Abstract
We report the first image of an intact, frozen hydrated eukaryotic cell using x-ray diffraction microscopy, or coherent x-ray diffraction imaging. By plunge freezing the specimen in liquid ethane and maintaining it below −170 °C, artifacts due to dehydration, ice crystallization, and radiation damage are greatly reduced. In this example, coherent diffraction data using 520 eV x rays were recorded and reconstructed to reveal a budding yeast cell at a resolution better than 25 nm. This demonstration represents an important step towards high resolution imaging of cells in their natural, hydrated state, without limitations imposed by x-ray optics.
X-ray microscopes allow high resolution microscopy of intact, hydrated biological specimens with thicknesses of many micrometers, beyond the limit of biological electron microscopy [1–3]. Radiation damage precludes repeated imaging of live specimens [4], but this can be mitigated by working at liquid nitrogen temperature [5,6]. In addition, single view flash imaging of cells using ultrabright sources has been proposed [7,8] as a way of capturing the image before radiolytical and thermal damage become evident.
In recent years, there has been much progress in developing zone plate microscopy for 3D imaging of frozen hydrated cells [9–13]. While there are demonstrations of x-ray optics with higher resolution [14–16], scientific applications using x-ray microscopes have mainly used Fresnel zone plate optics with 25–40 nm spatial resolution. These optics typically have a focusing efficiency in the 10% range [17] and the modulation transfer function for incoherent bright field imaging decreases the efficiency of utilization of higher spatial frequency information. As a result, while the practical advantages of lens-based microscopes will be the deciding factor for most studies, it is also worthwhile to consider alternative methods for high resolution x-ray imaging.
X-ray diffraction microscopy (XDM), also called coherent x-ray diffraction imaging, was proposed by Sayre as an imaging method that dispenses with the technological limits of lens efficiency and resolution [18]. Instead, the far-field diffraction pattern of an isolated object illuminated by a coherent x-ray beam is recorded. If the object is finite, and the diffraction pattern is sampled finely enough, the object can be reconstructed from the measured diffraction intensities alone [19,20]. In this manner one is able to eliminate limitations due to the efficiency and finite numerical aperture of x-ray optics [21]. Following a first demonstration by Miao et al. of imaging a nanofabricated test object [22], this and related approaches have been used by a growing number of groups, including demonstrations of imaging bacteria [23], yeast [24], herpes virions [25], malaria-infected erythrocytes [26], and chromosomes in 2D and 3D [27].
An important limitation applies to the demonstrations of x-ray diffraction microscopy of cells, chromosomes, and virions cited above: they have all involved dehydrated specimens at room temperature. Though Nishino et al. [27] have obtained a very exciting 3D XDM image of a dehydrated chromosome, they note significant resolution degradation due to accumulated radiation dose. In electron microscopy, stability against radiation damage has long been solved by imaging the specimen at cryogenic temperatures [28–30]. Electron microscopy studies of fixed and dried versus frozen hydrated blood platelets [31] reveal tremendous differences in structural preservation. However, because of the thickness limitations of electron microscopy on frozen hydrated specimens [1–3], to our knowledge only thin, peripheral regions of whole, unsectioned eukaryotic cells have been imaged in a frozen hydrated state using electron microscopy [32,33].
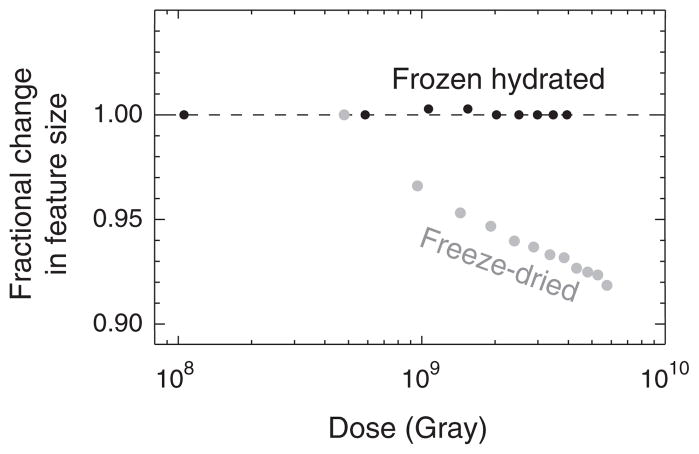
The benefits of cryo methods for XDM have been shown in studies by Shapiro [34] which demonstrate remarkable stability of the diffraction pattern out to doses well beyond the dose imparted in the work reported here (Fig. 1). In comparison, substantial specimen shrinkage was observed at room temperature. Two separate groups have estimated that cryo XDM has the potential to deliver sub-10 nm resolution 3D images of whole hydrated cells within the limits of radiation damage [35,36]. As an important step towards realizing this milestone, we report here the first use of x-ray diffraction microscopy to image a whole, unfixed, frozen hydrated eukaryotic cell.
