Abstract
Background
During transcription, the nontranscribed DNA strand becomes single-stranded DNA (ssDNA), which can form secondary structures. Unpaired bases in the ssDNA are less protected from mutagens and hence experience more mutations than do paired bases. These mutations are called transcription-associated mutations. Transcription-associated mutagenesis is increased under stress and depends on the DNA sequence. Therefore, selection might significantly influence protein-coding sequences in terms of the transcription-associated mutability per transcription event under stress to improve the survival of Escherichia coli.
Methodology/Principal Findings
The mutability index (MI) was developed by Wright et al. to estimate the relative transcription-associated mutability of bases per transcription event. Using the most stable fold of each ssDNA that have an average length n, MI was defined as (the number of folds in which the base is unpaired)/n×(highest –ΔG of all n folds in which the base is unpaired), where ΔG is the free energy. The MI values show a significant correlation with mutation data under stress but not with spontaneous mutations in E. coli. Protein sequence diversity is preferred under stress but not under favorable conditions. Therefore, we evaluated the selection pressure on MI in terms of the protein sequence diversity for all the protein-coding sequences in E. coli. The distributions of the MI values were lower at bases that could be substituted with each of the other three bases without affecting the amino acid sequence than at bases that could not be so substituted. Start codons had lower distributions of MI values than did nonstart codons.
Conclusions/Significance
Our results suggest that the majority of protein-coding sequences have evolved to promote protein sequence diversity and to reduce gene knockout under stress. Consequently, transcription-associated mutagenesis increases protein sequence diversity more effectively than does random mutagenesis under stress. Nonrandom transcription-associated mutagenesis under stress should improve the survival of E. coli.
Introduction
During transcription, the nontranscribed strand becomes single stranded, whereas the transcribed strand forms a complex with RNA polymerase and the nascent RNA transcript [1]. The ssDNA is much more vulnerable to most mutagens than is the double-stranded DNA [2] because it is not protected by pairing [3]. The resulting mutations include single-base substitutions [2], [4], [5], [6] and insertions/deletions (indels) [4], [7] and are called transcription-associated mutations [8] or transcription-induced mutations [9]. Therefore, transcription-associated mutagenesis should be active on the nontranscribed strands, in highly transcribed DNA regions, and in cells under stress where high levels of mutagens are active. The existence and significance of transcription-associated mutagenesis is widely supported. For example, increased mutations have been observed in highly transcribed regions in diverse species, such as Escherichia coli [9], [10], [11], yeast [8], and humans [12], [13]. The nontranscribed strand is thought to have greater numbers of mutations than the transcribed strand in E. coli [6], [9], [11] and humans [14], [15], [16]. The larger numbers of mutations on the nontranscribed strand are partly but not solely attributable to the activity of the transcription-coupled DNA repair system, which acts on the transcribed strand, in E. coli [6]. Transcription-associated mutations are considered to occur regardless of specific secondary structures [17]. Transcription-associated mutagenesis becomes active under stress [18], and occurs in both genomic DNA and plasmid DNA in E. coli [17]. Therefore, transcription-associated mutagenesis is considered to be an intrinsic source of mutations [5], [8], [11], [19]. Furthermore, transcription-associated mutations occur at a level that affects the genomic composition of T7 bacteriophage [20]. As transcription-associated mutations occurs asymmetrically on the transcribed and nontranscribed strands [6], when all the transcription-associated mutations that occur in a cell are not repaired, its two daughter cells have different genomic DNA. Transcription-coupled DNA repair act selectively on the transcribed strand [21]. Most, if not all, mutations on nontranscribed strands would be expressed only after replication. Therefore, transcription-associated mutations should predominantly exert their effects not on the transcribing cells but on their descendant cells. The mutation rate of E. coli is lower than one base pair per genome per replication [22]. Therefore, E. coli cells would often contain one transcription-associated mutation and no other types of mutations in a genomic strand and no mutations in the other genomic strand until cell division when transcription-associated mutagenesis operates. In such cases, the unmutated genomic DNA strand of the cell is inherited by one of its two daughter cells. Consequently, transcription-associated mutagenesis can be considered a safe way for dividing cells to rapidly increase the sequence diversity of the next generation.
Transcription-associated mutagenesis has often been investigated in reversion assays under stress [6], [23]. The nonrevertants in the reversion assays are often assumed to be nondividing cells. In reversion assays, organisms such as E. coli are engineered to divide actively and hence form large detectable viable colonies only when one or more of the requisite mutations occurs. E. coli can continue to divide slowly by living on the debris of other cells [24]. For example, E. coli can survive in batch cultures without any addition of nutrients for many months [25]. Therefore, the possibility that such nonrevertants divide as many times as they die cannot be excluded. If cells are assumed to be nondividing, it is difficult to see how E. coli could survive the high mutation rate experienced by revertants. However, such a high mutation rate can be explained if the E. coli cells live on cell debris and if transcription-associated mutagenesis plays a significant role. We describe here a possible scenario. After the nonrevertants are plated, the E. coli cells continue to divide slowly by living on the debris of other cells, perhaps following the death phase, in which about 99% of cells die. When the E. coli cells experience stress, transcription-associated mutagenesis increases [18]. E. coli safely increases its sequence diversity in the daughter cells whenever all the transcription-associated mutations that occur are not repaired correctly before cell division. Whenever detrimental mutations occur alone or in combination with preexisting mutations, the cells die and produce cell debris. The surviving cells gradually accumulate mutations as cell division recurs. While mutation and selection occur, transcription-associated mutations can occur at the sites that produce revertants. Therefore, the multiple mutations of the revertants can be interpreted as the result of mutation and selection through many generations, which would significantly reduce the estimated mutation rate. Revertants are usually counted 48 h after plating [6], [23], which is longer than the overnight or one day incubation typically required for colonies to appear when nonrevertant E. coli cells are plated on rich medium. This late appearance of revertants can be attributed to slow cell division, a relatively low mutation rate, and limited numbers of surviving cells per generation. The mismatched DNA base pairing caused by mutations can result in cross-strand deamination in vitro [26], [27], so it is possible for transcription-associated mutagenesis to rescue nondividing cells.
