen and Ogut 2006). Both naphthalene and diethyl maleate substantially deplete glutathione levels in airway epithelium, which may result in increased levels of glutathionylated actin. Reactive metabolites from naphthalene produce a double hit on actin and tropomyosin, resulting in both thiolation reactions (which are reversible) and covalent arylation reactions (which presumably are not). Ongoing work in our laboratory is evaluating the protein targets of diethyl maleate as a mechanism for distinguishing adducts that may be critical to cellular injury from those that are not.
We observed a potentially important group of protein targets in nasal olfactory epithelial incubations from the rat but not from the monkey; these proteins are responsible for protein folding and repair in the cell. Glutathione depletion is a necessary but insufficient prelude to naphthalene cytotoxicity. The loss of this key thiol leaves the cell vulnerable to intracellular reactive oxygen species that, with diethyl maleate, can be corrected by chaperones, protein disulfide isomerase, peroxiredoxins, and possibly several other proteins involved in antioxidant protection and protein folding. With naphthalene, several of these proteins, including the master regulator of the unfolded protein response, BiP/GRP78 (78 kDa glucose- regulated protein) (Townsend 2007), as well as the heat shock proteins HSP60 and HSP70, are adducted, thus potentially altering their ability to repair proteins undergoing thiol oxidation and unfolding during the early stages of naphthalene toxicity. This is consistent with findings showing marked up-regulation of the HSPs in mouse lung at early times after naphthalene treatment (Williams et al. 2003). An interesting but unproven link in possible mechanisms comes from recent work demonstrating that proteins involved in the unfolded protein response are substantially up-regulated in smokers compared with former smokers and nonsmokers (Kelsen et al. 2008).
The present study provides a list of potential protein targets and shows that some of these are similar in nasal epithelium of the rat (a known target for naphthalene toxicity) and that of the rhesus macaque. We derived several other adducted proteins from monkey nasal epithelial incubations, but these yielded unreliable identifications. We are working on methods for concentrating adducted proteins to provide greater sensitivity for these analyses. In the future, it will be important to gain additional insight regarding critical versus noncritical protein adduction. Part of this information will come from studies such as that published by Meier et al. (2007), which showed that metabolites of BHT alkylate key antioxidant enzymes and that the alkylation resulted in a decrement in enzyme function that, in turn, was consistent with the biological responses after BHT treatment.
In the present study we focused on protein adducts that are most likely related to acute cellular injury. Whether repeated cycles of injury and repair with subsequent hyperplasia are related to naphthalene tumorigenicity will need additional study. Finally, the possibility that exposure to naphthalene and other similar, volatile polycyclic aromatic hydrocarbons (PAHs) could produce lung disease other than cancer should be explored. In this regard, limited studies have reported correlations between serum levels of naphthalene and pyrene, but not other PAHs, with asthma incidence in children (Al-Daghri 2008). Moreover, there is a clear association of exposure to traffic-related pollution and asthma (Kim et al. 2005), and naphthalene is the most prevalent PAH in this mixture (Lu et al. 2005). Recent work showing that polymorphisms that result in alterations in enzymatic activities for two key enzymes involved in the metabolism of PAHs (microsomal epoxide hydrolase and glutathione transferase pi) lead to increased incidence of asthma in children living near freeways (Salam et al. 2007) provide an interesting but unsubstantiated link between PAHs in motor vehicle exhaust and asthma. The understanding of target proteins and their relationship to the pathology from naphthalene in animals should improve our ability to assess the possible risks of this compound in exposed human populations.
Conclusion
This study shows that the amounts of covalent adduct generated from naphthalene in incubations of rhesus nasal epithelium are similar to those formed using rat olfactory and septal nasal epithelium, two sites known to be susceptible to the cytotoxicity associated with naphthalene. Many of the adducted proteins identified are the same as those observed in incubations with other target tissues. Our results are consistent with the view that nasal epithelium of the rhesus macaque may be susceptible to naphthalene cytotoxicity.
Footnotes
Supplemental Material is available online (doi:10.1289/ehp.0901333 via http://dx.doi.org/).