FIG. 1.
Demonstration of the stability of frozen hydrated yeast cells to 520 eV x-ray exposure for various absorbed doses in Gray [34]. Shown here are the fractional radial shifts of the centers of speckles in coherent diffraction patterns of yeast cells. The top curve is of cells that were viewed while frozen hydrated at −170 °C, while the bottom curve is of cells that were freeze-dried and then exposed at room temperature. For the case of freeze-dried, room temperature cells, the center positions of speckles moved outwards from the diffraction pattern center, indicating a shrinkage of the object in real space.
The strain of Saccharomyces cerevisiae studied here carries a whi5 mutation. This mutation [37] produces smaller yeast cells than the wild type (3–4 μm for whi5 versus 6–7 μm for wild type). Following culture in a YPD solution at 30 °C, cells were rinsed and diluted with distilled water to proper concentration, and allowed to settle on a formvar coated rectangular electron microscope grid. The cells were plunge-frozen in liquid ethane to minimize ice crystal formation (we did not use glycerol or other cryo protectants, though they may be helpful in future work). Frozen hydrated specimen grids were loaded into a Gatan 630 side-entry cryo specimen holder which was then inserted into a goniometer stage in a custom-built XDM system [38] where specimens can be viewed in a frozen hydrated state at −170 °C.
In order to reconstruct an image from far-field or Fraunhofer coherent diffraction intensities, one must be able to supply some a priori information; in most cases this involves knowledge that the specimen occupies only a fraction 1/σo (where σo is known as the oversampling ratio) of the field of view corresponding to the recorded diffraction pattern [19,20]. When imaging frozen hydrated cells, this means that the cells must be surrounded by a region of sufficiently smooth ice so as not to produce significant x-ray scattering. In order to approach this condition, considerable care had to be taken to minimize frosting of the grid.
Apart from the fact that the specimen was in a frozen hydrated state, the diffraction data were acquired in a manner similar to what we have described previously for studies of freeze-dried yeast cells [24]. A zone plate monochromator [39] was used at undulator beam line 9.0.1 of the Advanced Light Source at Lawrence Berkeley National Laboratory to deliver a 520 eV x-ray beam to the experimental apparatus [38]. By using a 5 μm pinhole located 25 mm upstream of the specimen, we were able to provide illumination with both sufficient spatial coherence [40], and the required temporal coherence of λ/Δλ ≃ 500. By working at 520 eV, we were able to take advantage of the “water window” spectral region with low absorption in water and good contrast for organic materials [1,41]. For this particular specimen, photodiode measurements of the transmitted flux through the ice near the cell, versus measurements of the incident flux with the specimen removed, indicate a transmission through the ice of about 90%, corresponding to an ice thickness of about 1 μm. Diffraction data were recorded on a Roper Scientific (Trenton, NJ) MTE-2 in-vacuum CCD detector with 1340 × 1300 pixels of 20 μm size each, located 17.5 cm downstream of the specimen. By using a movable beamstop and multiple summed recordings, we were able to record x-ray diffraction data over a large dynamic range spanning 5 orders of magnitude; however, we were not able to collect data from about a 20 × 20 pixel region at the center of the diffraction pattern due to detector saturation limits. The full 2D data set was assembled from 120 exposures with a total illumination time of 215 sec and an estimated total dose to the specimen of about 1.7 × 108 Gray.
For diffraction data analysis and image reconstruction, a 1100 × 1100 array was extracted from the assembled set of diffraction recordings. The assembled diffraction pattern extends to 37 μm−1 spatial frequency at corners, which corresponds to a half-period size of 14 nm. The pixel size of the real space image reconstruction array is about 17 nm. Once the tight support was found by the shrink wrap algorithm [42] and manual adjustments, the final result was obtained by averaging a total of 10 reconstructions from independent random phase starts. Each of these reconstructions was run for 104 iterations using the difference map algorithm [43], averaging every 20th iteration after the first 8000 iterations.