The ssDNA of the nontranscribed strand forms secondary structures [18], which have different stability. Therefore, some ssDNA sequences are sustained for a longer time than others. Consequently, individual bases in the nontranscribed strand display different transcription-associated mutability per transcription event, depending on the period during which the base is unpaired [28]. Transcription-associated mutability per transcription event has been estimated by a few methods [3], [28]. Among them (see “Materials and Methods” for details), the mutability index (MI), developed by Wright et al., focuses on transcription-associated mutagenesis under stress conditions [28]. The following is a description of the method of Wright et al. [28]. Because the length of ssDNA can vary, the length of the ssDNA was simplified to an average value n [28]. Therefore, any given base is assumed to belong to n ssDNA. The most stable fold of each ssDNA was identified, together with its –ΔG (the negative free energy) and pairing information. Using these n folds containing a given base, the MI of the given base was defined as (number of folds in which the base is unpaired)/n×(highest −ΔG of all n folds in which the base is unpaired). After scanning the average ssDNA length (n) for high −ΔG to best match known in vivo hotspot data for E. coli, Wright et al. set the average length of ssDNA (n) to 30 nt. The calculated MI values showed a positive significant correlation with the in vivo mutation data from reversion assays, but not with spontaneous mutation data [28]. This result suggests that MI can represent the relative transcription-associated mutability per transcription event in E. coli under the conditions of the reversion assays. Nontranscription-associated mutations were not excluded from either of the mutation data sets used for the validation tests. Therefore, the validation results imply that transcription-associated mutations constitute a large fraction of the total mutations in highly transcribed regions under the conditions of the reversion assays. Conversely, the invalidation of MI by the spontaneous mutation data might be attributable to either or both of the following two causes. First, transcription-associated mutations might not constitute a large enough fraction of the total spontaneous mutations to show a correlation. Second, the MI for the 30-nt ssDNA might not represent transcription-associated mutability per transcription event well under favorable conditions. This may occur because the average length of ssDNA was set to 30 nt by screening hotspot data that were obtained under stress [28].
Transcription-associated mutability per transcription event depends on the secondary structures formed by the DNA sequence [23], [28], [29] and can therefore be influenced by selection [3]. Every mutation does not exert the same effect. In protein-coding sequences, silent mutations circumvent potentially deleterious effects but do not increase the protein sequence diversity. Protein sequence diversity is advantageous under stress but not under conditions of spontaneous mutation. Interestingly, MI showed a positive significant correlation with mutation data under stress but not under conditions of spontaneous mutation [28]. Transcription-associated mutagenesis has been strongly suggested to play important roles under stress. For example, transcription-associated mutations are abundant under stress [18], [28] and in highly transcribed regions [10], [11]. Transcribed regions under a given stress might be better targets for beneficial mutations under that stress [30]. The MI validation by Wright et al. [28] suggested that transcription-associated mutagenesis is responsible for a large fraction of total mutations. Therefore, protein-coding sequences might have evolved to effectively increase protein sequence diversity by controlling transcription-associated mutability per transcription event under stress and hence MI values. If nonrandom MI values have been shaped within protein-coding sequences, it would provide a clear advantage for the survival of E. coli. In the present study, we analyzed the effect of selection on MI values using 4,132 protein-coding sequences from E. coli K12 MG1655, a fully sequenced E. coli strain. Our results show that bases have higher MI values when one or more of the three possible single-base substitutions at that base changes the encoded amino acid than when none of the three single-base substitutions at the base changes the encoded amino acid. Start codons have evolved to have lower MI values than nonstart codons. The selection pressure is different on different base groups in individual proteins. Our results suggest that the majority of protein-coding sequences have evolved in E. coli to produce transcription-associated mutations in such a way as to reduce gene knockout, while increasing protein sequence diversity, under stress. Such nonrandom mutagenesis would provide better sets of mutations even before the mutations are exposed to selection. Different selection pressures on MI allow each of the protein-coding sequences in the genome to have different evolvability. We discuss the biological benefits of nonrandom MI values, how selection shapes MI values, and the variation in the selection pressures on MI values.
Results
Calculation of MI
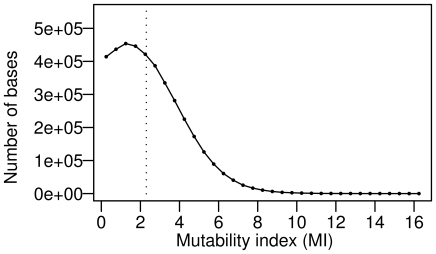
Transcription-associated mutability per transcription event should be affected by the local secondary structures of the RNA transcript, on which the functions of noncoding genes depend. Therefore, noncoding genes were excluded from this analysis. MI values for the 3,958,572 bases in the 4,132 protein-coding sequences of the E. coli K12 MG1655 genome were calculated according to the method described by Wright et al. [28], and the negative MI values were then converted to zero (see the “Materials and Methods” for details). The mass distribution of the MI values was concentrated on smaller MI values (0≤MI≤2) and exhibited a longer tail at the larger MI values (8≤MI, skewness = 0.85; Figure 1).
Figure 1. Distribution of the mutability index (MI) values of bases located in protein-coding sequences of E. coli K12 MG1655.
Closed circles indicate the number of bases in each MI value range (0.5). The vertical dotted line indicates the mean value (2.54).
Evaluation of selection pressures in terms of protein sequence diversity
Sequence properties affecting protein sequence diversity
To analyze the selection pressure on MI in terms of the generation of protein sequence diversity, or protein evolvability, all the bases in each protein-coding sequence, with the exception of those comprising the start and stop codons, were divided into three groups. The groups were based on the results of the three possible single-base substitutions at that base. The three groups were defined as follows: 1) all three single-base substitutions resulted in silent mutations, which do not change the encoded amino acid (Basesilent); 2) all three single-base substitutions resulted in missense mutations that changed the encoded amino acid (Basemissense); and 3) the three single-base substitutions produced two or three types of mutations: silent mutation, missense mutation, or nonsense mutation (Baseother; Tables 1–2).