We thank W. Jewell for assistance with adducted protein identifications.
This work was supported the Electric Power Research Institute (agreement EP-P25766/C12455) and by the grants 04311 and 04699 from the National Institute of Environmental Health Sciences. Nasal tissues were from the California Regional Primate Research Center, which is supported by grant 0169 from the National Center for Research Resources.
References
- Abdo K, Eustis S, McDonald M, Joiken M, Adkins B, Haseman J. Naphthalene: a respiratory tract toxicant and carcinogen for mice. Inhal Toxicol. 1992;4:393–409. [Google Scholar]
- Abdo K, Grumbein S, Chou BJ, Herbert R. Toxicity and carcinogenicity study in F344 rats following 2 years of whole-body exposure to naphthalene vapors. Inhal Toxicol. 2001;13:931–950. doi: 10.1080/089583701752378179. [DOI] [PubMed] [Google Scholar]
- Al-Daghri NM. Serum polycyclic aromatic hydrocarbons among children with and without asthma: correlation to environmental and dietary factors. Int J Occup Med Environ Health. 2008;21:211–217. doi: 10.2478/v10001-008-0021-0. [DOI] [PubMed] [Google Scholar]
- Baldwin RM, Shultz MA, Buckpitt AR. Bioactivation of the pulmonary toxicants naphthalene and 1-nitronaphthalene by rat CYP2F4. J Pharmacol Exp Ther. 2005;312:857–865. doi: 10.1124/jpet.104.075440. [DOI] [PubMed] [Google Scholar]
- Boland B, Lin CY, Morin D, Miller L, Plopper CG, Buckpitt A. Site specific metabolism of naphthalene and 1-nitronaphthalene in dissected airways of rhesus macaques. J Pharmacol Exp Ther. 2004;310:546–554. doi: 10.1124/jpet.103.063669. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brodie BB, Reid WD, Cho AK, Sipes G, Krishna G, Gillette JR. Possible mechanism of liver necrosis caused by aromatic organic compounds. Proc Natl Acad Sci USA. 1971;68:160–164. doi: 10.1073/pnas.68.1.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brusick D. Critical assessment of the genetic toxicity of naphthalene. Regul Toxicol Pharmacol. 2008;51:S37–S42. doi: 10.1016/j.yrtph.2007.08.013. [DOI] [PubMed] [Google Scholar]
- Buckpitt A, Bahnson L. Naphthalene metabolism by human lung microsomal enzymes. Toxicology. 1986;41:333–341. doi: 10.1016/0300-483x(86)90186-1. [DOI] [PubMed] [Google Scholar]
- Buckpitt A, Boland B, Isbell M, Morin D, Shultz M, Baldwin R, et al. Naphthalene-induced respiratory tract toxicity: metabolic mechanisms of toxicity. Drug Metab Rev. 2002;34:791–820. doi: 10.1081/dmr-120015694. [DOI] [PubMed] [Google Scholar]
- Buckpitt A, Buonarati M, Avey LB, Chang AM, Morin D, Plopper CG. Relationship of cytochrome P450 activity to Clara cell cytotoxicity. II. Comparison of stereoselectivity of naphthalene epoxidation in lung and nasal mucosa of mouse, hamster, rat and rhesus monkey. J Pharmacol Exp Ther. 1992;261:364–372. [PubMed] [Google Scholar]
- Buckpitt A, Chang AM, Weir A, Van Winkle L, Duan X, Philpot R, et al. Relationship of cytochrome P450 activity to Clara cell cytotoxicity. IV. Metabolism of naphthalene and naphthalene oxide in microdissected airways from mice, rats, and hamsters. Mol Pharmacol. 1995;47:74–81. [PubMed] [Google Scholar]