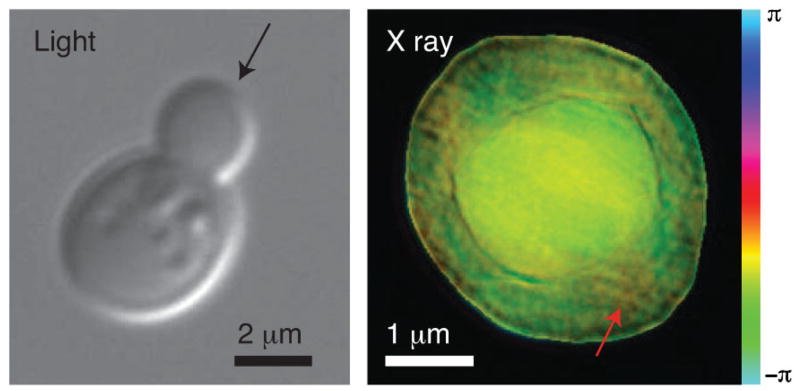
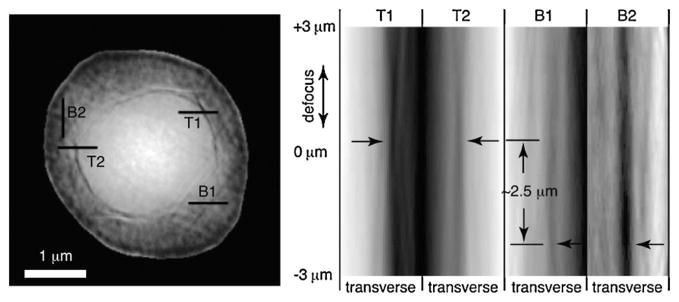
The right side of Fig. 2 shows the reconstructed complex image of a frozen hydrated yeast cell obtained using the method described above. For reference, a visible light micrograph of a budding yeast is also shown (obtained using a a Zeiss Axioplan2 microscope with a 100 ×, NA = 1.3 immersion objective with DIC optics, and a mRM Axiocam with Zeiss AXIOVISION 7.1 software). In the x-ray diffraction micrograph, we see what appears to be two cell bodies lying on top of each other. Since this inner cell body is too large to be a nucleus in this cell type, we believe that the image shows a new-born bud produced as part of the division process in these cells, viewed from directly above the bud with the parent cell underneath (see the assumed illuminating beam orientation in Fig. 2 as indicated by the black arrow). We have also collected a limited data set with the sample rotated at 15°. While we have not obtained a reconstructed image from this data set (possibly due to ice scattering, as discussed below), it provides useful autocorrelation data (obtained by squaring the Fourier transform of the recorded data intensities and applying a high-pass Fourier filter to the result) [44]. The autocorrelation agrees well with expectations for a budding yeast cell viewed in the assumed orientation. Additional evidence is available by “focusing through” the reconstructed image. Since we have reconstructed a complex wave field at a depth plane where the outermost edges of the object within its support are sharpest [45,46], we are able to use Fresnel propagation to generate the reconstructed wave field at nearby depth planes. In Fig. 3 we show a series of transverse (abscissa) versus longitudinal (ordinate) “images” of this through-focus procedure, where one can see sharp edges at depth planes that are separated by about 2.5 μm.
FIG. 2.
(color). Images of Saccharomyces cerevisiae yeast cells. At left is shown a visible light DIC image of a budding yeast taken using a 100× NA = 1.3 immersion objective. The arrow indicates the assumed beam direction for the x-ray diffraction micrograph at right, which is of a different yeast cell. The x-ray diffraction micrograph is a complex wave reconstruction at 0 μm defocus, where the magnitude is represented by brightness and the phase by hue. A possible mitochondrion is indicated with the red arrow.
FIG. 3.
Through-focus imaging using the complex wave front reconstructed in XDM. Four selected transverse lines are shown on the magnitude-only representation of the 0 μm defocus reconstructed wave front at left. The image at right shows transverse line profiles of the reconstructed image magnitude as the wave field is defocused by propagation from the reconstruction plane. In-focus edges look like the waist of an hourglass in such a representation; the line profiles from the inner and outer spherical objects in the reconstruction appear to be at different focal planes, consistent with the interpretation that the larger parent cell is at lines B1 and B2, and the bud is at lines T1 and T2.