Table 1. Mutations according to position in the protein-coding sequence.
| Bases | Single-base substitution | Insertion or deletion | ||
| Main results | Main effect | Main result | Effects | |
| Start codon | Disruption of start codon | Gene knockout | Frameshift mutations (unless multiples of three bases are inserted or deleted) | Gene knockout or protein truncation, with some functional impairment when it causes a frameshift |
| Codons occurring between the start and stop codons | Only silent mutations (Basesilent) | Neutral effect | ||
| Only missense mutations (Basemissense) | Single amino-acid substitution | |||
| Mixture of silent, missense, and nonsense mutations (Baseother) | Various effects | |||
Table 2. Compositions of base types in protein-coding sequences.
| Base type | Mean | 95% CI |
| Basesilent | 16.63 | 16.57–16.69 |
| Basemissense | 56.96 | 56.90–57.02 |
| Baseother | 26.42 | 26.32–26.52 |
| Sum | 100.0 |
Generation of control sequences
To analyze the selection pressure on MI in terms of the generation of protein-coding sequence diversity, control sequences with the same related features were required. Protein-coding sequences exert most of their effects via their encoded proteins. The compositional ratios of Basesilent, Basemissense, and Baseother would also affect the potential to generate protein sequence diversity. To comport the same product protein sequence and the same compositional ratios of Basesilent, Basemissense, and Baseother, we generated 100 control sequences per protein-coding sequence by shuffling the positions of the synonymous codons within the protein-coding sequence, except for the start codons and stop codons (Figure S1). The resulting control sequences also had the same GC content as the corresponding protein-coding sequence, which might affect the local secondary structures [3], because they had the same codon usage. The MI values of the control sequences were calculated as described above for the protein-coding sequences.
Evaluation of the selection pressure on MI (SMI)
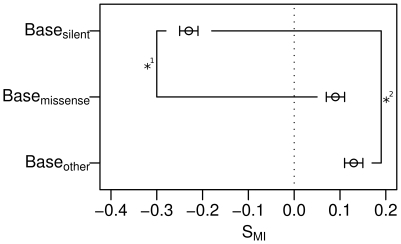
The protein-coding sequences and their control sequences have different average potentials to produce transcription-associated mutagenesis per transcription event [3]. By our calculation, the lacI gene, for example, has an average MI of 2.08 but its control sequences have average MI values between 1.78 and 2.42. Transcription-associated mutability is also affected by changes in the transcription level [10]. Therefore, to compare the relative MI values of individual bases in each of the protein-coding sequences and the control sequences, standard-normalized MI values were introduced into this analysis. The selection pressure on MI at Basesilent, for example, in a protein-coding sequence was evaluated in the following way (Figure S2). All MI values were standard normalized within the protein-coding sequence or in each control sequence. All the resulting z-scores of the MI values for Basesilent, for example, were averaged within each protein-coding sequence or within each control sequence. The average z-score for Basesilent in the protein-coding sequence was ranked against those in 100 control sequences. The resulting rank values were linearly transformed to “SMI” values (−1≤SMI≤1), which indicates the selection pressure on MI under stress. As a consequence, each protein-coding sequence had one SMI value for Basesilent. A negative or positive SMI value for Basesilent in a protein-coding sequence indicated that the protein-coding sequence had a lower or higher average z-score for the MI values at Basesilent, respectively, than did 50% of the random control sequences. For example, a negative SMI value for a protein-coding sequence at Basesilent indicates that the protein-coding sequence has evolved to have lower transcription-associated mutability per transcription event at Basesilent under stress. This method of calculating SMI values was also applied to Basemissense, Baseother, and all other base groups in the subsequent analyses described in this manuscript. Basesilent exhibited lower SMI values than those of Basemissense and Baseother (Wilcoxon signed-rank test, P<1e−90 and P<1e−90, respectively; see “Materials and Methods” for the choice of statistical methods). Basesilent and Basemissense exhibited distributions of SMI lower and higher than zero, respectively (Wilcoxon rank-sum test, P<1e−100 and P<1e−100, respectively; Figure 2). These results suggest that protein-coding sequences have evolved to increase the ratio of missense mutations to silent mutations among transcription-associated mutations under stress. In short, our results suggest that the genome sequence has evolved to increase protein evolvability under stress.
Figure 2. SMI of the base groups according to the mutation types generated by a single-base substitution.
The bases were grouped according to the type of mutations caused by single-base substitutions. “Basesilent” and “Basemissense” indicate bases where all single-base substitutions at the bases result in silent and missense mutations, respectively. “Baseother” indicates all bases other than Basesilent, Basemissense, and bases that occur in start and stop codons in protein-coding sequences. Circles and error bars indicate the pseudomedian and the 95% confidence interval (CI), respectively, as assessed using the Wilcoxon rank-sum test. The vertical dotted line represents the 0 value for SMI. Calculations of the Wilcoxon signed-rank test: *1 P<1e−90; *2 P<1e−90.