- Chen FC, Ogut O. Decline of contractility during ischemia-reperfusion injury: actin glutathionylation and its effect on allosteric interaction with tropomyosin. Am J Physiol Cell Physiol. 2006;290:C719–C727. doi: 10.1152/ajpcell.00419.2005. [DOI] [PubMed] [Google Scholar]
- Cho M, Chichester C, Morin D, Plopper C, Buckpitt A. Covalent interactions of reactive naphthalene metabolites with proteins. J Pharmacol Exp Ther. 1994;269:881–889. [PubMed] [Google Scholar]
- Cho TM, Rose RL, Hodgson E. In vitro metabolism of naphthalene by human liver microsomal cytochrome P450 enzymes. Drug Metab Dispos. 2006;34:176–183. doi: 10.1124/dmd.105.005785. [DOI] [PubMed] [Google Scholar]
- Dalle-Donne I, Giustarini D, Rossi R, Colombo R, Milzani A. Reversible S-glutathionylation of Cys374 regulates actin filament formation by inducing structural changes in the actin molecule. Free Radic Biol Med. 2003;34:23–32. doi: 10.1016/s0891-5849(02)01182-6. [DOI] [PubMed] [Google Scholar]
- Dennehy MK, Richards KA, Wernke GR, Shyr Y, Liebler DC. Cytosolic and nuclear protein targets of thiol-reactive electrophiles. Chem Res Toxicol. 2006;19:20–29. doi: 10.1021/tx050312l. [DOI] [PubMed] [Google Scholar]
- Ding X, Kaminsky LS. Human extrahepatic cytochromes P450: function in xenobiotic metabolism and tissue-selective chemical toxicity in the respiratory and gastrointestinal tracts. Annu Rev Pharmacol Toxicol. 2003;43:149–173. doi: 10.1146/annurev.pharmtox.43.100901.140251. [DOI] [PubMed] [Google Scholar]
- Dodd DE, Gross EA, Miller RA, Wong BA. Nasal olfactory epithelial lesions in F344 and SD rats following 1- and 5-day inhalation exposure to naphthalene vapor. Int J Toxicol. 2010;29(2):175–184. doi: 10.1177/1091581809357955. [DOI] [PubMed] [Google Scholar]
- Fang J, Koen Y, Hanzlik R. Bioinformatic analysis of xenobiotic reactive metabolite target proteins and their interacting partners. BMC Chemical Biology. 2009;9:5. doi: 10.1186/1472-6769-9-5. [Online 12 June 2009] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukami T, Katoh M, Yamazaki H, Yokoi T, Nakajima M. Human cytochrome p450 2A13 efficiently metabolizes chemicals in air pollutants: naphthalene, styrene, and toluene. Chem Res Toxicol. 2008;21:720–725. doi: 10.1021/tx700325f. [DOI] [PubMed] [Google Scholar]
- Green T, Toghill A, Foster JR. The role of cytochromes P-450 in styrene induced pulmonary toxicity and carcinogenicity. Toxicology. 2001;169:107–117. doi: 10.1016/s0300-483x(01)00488-7. [DOI] [PubMed] [Google Scholar]
- Hanzlik RP, Fang J, Koen YM. Filling and mining the reactive metabolite target protein database. Chem Biol Interact. 2009;179:38–44. doi: 10.1016/j.cbi.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikehata K, Duzhak TG, Galeva NA, Ji T, Koen YM, Hanzlik RP. Protein targets of reactive metabolites of thiobenzamide in rat liver in vivo. Chem Res Toxicol. 2008;21:1432–1442. doi: 10.1021/tx800093k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Agency for Research on Cancer. Naphthalene. IARC Monogr Eval Carcinog Risks Hum. 2002;82:367–436. [Google Scholar]