The reconstructed x-ray diffraction micrograph shown in Fig. 2 does not have the same resolution or degree of image contrast as we have been able to demonstrate when imaging freeze-dried yeast [24]; we attribute this to the fact that the ice layer outside the cell is contributing some weak scattering which slightly violates the conditions required for imposition of a finite support constraint [47]. As the experiment proceeded, ice buildup continued which precluded reconstruction at additional tilt angles; this limitation is being addressed for future experiments through improved vacuum conditions and the use of an electron-microscope-type anticontaminator [48]. In spite of these limitations in our first frozen hydrated XDM demonstration, we are able to recognize subcelluar features including what may be a mitochondrion in the parent cell (indicated by the red arrow in Fig. 2).
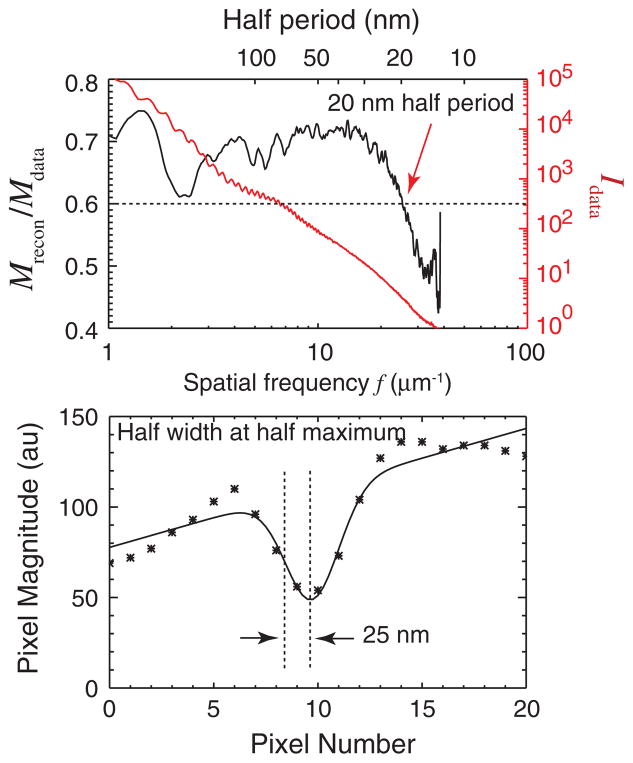
The resolution can be estimated to 25 nm or better using two independent measures. While the diffraction data extend to the edge of our CCD detector at a spatial frequency of 37 μm−1, we do not claim this as representing the resolution of our reconstructed image. Instead, the magnitude ratio Mrecon/Mdata (known as the phase retrieval transfer function or PRTF [24,45,49]) provides a good metric of the resolution of the reconstructed image. It does so by measuring the reproducibility of the phases recovered in the Fourier plane as the iterative reconstruction proceeds; phases that are reproducible lead to constructive interference when complex iterates are added together, while less reproducible phases lead to a lower value in the average. The spatial-frequency-dependent magnitude ratio Mrecon/Mdata plot is shown at left in Fig. 4. The resolution cutoff is estimated at the spatial frequency where the PRTF drops below a value of 0.6. This provides one estimate of the half-period resolution of 20 nm for the reconstructed x-ray diffraction micrograph.
FIG. 4.
(color). Estimation of the resolution of the x-ray diffraction micrograph. At top is shown both the azimuthal average of the diffraction intensity signal Idata (red), and the magnitude ratio (or PRTF) Mrecon/Mdata (black). The PRTF measures the reproducibility of reconstructed pixel values as a function of spatial frequency; its decline at higher frequency provides an indication of the resolution. The resolution cutoff is estimated at the spatial frequency where the PRTF drops below a value of 0.6. This provides one estimate of the half-period resolution of 20 nm. At bottom is shown a line scan across T2 in Fig. 3 at the plane of sharpest focus of this object. Also shown is a Gaussian-smoothed fit which indicates a half width at half maximum of 25 nm (or σ = 21 nm in exp[−0.5x2/σ2]) for the combination of feature size and imaging resolution.
Another measure of the resolution can be obtained by looking at the minimum width of features in the reconstructed image. Since the appearance of features in the image represents a convolution of their true shape with the point spread function of the imaging system, this measure is imperfect. Even so, in Fig. 4 we show a plot of the reconstructed intensity across a line in its in-focus plane, along with a Gaussian fit to the sharp feature seen. The Gaussian fit exp[−0.5x2/σ2] has σ = 21 nm, again indicating that a high resolution image was obtained.