Selection pressure on MI in terms of gene knockout
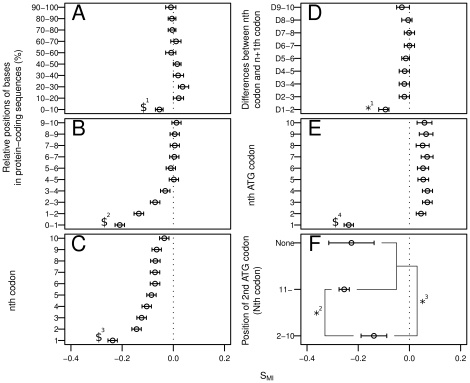
Start codons are the sites most likely to be involved in mutation-based gene knockout within protein-coding sequences. To assess the overall distribution of SMI in protein-coding sequences, we grouped the primary positions of protein-coding sequences into deciles, in a 5′ to 3′ direction. Among the 10 base groups, the base group of the first decile showed the lowest distribution of SMI values (Wilcoxon signed-rank test with the division having the second-lowest SMI, P<0.01; Figure 3A). To analyze this result in greater detail, the first decile was further divided into 1% divisions. Among the resulting 10 groups, the base group of the first percentile showed the lowest distribution of SMI values (Wilcoxon rank-sum test with the division of the second-lowest SMI, P<1e−10; Figure 3B). Start codons occupy the 5′-most positions in protein-coding sequences. This may explain why the base group at the 5′-most position had the lowest SMI value among the groups tested. To confirm this, the bases were grouped for each nth codon and the SMI were compared. The start codons displayed the lowest SMI values among the codons located in the 5′ regions of the protein-coding sequences (Wilcoxon signed-rank test with the division of the second-lowest SMI, P<1e−20; Figure 3C). The tendency to form secondary structures is often affected by the primary position on the protein-coding sequence. For instance, bases at the 5′ end are affected by the efficiency of translation initiation and bases at the 3′ end by Rho-independent transcription termination. The base groups of the first four percentiles showed a reduction in SMI values with decreasing percentile (Figure 3B). Therefore, the lowest SMI values observed for the start codons may be caused by this tendency toward low SMI values in the first four percentiles. To exclude this possibility, we compared the results for the subtraction of the SMI values of adjacent codons among the first 10 codons. When the SMI values for the second codons were subtracted from the SMI values for the start codons, the results were less than zero (Wilcoxon rank-sum test, P<1e−20; Figure 3D). However, the other adjacent codon pairs tested did not yield values significantly less than zero (Wilcoxon rank-sum test, P>0.01; Figure 3D). Therefore, the lower SMI values obtained for the start codons are not just a tendency found at the 5′ ends of protein-coding sequences. Start codons exhibit lower SMI values than those of other ATG codons (Wilcoxon rank-sum test, P<1e−60; Figure 3E). This result excludes the possibility of a fitness effect exerted by the mutation of ATG codons. To confirm that protein-coding gene knockout influences the effects of the selection pressure on MI values, the SMI values for the start codons were compared according to the position of the first nonstart ATG codon. The presence of a closely following nonstart ATG codon at the 5′ end of the protein-coding sequence implies that single-base substitutions at start codons cause the deletion of several N-terminal amino acid residues. These N-terminal deletions have a lower potential to cause protein-coding gene knockout. The SMI values of start codons were higher in the presence of nonstart ATG codons in the first 10 codons than in the presence of nonstart ATG codons after the first 10 codons (Wilcoxon rank-sum test, P<1e−4; Figure 3F) and in the absence of nonstart ATG codons within the first 10 codons (Wilcoxon rank-sum test, P<1e−4; Figure 3F). These results demonstrate that selection acts to reduce the MI values of start codons when start codon mutations cause gene knockout. These results suggest that protein-coding sequences have evolved to reduce the proportion of gene knockouts by transcription-associated mutations under stress.
Figure 3. SMI of the base groups according to their relative positions within the protein-coding sequences.
(A) The relative positions were grouped into deciles from 0 to 100%. (B) The relative positions were then grouped into percentiles from 0 to 10%. (C) Start codons and their adjacent downstream codons. The 1 on the Y-axis indicates the start codon. (D) Difference in the SMI values for the n th codon and the n+1th codon. For example, “D1–2” indicates the difference between the SMI values of the first codons (start codons) and those of the second codons. (E) Start codons and their following nonstart ATG codons. The 1 on the Y-axis indicates the start codon. (F) SMI values for the start codons, according to the positions of the first nonstart ATG codons. $ indicates the largest P value among those calculated between a base group marked $ and all the other base groups: $1 P<0.01; $2 P<1e−10; $3 P<1e−20; $4 P<1e−60; *1 P<1e−20; *2 P<1e−4; *3 P<1e−4 ($1, $3: Wilcoxon signed-rank test; $2, $4, *1, *2, *3: Wilcoxon rank-sum test). Circles and error bars indicate the pseudomedian and the 95% CI, respectively, as assessed using the Wilcoxon rank-sum and signed-rank test. The vertical dotted lines represent the 0 value for SMI.
Correlation between before and after single-base substitutions
A single-base substitution in ssDNA often changes the most stable secondary structure of the ssDNA. Therefore, a single-base substitution of a base may change the MI value of the base itself and of its 58 neighboring bases (29 bases upstream and 29 bases downstream; Figure S3). The altered MI value would affect the positive and negative selection that shapes the nonrandom MI. To examine how nonrandom MI distributions are shaped, it is important to assess the mutational effect of a specific base on its own MI value and on the MI values of neighboring bases. Therefore, we repeated 10,000 random single-base substitutions 100 times and analyzed the correlation values of the MI values for the mutated bases themselves and for their neighboring bases before and after single-base substitutions (see “Materials and Methods”). The MI values showed significant positive correlations at both the mutated bases (R2 = 0.29, P<1e−100) and the neighboring bases (R2 = 0.79, P<1e−100). The bases with high MI values tended to retain similar MI values after single-base substitutions (Figure S4). These results demonstrate that the MI values of bases are usually subject to larger variation when mutations occur at the base itself than when mutations occur at neighboring bases, and that bases with high MI values are more tolerant of variation in their MI values after single-base substitutions at those bases than are bases with low MI values.
Discussion
Biological benefits of the nonrandom distribution of MI values under stress
In this study, we have demonstrated that selection acts on protein-coding sequences to lower the MI values at Basesilent and to increase the MI values at Basemissense and Baseother under stress (Figure 2). Under stress, protein sequence diversity is preferred and some of this diversity is beneficial. Most beneficial mutations are missense mutations rather than indels [31]. Missense mutations can occur at Basemissense and Baseother but not at Basesilent. However, some missense mutations can have detrimental effects. Therefore, under stress, high MI at Basemissense and Baseother and low MI at Basesilent can increase the chance of beneficial mutations at the cost of increasing detrimental mutations. The proportion of detrimental mutations among indels is larger than the proportion of detrimental mutations among single-base substitutions. Therefore, the proportion of detrimental mutations among indels is larger than the proportion of detrimental mutations among indels plus single-base substitutions. A protein sequence can be mutated only by indels at Basesilent and by both indels and single-base substitutions at Basemissense and Baseother. Therefore, the proportion of detrimental mutations among mutations that result in a change in the protein sequence is larger at Basesilent than at Basemissense and Baseother.