- Kelsen SG, Duan X, Ji R, Perez O, Liu C, Merali S. Cigarette smoke induces an unfolded protein response in the human lung: a proteomic approach. Am J Respir Cell Mol Biol. 2008;38:541–550. doi: 10.1165/rcmb.2007-0221OC. [DOI] [PubMed] [Google Scholar]
- Kim JH, Kim JK, Son BK, Oh JE, Lim DH, Lee KH, et al. Effects of air pollutants on childhood asthma. Yonsei Med J. 2005;46:239–244. doi: 10.3349/ymj.2005.46.2.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanza DL, Code E, Crespi CL, Gonzalez FJ, Yost GS. Specific dehydrogenation of 3-methylindole and epoxidation of naphthalene by recombinant human CYP2F1 expressed in lymphoblastoid cells. Drug Metab Dispos. 1999;27:798–803. [PubMed] [Google Scholar]
- Lee MG, Phimister A, Morin D, Buckpitt A, Plopper C. In situ naphthalene bioactivation and nasal airflow cause region-specific injury patterns in the nasal mucosa of rats exposed to naphthalene by inhalation. J Pharmacol Exp Ther. 2005;314:103–110. doi: 10.1124/jpet.105.084517. [DOI] [PubMed] [Google Scholar]
- Li Z, Sandau CD, Romanoff LC, Caudill SP, Sjodin A, Needham LL, et al. Concentration and profile of 22 urinary polycyclic aromatic hydrocarbon metabolites in the US population. Environ Res. 2008;107:320–331. doi: 10.1016/j.envres.2008.01.013. [DOI] [PubMed] [Google Scholar]
- Lin CY, Boland BC, Lee YJ, Salemi MR, Morin D, Miller LA, et al. Identification of proteins adducted by reactive metabolites of naphthalene and 1-nitronaphthalene in dissected airways of rhesus macaques. Proteomics. 2006;6:972–982. doi: 10.1002/pmic.200500170. [DOI] [PubMed] [Google Scholar]
- Lin CY, Isbell MA, Morin D, Boland BC, Salemi MR, Jewell WT, et al. Characterization of a structurally intact in situ lung model and comparison of naphthalene protein adducts generated in this model vs lung microsomes. Chem Res Toxicol. 2005;18:802–813. doi: 10.1021/tx049746r. [DOI] [PubMed] [Google Scholar]
- Long PH, Herbert RA, Peckham JC, Grumbein SL, Shackelford CC, Abdo K. Morphology of nasal lesions in F344/N rats following chronic inhalation exposure to naphthalene vapors. Toxicol Pathol. 2003;31:655–664. doi: 10.1080/01926230390242016. [DOI] [PubMed] [Google Scholar]
- Lu R, Wu J, Turco RP, Winer AM, Atkinson R, Arey J, et al. Naphthalene distributions and human exposure in Southern California. Atmos Environ. 2005;39:489–507. [Google Scholar]
- Meier BW, Gomez JD, Kirichenko OV, Thompson JA. Mechanistic basis for inflammation and tumor promotion in lungs of 2,6-di-tert-butyl-4-methylphenol-treated mice: electrophilic metabolites alkylate and inactivate antioxidant enzymes. Chem Res Toxicol. 2007;20:99–107. doi: 10.1021/tx060214f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier BW, Gomez JD, Zhou A, Thompson JA. Immunochemical and proteomic analysis of covalent adducts formed by quinone methide tumor promoters in mouse lung epithelial cell lines. Chem Res Toxicol. 2005;18:1575–1585. doi: 10.1021/tx050108y. [DOI] [PubMed] [Google Scholar]
- National Toxicology Program. Naphthalene, CAS No. 91-20-3. Report on Carcinogens. Eleventh Edition. 2004. [accessed 12 August 2009]. Available: http://ntp.niehs.nih.gov/ntp/roc/eleventh/profiles/s116znph.pdf.