The data reported here represent the first use of x-ray diffraction microscopy to image a frozen hydrated eukaryotic cell. It was obtained using a beam line and apparatus that is far from optimal for this purpose. Based on this experience we are now implementing improvements aimed at improving the apparatus, to be able to move from 2D to 3D imaging. We are also planning to construct an optimized beam line that will provide the required coherent flux to allow routine collection of a 3D data set in minutes, rather than hours. While x-ray free electron laser sources will also open up exciting new opportunities in XDM [8,50], for true 3D imaging it is necessary to acquire data from an object that remains unchanged as it is rotated so that one can populate 3D Fourier space [49,51]; this means the exposure in each 2D view must be delivered slowly so that the temperature of the specimen does not rise. For this reason, we feel that cryo XDM of frozen hydrated whole cells using synchrotron sources has considerable potential.
Acknowledgments
We wish to thank the National Institute for General Medical Services at the National Institutes for Health for support of the application of XDM to biological imaging under Contract No. 5R21EB6134. We also wish to thank the Division of Materials Sciences and Engineering, Office of Basic Energy Sciences, at the Department of Energy for support of XDM methods and instrumentation development under Contract No. DE-FG02-07ER46128. We thank the ALS staff for their excellent support. The ALS is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231. Finally, we thank David Sayre for helpful suggestions and discussions, and thank Ken Downing and Eva Nogales for their help with specimen freezing.
References
- 1.Sayre D, et al. Science. 1977;196:1339. doi: 10.1126/science.867033. [DOI] [PubMed] [Google Scholar]
- 2.Grimm R, et al. Biophys J. 1997;72:482. doi: 10.1016/S0006-3495(97)78689-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jacobsen C, et al. In: X-ray Microscopy and Spectromicroscopy. Thieme J, et al., editors. II. Springer-Verlag; Berlin: 1998. pp. 93–102. [Google Scholar]
- 4.Kirz J, et al. Q Rev Biophys. 1995;28:33. doi: 10.1017/s0033583500003139. [DOI] [PubMed] [Google Scholar]
- 5.Schneider G. Ultramicroscopy. 1998;75:85. doi: 10.1016/s0304-3991(98)00054-0. [DOI] [PubMed] [Google Scholar]
- 6.Maser J, et al. J Microsc. 2000;197:68. doi: 10.1046/j.1365-2818.2000.00630.x. [DOI] [PubMed] [Google Scholar]
- 7.Solem JC, Baldwin GC. Science. 1982;218:229. doi: 10.1126/science.218.4569.229. [DOI] [PubMed] [Google Scholar]
- 8.Bergh M, et al. Q Rev Biophys. 2008;41:181. doi: 10.1017/S003358350800471X. [DOI] [PubMed] [Google Scholar]
- 9.Wang Y, et al. J Microsc. 2000;197:80. doi: 10.1046/j.1365-2818.2000.00629.x. [DOI] [PubMed] [Google Scholar]
- 10.Weiß D, et al. Ultramicroscopy. 2000;84:185. doi: 10.1016/s0304-3991(00)00034-6. [DOI] [PubMed] [Google Scholar]
- 11.Schneider G, et al. Surf Rev Lett. 2002;9:177. [Google Scholar]
- 12.Larabell C, Le Gros M. Mol Biol Cell. 2004;15:957. doi: 10.1091/mbc.E03-07-0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parkinson D, et al. J Struct Biol. 2008;162:380. doi: 10.1016/j.jsb.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chao W, et al. Nature (London) 2005;435:1210. doi: 10.1038/nature03719. [DOI] [PubMed] [Google Scholar]
- 15.Jefimovs K, et al. Phys Rev Lett. 2007;99:264801. doi: 10.1103/PhysRevLett.99.264801. [DOI] [PubMed] [Google Scholar]
- 16.Kang H, et al. Appl Phys Lett. 2008;92:221114. [Google Scholar]
- 17.Kirz J. J Opt Soc Am. 1974;64:301. [Google Scholar]