These results imply that protein-coding sequences have evolved to reduce the proportion of detrimental mutations produced while increasing protein sequence diversity under stress. Supporting this proposition, the MI values at start codons are low when mutations at start codons are likely to cause gene knockouts (Figure 3). Gene knockouts often result in detrimental mutations, for example in essential genes, and often exert disadvantageous effects. In prokaryotes such as E. coli, selective pressure is exerted on the genome length, so they have very compact genomes [32]. This implies that most preexisting genes recently contributed to the fitness of the cell before they were lost in response to the selection pressure on genome size. Consequently, our data suggest that protein-coding sequences have evolved to control the transcription-associated mutability per transcription event to increase protein evolvability and to reduce the proportion of detrimental mutations produced under stress.
Selection type that shapes nonrandom MI values within protein-coding sequences
The reduction of MI values at Basesilent and start codons can be achieved by purifying selection. Detrimental mutations are not passed on to the descendant cells. We have demonstrated that mutations alter the MI values of neighboring bases as well as that of the mutated base itself. Therefore, the different MI values of start codons and Basesilent can be caused by mutations at neighboring bases. E. coli cells and their descendants that have start codons and Basesilent with high MI values would frequently suffer detrimental mutations because mutations occur frequently at these sites. Increases in the MI values at Basemissense can be achieved by positive selection. Positive selection implies that the mutated bases are inherited by the descendant cells and become dominant. The new bases at these sites will have different MI values. However, as we have demonstrated, new MI values correlate positively with the old MI values at the mutated sites. In particular, bases with high MI values tend to retain these high MI values after single-base substitutions (Figure S4). Therefore, high MI values at Basemissense are frequently retained, although some degree of fluctuation cannot be avoided. The distribution of SMI values at Baseother was closer to that at Basemissense than to that at Basesilent (Figure 2). Single-base substitutions at Baseother are made up of two or more types of mutations: silent, missense, and nonsense mutations. The compositions of silent mutations, missense mutations, and nonsense mutations at Baseother were 26%, 60%, and 14%, respectively, throughout all protein-coding sequences. Because the proportion of missense mutations is highest at Baseother, the MI values at Baseother will be influenced by selection in a similar way to those at Basemissense rather than to those at Basesilent.
Variation in the selection pressure on MI values
Transcription-associated mutations increase under stress and in highly transcribed regions. Individual protein-coding sequences are repressed or derepressed under different stresses and have different transcription levels under specific stresses. Therefore, each protein-coding sequence has a different SMI value for each of its base groups (Table 3). This result suggests that the protein products of individual protein-coding sequences have different evolvability. Interestingly, about one third of protein-coding sequences have SMI values that are larger than zero at Basesilent (Table 3). This suggests that about one third of protein-coding sequences have evolved to reduce protein sequence diversity under stress. Reduced protein evolvability is beneficial under favorable conditions. Therefore, the high SMI values at Basesilent and the low SMI values at Basemissense and Baseother might be attributable to the activity of transcription-associated mutagenesis under favorable conditions. However, the development of an index that predicts transcription-associated mutability per transcription event under favorable conditions and its subsequent analysis will be necessary to determine the mechanism underlying this phenomenon.
Table 3. Distributions of SMI values.
| −1.0≤SMI<−0.5 | −0.5≤SMI<0 | 0≤SMI<0.5 | 0.5≤SMI≤1 | Sum | |
| Basemissense | 21.25% | 21.03% | 25.46% | 32.26% | 100.0% |
| Baseother | 19.53% | 21.93% | 24.25% | 34.29% | 100.0% |
| Basesilent | 38.53% | 25.62% | 20.57% | 15.27% | 100.0% |
Conclusions
In this study, we evaluated the selection pressure on MI, which represents the relative potential for transcription-associated mutagenesis per transcription event under stress. Our results suggest that the majority of protein-coding sequences have evolved to increase protein sequence diversity by controlling transcription-associated mutagenesis under stress and that transcription-associated mutagenesis produces protein sequence diversity more effectively than does random mutagenesis. Therefore, transcription-associated mutagenesis will confer faster protein evolvability under stress, and will improve the chance of survival of E. coli.
Materials and Methods
Sequence data
The sequence of E. coli K12 MG1655 was downloaded from the NCBI ftp site (ftp://ftp.ncbi.nih.gov/genomes/Bacteria/Escherichia_coli_K12). The sequences of the 4,132 E. coli protein-coding genes were used in this analysis. For each protein-coding sequence, 100 control sequences were generated by synonymous codon shuffling; the codon position for any given amino acid was shuffled, whereas the positions of the start and stop codons were maintained (see Figure S1). This step was repeated for each of the 20 amino acids. Random shuffling was performed using the “random” module of the standard library of the Python program, version 2.6 (http://www.python.org).
Sequence features
Relative base positions in protein-coding sequences
The base pairs in a protein-coding sequence with a length of n base pairs were numbered from 1 to n. For the k th base, the length ratio was defined as ([k–1]/[n–1])×100. For example, the first base of the start codon and the last base of the stop codon were assigned positions 0 and 100, respectively.
n th codon
In a protein-coding sequence that produces a protein of n amino acids, the codons were numbered from 1 to n+1. For example, the start and stop codons were assigned positions 1 and n+1, respectively.
n th ATG codon
In a protein-coding sequence, the start codon was assigned position 1, even when it was not an ATG. The subsequent k ATG codons were sequentially assigned positions from 2 to k+1.