- Nhamburo PT, Kimura S, McBride OW, Kozak CA, Gelboin HV, Gonzalez FJ. The human CYP2F gene subfamily: identification of a cDNA encoding a new cytochrome P450, cDNA-directed expression, and chromosome mapping. Biochemistry. 1990;29:5491–5499. doi: 10.1021/bi00475a012. [DOI] [PubMed] [Google Scholar]
- North DW, Abdo KM, Benson JM, Dahl AR, Morris JB, Renne R, et al. A review of whole animal bioassays of the carcinogenic potential of naphthalene. Regul Toxicol Pharmacol. 2008;51:S6–S14. doi: 10.1016/j.yrtph.2007.09.022. [DOI] [PubMed] [Google Scholar]
- Odum J, Foster JR, Green T. A mechanism for the development of Clara cell lesions in the mouse lung after exposure to trichloroethylene. Chem Biol Interact. 1992;83:135–153. doi: 10.1016/0009-2797(92)90042-j. [DOI] [PubMed] [Google Scholar]
- Phimister AJ, Williams KJ, Van Winkle LS, Plopper CG. Consequences of abrupt glutathione depletion in murine Clara cells: ultrastructural and biochemical investigations into the role of glutathione loss in naphthalene cytotoxicity. J Pharmacol Exp Ther. 2005;314:506–513. doi: 10.1124/jpet.105.084533. [DOI] [PubMed] [Google Scholar]
- Plopper CG, Chang A, Pang A, Buckpitt A. Use of microdissected airways to better define metabolism and cytotoxicity in murine bronchiolar epithelium. Exp Lung Res. 1991;17:181–196. doi: 10.3109/01902149109064411. [DOI] [PubMed] [Google Scholar]
- Plopper CG, Macklin J, Nishio S, Hyde D, Buckpitt A. Relationship of cytochrome P450 activity to Clara cell cytotoxicity. III. Morphometric comparison of changes in the epithelial populations of terminal bronchioles and lobar bronchi in mice, hamsters and rats after parenteral administration of naphthalene. Lab Invest. 1992;67:553–565. [PubMed] [Google Scholar]
- Saeed M, Higginbotham S, Gaikwad N, Chakravarti D, Rogan E, Cavalieri E. Depurinating naphthalene-DNA adducts in mouse skin related to cancer initiation. Free Radic Biol Med. 2009;47:1075–1081. doi: 10.1016/j.freeradbiomed.2009.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed M, Higginbotham S, Rogan E, Cavalieri E. Formation of depurinating N3adenine and N7guanine adducts after reaction of 1,2-naphthoquinone or enzyme-activated 1,2-dihydroxynaphthalene with DNA. Implications for the mechanism of tumor initiation by naphthalene. Chem Biol Interact. 2007;165:175–188. doi: 10.1016/j.cbi.2006.12.007. [DOI] [PubMed] [Google Scholar]
- Salam MT, Lin PC, Avol EL, Gauderman WJ, Gilliland FD. Microsomal epoxide hydrolase, glutathione S-transferase P1, traffic and childhood asthma. Thorax. 2007;62:1050–1057. doi: 10.1136/thx.2007.080127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shultz MA, Choudary PV, Buckpitt AR. Role of murine cytochrome P-450 2F2 in metabolic activation of naphthalene and metabolism of other xenobiotics. J Pharmacol Exp Ther. 1999;290:281–288. [PubMed] [Google Scholar]
- Townsend DM. S-Glutathionylation: indicator of cell stress and regulator of the unfolded protein response. Mol Interv. 2007;7:313–324. doi: 10.1124/mi.7.6.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Winkle L, Johnson Z, Nishio S, Brown C, Plopper CG. Early events in naphthalene-induced acute Clara cell toxicity. Comparison of membrane permeability and ultrastructure. Am J Respir Cell Mol Biol. 1999;21:44–53. doi: 10.1165/ajrcmb.21.1.3630. [DOI] [PubMed] [Google Scholar]
- West J, Pakenham G, Morin D, Fleschner C, Buckpitt A, Plopper C. Inhaled naphthalene causes dose dependent Clara cell toxicity in mice but not in rats. Toxicol Appl Pharmacol. 2001;173:114–119. doi: 10.1006/taap.2001.9151. [DOI] [PubMed] [Google Scholar]
- Williams KE, Carver TA, Miranda JJ, Kautiainen A, Vogel JS, Dingley K, et al. Attomole detection of in vivo protein targets of benzene in mice: evidence for a highly reactive metabolite. Mol Cell Proteomics. 2002;1:885–895. doi: 10.1074/mcp.m200067-mcp200. [DOI] [PubMed] [Google Scholar]
- Williams KJ, Cruikshank MK, Plopper CG. Pulmonary heat shock protein expression after exposure to a metabolically activated Clara cell toxicant: relationship to protein adduct formation. Toxicol Appl Pharmacol. 2003;192:107–118. doi: 10.1016/s0041-008x(03)00302-8. [DOI] [PubMed] [Google Scholar]