- 18.Sayre D. In: Imaging Processes and Coherence in Physics. Schlenker M, et al., editors. Springer-Verlag; Berlin: 1980. pp. 229–235. [Google Scholar]
- 19.Fienup J. Opt Lett. 1978;3:27. doi: 10.1364/ol.3.000027. [DOI] [PubMed] [Google Scholar]
- 20.Miao J, et al. J Opt Soc Am A. 1998;15:1662. [Google Scholar]
- 21.Huang X, et al. Opt Express. 2009;17:13541. doi: 10.1364/oe.17.013541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miao J, et al. Nature (London) 1999;400:342. [Google Scholar]
- 23.Miao J, et al. Proc Natl Acad Sci USA. 2003;100:110. [Google Scholar]
- 24.Shapiro D, et al. Proc Natl Acad Sci USA. 2005;102(15):343. [Google Scholar]
- 25.Song C, et al. Phys Rev Lett. 2008;101:158101. doi: 10.1103/PhysRevLett.101.158101. [DOI] [PubMed] [Google Scholar]
- 26.Williams G, et al. Cytometry Part A. 2008;73a:949. doi: 10.1002/cyto.a.20616. [DOI] [PubMed] [Google Scholar]
- 27.Nishino Y, et al. Phys Rev Lett. 2009;102:018101. [Google Scholar]
- 28.Taylor K, Glaeser R. Science. 1974;186:1036. doi: 10.1126/science.186.4168.1036. [DOI] [PubMed] [Google Scholar]
- 29.Taylor K, Glaeser R. J Ultrastruct Res. 1976;55:448. doi: 10.1016/s0022-5320(76)80099-8. [DOI] [PubMed] [Google Scholar]
- 30.Steinbrecht RA, Zierold K, editors. Cryotechniques in Biological Electron Microscopy. Springer-Verlag; Berlin: 1987. [Google Scholar]
- 31.O’Toole E, et al. J Struct Biol. 1993;110:55. doi: 10.1006/jsbi.1993.1004. [DOI] [PubMed] [Google Scholar]
- 32.Medalia O, et al. Science. 2002;298:1209. doi: 10.1126/science.1076184. [DOI] [PubMed] [Google Scholar]
- 33.Lučić V, et al. J Struct Biol. 2007;160:146. doi: 10.1016/j.jsb.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 34.Shapiro D. PhD thesis. Stony Brook University; 2004. [Google Scholar]
- 35.Shen Q, et al. J Synchrotron Radiat. 2004;11:432. doi: 10.1107/S0909049504016772. [DOI] [PubMed] [Google Scholar]
- 36.Howells M, et al. J Electron Spectrosc Relat Phenom. 2009;170:4. doi: 10.1016/j.elspec.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tyers M, et al. EMBO J. 1993;12:1955. doi: 10.1002/j.1460-2075.1993.tb05845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beetz T, et al. Nucl Instrum Methods Phys Res, Sect A. 2005;545:459. doi: 10.1016/j.nima.2005.01.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Howells M, et al. In: Design and Microfabrication of Novel X-Ray Optics. Mancini D, editor. Vol. 4783. SPIE; Bellingham, WA: 2002. pp. 65–73. [Google Scholar]
- 40.Miao H. PhD thesis. Stony Brook University; 2008. [Google Scholar]
- 41.Wolter H. Ann Phys (Leipzig) 1952;10:94. [Google Scholar]
- 42.Marchesini S, et al. Phys Rev B. 2003;68:140101. [Google Scholar]
- 43.Elser V. J Opt Soc Am A. 2003;20:40. doi: 10.1364/josaa.20.000040. [DOI] [PubMed] [Google Scholar]
- 44.See EPAPS Document No. E-PRLTAO-103-029941 for supplementary autocorrelation figures. For more information on EPAPS, see http://www.aip.org/pubservs/epaps.html.
- 45.Thibault P, et al. Acta Crystallogr Sect A. 2006;62:248. doi: 10.1107/S0108767306016515. [DOI] [PubMed] [Google Scholar]
- 46.Spence J, et al. Phil Trans R Soc A. 2002;360:875. doi: 10.1098/rsta.2001.0972. [DOI] [PubMed] [Google Scholar]
- 47.Miao H, et al. J Phys Conf Ser. 2009;186:012055. [Google Scholar]
- 48.Cheng A, et al. J Struct Biol. 2006;154:303. doi: 10.1016/j.jsb.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 49.Chapman H, et al. J Opt Soc Am A. 2006;23:1179. doi: 10.1364/josaa.23.001179. [DOI] [PubMed] [Google Scholar]
- 50.Chapman H, et al. Nature Phys. 2006;2:839. [Google Scholar]
- 51.Miao J, et al. Phys Rev Lett. 2002;89:088303. [Google Scholar]