Selection of a method to predict transcription-associated mutability per transcription event
Two methods have been developed to predict transcription-associated mutagenesis potentials. The first is the MI of Wright et al., which focuses on transcription-associated mutations and was validated using in vivo mutation data from reversion assays. Because the purpose of this study was to investigate the selection pressure on transcription-associated mutability per transcription event under stress, Wright et al.'s MI was adequate. Hoede et al. [3] developed another index, the transcription-driven mutability index (TDMI). To validate these indices, these authors used conservation data taken from the alignment of orthologous gene sequences among E. coli strains. The mutations that generated these variable sites might be originated under stress, under favorable conditions, or under a mixture of both types of conditions. The variable sites in the sequence alignments were the results of mutations in the ancestral sequences. Therefore, the TDMI values for the ancestral sequences should have been compared with the conservation data in the validation process. However, the TDMI values were calculated from the sequence of an extant E. coli strain. Therefore, TDMI was not adequate for the present analysis.
Calculation of MI values
We initially calculated the MI values according to the method described by Wright et al. [28] using the hybrid-ss-min program of the UNAFold package [33], which is the local program from the DINAMelt web server, version 3.6 [34], used by Wright et al. The calculation of MI with an average ssDNA length of 30 nt was described here. The protein-coding sequences were extended to include the 29-nt (that is [30]–[1]-nt) upstream and downstream sequences. Subsequences were then generated by sliding a window of 30 nt along the resulting extended sequence. Consequently, each individual base belonged to 30 subsequences. All subsequences were then folded using the hybrid-ss-min program [33] and the ΔG values and paired/unpaired status of the most stable folds of each given subsequence were determined. The MI value for each base was calculated using the equation proposed by Wright et al. [28]: (number of folds in which the base is unpaired)/30×(highest –ΔG of all 30 folds in which the base is unpaired). A small proportion of MI values (0.4%) was smaller than zero when the first factor, ‘number of folds in which the base is unpaired’, was greater than zero but the second factor, ‘highest –ΔG of all n folds in which the base is unpaired’, was smaller than zero. However, when the ‘number of folds in which the base is unpaired’ is zero, the MI value becomes zero. This contradicts the assumption that bases that remain unpaired for a longer time have higher MI values. Stable folds are not formed when the –ΔG of the most stable fold is less than zero. Therefore, these values were converted to zero.
Evaluation of the effect of selection on MI (SMI)
For protein-coding sequences and their control sequences, the MI value of each base was standard normalized within the protein-coding sequence. To compare the transcription-associated mutability per transcription event among the base groups, the standard-normalized MI values for each base group were averaged for the protein-coding sequence and for each of the control sequences. The rank of a gene sequence at a given base group was calculated from these averaged values against the corresponding values of 100 control sequences. The rank was then transformed to SMI values using the equation (2×rank – 102)/100. As a result, each protein-coding sequence had a value between −1 and 1 (−1≤SMI≤1) for each base group.
Correlation of MI values before and after mutation
Ten thousand single bases selected randomly from protein-coding sequences were randomly mutated to one of the three other possible bases. The MI values of the base targeted by the mutation and of the 29 bases located upstream and downstream from this position were calculated before and after the mutation. The neighboring bases that were not within the protein-coding sequences were excluded from the calculation of the correlation. The MI values of the same bases before and after mutation were used to calculate Pearson's correlation values. These steps were repeated 100 times. The mean Pearson's correlation values were used for this study.
Selection of statistical methods
Wilcoxon signed-rank test and Wilcoxon rank-sum test are nonparametric statistical methods and are alternatives to paired Student's t test and Student's t test, respectively. Nonparametric statistical methods are robust and can be used even when a normal distribution cannot be assumed. To compare the difference of groups, the Wilcoxon signed-rank test and Wilcoxon rank-sum test were used. We used paired comparisons (and hence the Wilcoxon signed-rank test) wherever possible. For example, in Figure 3C, the Wilcoxon signed-rank test was used because every protein-coding sequence had more than 10 codons, so a paired comparison of the start codons and ith codons (2≤i≤10) inside each of the protein-coding sequences was possible. Conversely, in Figure 3E, the Wilcoxon rank-sum test was used because some protein-coding sequences did not have 10 ATG codons and paired comparisons were not possible in some protein-coding sequences.
All statistical analyses were performed using the R statistical package [35]. All rankings were made in increasing order.
Supporting Information
Schematic representation of synonymous codon shuffling. (A) An imaginary sequence encoding six amino acids. The start codon and stop codon were excluded from shuffling. The arrows indicate random shuffling. The positions of synonymous codons (encoding the same amino acid) were shuffled randomly. This shuffling was repeated for each of the 20 amino acids. In the example sequence, there is one codon for serine, so it was self-shuffled. (B) All the control sequences generated from the sequence in (A). The control sequences can include the protein-coding sequence if the synonymous-codon-shuffled sequences happen to include the same sequence as the protein-coding sequence.
(0.01 MB PDF)
Example of calculating SMI from arbitrary MI values. (A) MI values for an imaginary protein-coding sequence and its control sequences. The control sequences were generated with synonymous codon shuffling. The z-score values for MI were arbitrarily assigned for the demonstration. Base groups START, O, M, S, and STOP represent the start codon, Baseother, Basemissense, Basesilent, and the stop codon, respectively. (B) The average z-scores for MI for the individual base groups of individual sequences. For example, Basesilent of the protein-coding sequence has z-scores for the MI values of − 2.01, 2.95, and 0.42 in (A), so its average value (0.45) is written as the corresponding value in (B). (C) The z-score average for MI of the protein-coding sequence was ranked among the corresponding values for the control sequences. The rank values (1≤rank≤N+1) obtained were linearly transformed to SMI (− 1≤SMI≤1) using the equation (2×rank − 2 − N)/N, where N is the number of control sequences (which is four in this example and 100 in the main text). In this example, four control sequences were used for clarity and the values were rounded to the nearest two decimal places, although we used 100 control sequences and calculated them to 15 decimal places in the actual calculation in the main text.
(0.34 MB PDF)
An example of the effect of a single-base substitution on secondary structure. (A) A single-base substitution can affect the most stable secondary structures of all ssDNA sequences that contain the mutated site. Each underlined sequence is 30 nt. (B) The sequence underlined by third line in (A). (C) The resulting sequence of G-to-C single-base substitution at the base marked with a star in the sequence shown in (B). The secondary structures of (B) and (C) indicate the most stable secondary structures of these sequences. In this example, the G-to-C mutation changes the most stable secondary structure and hence the paired/unpaired state and the −ΔG value of the bases, which determine their MI values.
(0.54 MB TIF)
Correlation of MI values before and after single-base substitutions. The correlations of the MI values before and after mutation were plotted. The MI values were divided into 10 groups according to the rank of each before and after a single-base substitution. The diameter of each circle is proportional to the number in each division.
(0.57 MB TIF)
Acknowledgments
We thank Dr Rintaro Saito of Keio University, Japan, for reviewing the manuscript and commenting on its statistical aspects. We also thank Dr Kochiwa Hiromi and Dr Douglas Murray of Keio University, Japan, for their comments on the manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This research was supported in part by a Grant-in-Aid from Keio University Global Center of Excellence Program and Mori memorial grant and by research funds from the Yamagata prefectural government and Tsuruoka City, Japan. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Korzheva N, Mustaev A, Kozlov M, Malhotra A, Nikiforov V, et al. A structural model of transcription elongation. Science. 2000;289:619–625. doi: 10.1126/science.289.5479.619. [DOI] [PubMed] [Google Scholar]
- 2.Singer B, Kusmierek JT. Chemical Mutagenesis. Annual Review of Biochemistry. 1982;51:655–691. doi: 10.1146/annurev.bi.51.070182.003255. [DOI] [PubMed] [Google Scholar]
- 3.Hoede C, Denamur E, Tenaillon O. Selection Acts on DNA Secondary Structures to Decrease Transcriptional Mutagenesis. PLoS Genetics. 2006;2:e176. doi: 10.1371/journal.pgen.0020176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Boer JG, Ripley LS. Demonstration of the production of frameshift and base-substitution mutations by quasipalindromic DNA sequences. Proceedings of the National Academy of Sciences of the United States of America. 1984;81:5528–5531. doi: 10.1073/pnas.81.17.5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klapacz J, Bhagwat AS. Transcription-dependent increase in multiple classes of base substitution mutations in Escherichia coli. Journal of Bacteriology. 2002;184:6866–6872. doi: 10.1128/JB.184.24.6866-6872.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klapacz J, Bhagwat AS. Transcription promotes guanine to thymine mutations in the non-transcribed strand of an Escherichia coli gene. DNA Repair. 2005;4:806–813. doi: 10.1016/j.dnarep.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 7.Ripley LS. Model for the Participation of Quasi-Palindromic DNA Sequences in Frameshift Mutation. Proceedings of the National Academy of Sciences of the United States of America. 1982;79:4128–4132. doi: 10.1073/pnas.79.13.4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Datta A, Jinks-Robertson S. Association of increased spontaneous mutation rates with high levels of transcription in yeast. Science. 1995;268:1616–1619. doi: 10.1126/science.7777859. [DOI] [PubMed] [Google Scholar]
- 9.Beletskii A, Bhagwat AS. Transcription-induced mutations: increase in C to T mutations in the nontranscribed strand during transcription in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:13919–13924. doi: 10.1073/pnas.93.24.13919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reimers JM, Schmidt KH, Longacre A, Reschke DK, Wright BE. Increased transcription rates correlate with increased reversion rates in leuB and argH Escherichia coli auxotrophs. Microbiology. 2004;150:1457–1466. doi: 10.1099/mic.0.26954-0. [DOI] [PubMed] [Google Scholar]
- 11.Fix D, Canugovi C, Bhagwat AS. Transcription increases methylmethane sulfonate-induced mutations in alkB strains of Escherichia coli. DNA Repair (Amst) 2008;7:1289–1297. doi: 10.1016/j.dnarep.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bachl J, Carlson C, Gray-Schopfer V, Dessing M, Olsson C. Increased Transcription Levels Induce Higher Mutation Rates in a Hypermutating Cell Line. J Immunol. 2001;166:5051–5057. doi: 10.4049/jimmunol.166.8.5051. [DOI] [PubMed] [Google Scholar]
- 13.Da Sylva TR, Gordon CS, Wu GE. A genetic approach to quantifying human in vivo mutation frequency uncovers transcription level effects. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis. 2009;670:68–73. doi: 10.1016/j.mrfmmm.2009.07.008. [DOI] [PubMed] [Google Scholar]
- 14.Skandalis A, Ford BN, Glickman BW. Strand bias in mutation involving 5-methylcytosine deamination in the human hprt gene. Mutat Res. 1994;314:21–26. doi: 10.1016/0921-8777(94)90057-4. [DOI] [PubMed] [Google Scholar]
- 15.Green P, Ewing B, Miller W, Thomas PJ, Program NCS, et al. Transcription-associated mutational asymmetry in mammalian evolution. Nat Genet. 2003;33:514–517. doi: 10.1038/ng1103. [DOI] [PubMed] [Google Scholar]
- 16.Mugal CF, von Grünberg H-H, Peifer M. Transcription-induced mutational strand bias and its effect on substitution rates in human genes. Mol Biol Evol. 2009;26:131–142. doi: 10.1093/molbev/msn245. [DOI] [PubMed] [Google Scholar]
- 17.Beletskii A, Bhagwat AS. Transcription-induced cytosine-to-thymine mutations are not dependent on sequence context of the target cytosine. Journal of Bacteriology. 2001;183:6491–6493. doi: 10.1128/JB.183.21.6491-6493.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wright BE. Stress-directed adaptive mutations and evolution. Mol Microbiol. 2004;52:643–650. doi: 10.1111/j.1365-2958.2004.04012.x. [DOI] [PubMed] [Google Scholar]
- 19.Hudson RE, Bergthorsson U, Ochman H. Transcription increases multiple spontaneous point mutations in Salmonella enterica. Nucl Acids Res. 2003;31:4517–4522. doi: 10.1093/nar/gkg651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beletskii A, Grigoriev A, Joyce S, Bhagwat AS. Mutations induced by bacteriophage T7 RNA polymerase and their effects on the composition of the T7 genome. Journal of Molecular Biology. 2000;300:1057–1065. doi: 10.1006/jmbi.2000.3944. [DOI] [PubMed] [Google Scholar]
- 21.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 22.Drake JW, Charlesworth B, Charlesworth D, Crow JF. Rates of Spontaneous Mutation. Genetics. 1998;148:1667–1686. doi: 10.1093/genetics/148.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burkala E, Reimers JM, Schmidt KH, Davis N, Wei P, et al. Secondary structures as predictors of mutation potential in the lacZ gene of Escherichia coli. Microbiology. 2007;153:2180–2189. doi: 10.1099/mic.0.2007/005470-0. [DOI] [PubMed] [Google Scholar]
- 24.Finkel SE. Long-term survival during stationary phase: evolution and the GASP phenotype. Nat Rev Microbiol. 2006;4:113–120. doi: 10.1038/nrmicro1340. [DOI] [PubMed] [Google Scholar]
- 25.Finkel SE, Kolter R. Evolution of microbial diversity during prolonged starvation. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:4023–4027. doi: 10.1073/pnas.96.7.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sowers LC, Shaw BR, Veigl ML, Sedwick WD. DNA base modification: ionized base pairs and mutagenesis. Mutation Research. 1987;177:201–218. doi: 10.1016/0027-5107(87)90003-0. [DOI] [PubMed] [Google Scholar]
- 27.Williams LD, Shaw BR. Protonated base pairs explain the ambiguous pairing properties of O6-methylguanine. Proceedings of the National Academy of Sciences of the United States of America. 1987;84:1779–1783. doi: 10.1073/pnas.84.7.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wright BE, Reschke DK, Schmidt KH, Reimers JM, Knight W. Predicting mutation frequencies in stem-loop structures of derepressed genes: implications for evolution. Mol Microbiol. 2003;48:429–441. doi: 10.1046/j.1365-2958.2003.t01-1-03436.x. [DOI] [PubMed] [Google Scholar]
- 29.Wright BE, Schmidt KH, Davis N, Hunt AT, Minnick MF. II. Correlations between secondary structure stability and mutation frequency during somatic hypermutation. Mol Immunol. 2008;45:3600–3608. doi: 10.1016/j.molimm.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Galhardo RS, Hastings PJ, Rosenberg SM. Mutation as a stress response and the regulation of evolvability. Crit Rev Biochem Mol Biol. 2007;42:399–435. doi: 10.1080/10409230701648502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maclean RC, Buckling A. The distribution of fitness effects of beneficial mutations in Pseudomonas aeruginosa. PLoS Genetics. 2009;5:e1000406. doi: 10.1371/journal.pgen.1000406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Day MJ, Miller RV. WHY ARE GENES LOST? WHY DO GENES PERSIST? In: Miller RV, Day MJ, editors. Microbial evolution: gene establishment, survival, and exchange. Washington, DC: American Society Microbiology; 2004. pp. 290–294. [Google Scholar]
- 33.Markham NR, Zuker M. UNAFold: software for nucleic acid folding and hybridiziation. In: Keith JM, editor. Bioinformatics. Totowa, NJ: Humana Press; 2008. pp. 3–31. [DOI] [PubMed] [Google Scholar]
- 34.Markham NR, Zuker M. DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Res. 2005;33:W577–581. doi: 10.1093/nar/gki591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.R Development Core Team. R: A Language and Environment for Statistical Computing. 2008. Vienna, Austria.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Schematic representation of synonymous codon shuffling. (A) An imaginary sequence encoding six amino acids. The start codon and stop codon were excluded from shuffling. The arrows indicate random shuffling. The positions of synonymous codons (encoding the same amino acid) were shuffled randomly. This shuffling was repeated for each of the 20 amino acids. In the example sequence, there is one codon for serine, so it was self-shuffled. (B) All the control sequences generated from the sequence in (A). The control sequences can include the protein-coding sequence if the synonymous-codon-shuffled sequences happen to include the same sequence as the protein-coding sequence.
(0.01 MB PDF)
Example of calculating SMI from arbitrary MI values. (A) MI values for an imaginary protein-coding sequence and its control sequences. The control sequences were generated with synonymous codon shuffling. The z-score values for MI were arbitrarily assigned for the demonstration. Base groups START, O, M, S, and STOP represent the start codon, Baseother, Basemissense, Basesilent, and the stop codon, respectively. (B) The average z-scores for MI for the individual base groups of individual sequences. For example, Basesilent of the protein-coding sequence has z-scores for the MI values of − 2.01, 2.95, and 0.42 in (A), so its average value (0.45) is written as the corresponding value in (B). (C) The z-score average for MI of the protein-coding sequence was ranked among the corresponding values for the control sequences. The rank values (1≤rank≤N+1) obtained were linearly transformed to SMI (− 1≤SMI≤1) using the equation (2×rank − 2 − N)/N, where N is the number of control sequences (which is four in this example and 100 in the main text). In this example, four control sequences were used for clarity and the values were rounded to the nearest two decimal places, although we used 100 control sequences and calculated them to 15 decimal places in the actual calculation in the main text.
(0.34 MB PDF)
An example of the effect of a single-base substitution on secondary structure. (A) A single-base substitution can affect the most stable secondary structures of all ssDNA sequences that contain the mutated site. Each underlined sequence is 30 nt. (B) The sequence underlined by third line in (A). (C) The resulting sequence of G-to-C single-base substitution at the base marked with a star in the sequence shown in (B). The secondary structures of (B) and (C) indicate the most stable secondary structures of these sequences. In this example, the G-to-C mutation changes the most stable secondary structure and hence the paired/unpaired state and the −ΔG value of the bases, which determine their MI values.
(0.54 MB TIF)
Correlation of MI values before and after single-base substitutions. The correlations of the MI values before and after mutation were plotted. The MI values were divided into 10 groups according to the rank of each before and after a single-base substitution. The diameter of each circle is proportional to the number in each division.
(0.57 MB TIF)