Abstract
Cells receive and interpret extracellular signals to regulate cellular responses such as proliferation, cell survival and differentiation. However, proper inactivation of these signals is critical for appropriate homeostasis. Cbl proteins are E3-ubiquitin ligases that restrict receptor tyrosine kinase (RTK) signaling, most notably EGFR (Epidermal Growth Factor Receptor), via the endocytic pathway. Consistently, many mutant phenotypes of Drosophila cbl (D-cbl) are due to inappropriate activation of EGFR signaling. However, not all D-cbl phenotypes can be explained by increased EGFR activity. Here, we report that D-Cbl also negatively regulates Notch activity during eye and wing development. D-cbl produces two isoforms by alternative splicing. The long isoform, D-CblL, regulates the EGFR. We found that the short isoform, D-CblS, preferentially restricts Notch signaling. Specifically, our data imply that D-CblS controls the activity of the Notch ligand Delta. Taken together, these data suggest that D-Cbl controls the EGFR and Notch/Delta signaling pathways through production of two alternatively spliced isoforms during development in Drosophila.
Keywords: Drosophila, cbl, D-cbl, RTK signaling, EGFR, Notch, Delta
Introduction
Proper development of multi-cellular organisms requires coordinated cell proliferation, cell survival and cell fate specification through the action of several common signaling pathways. Signaling mediated by the Epidermal Growth Factor Receptor (EGFR) and Notch play fundamental roles in these processes in organisms ranging from nematodes to mammals. Even minor defects in these pathways have been linked to developmental abnormalities and human disease.
Studies in the Drosophila compound eye have provided important insights into how these signaling pathways generate precise developmental patterns. The compound eye is composed of ~800 ommatidia, a repetitive unit containing a precise number of different cell types. Retinal specification starts during 3rd instar larval stage when the morphogenetic furrow (MF) moves from posterior to anterior across the eye disc (Ready et al., 1976) (reviewed by Voas and Rebay, 2004). Cells located in and posterior to the MF are specified to adopt cell fate in a strict sequence to form the ommatidium. Each ommatidium contains eight photoreceptor neurons (‘R’ cells), four cone cells and a number of pigment cells. R8 is the first specified neuron and serves as the founder cell for recruitment of the remaining R cells, R1–R7 (Cagan and Ready, 1989; Ready et al., 1976). The EGFR pathway controls the specification of all cell types except R8 (Freeman, 1994, 1996; Kumar et al., 1998).
Notch signaling is used several times during eye development (Cagan and Ready, 1989; Parks et al., 1995). In the MF, Notch controls lateral inhibition, a process through which individual R8 cells are selected from a group of cells with equivalent developmental potential. Lateral inhibition also ensures that the R8 cells are precisely spaced (Baker and Zitron, 1995; Cagan and Ready, 1989; Li and Baker, 2001). Later in eye development, Notch contributes to R7, cone and pigment cell specification (Cooper and Bray, 2000; Nagaraj and Banerjee, 2007; Tomlinson and Struhl, 2001; Tsuda et al., 2002).
Notch and its ligands Delta and Serrate are transmembrane domain proteins which signal in a juxtacrine manner. The activity of Notch, Delta and Serrate is controlled by endocytosis and endosomal protein sorting (reviewed in Le Borgne, 2006). These processes are regulated by several E3-ubiquitin ligases. Ubiquitination by the HECT-type E3-ubiquitin ligases Suppressor of deltex (Su(dx)/Itch) and Nedd4 routes Notch into the endocytic pathway for lysosomal degradation (Fostier et al., 1998; Sakata et al., 2004; Wilkin et al., 2004). In contrast, the RING finger E3-ubiqitin ligase Deltex (Dx) positively regulates Notch signaling (Hori et al., 2004; Matsuno et al., 1995). It is unclear how ubiquitination by Dx prevents Notch from Su(dx)- or Nedd4-induced degradation. It is possible that Dx keeps Notch in a different endosomal microenvironment which may allow processing and activation of Notch by γ-Secretase. This view is further supported by the recent suggestion that γ-Secretase cleavage of Notch occurs at the endosome (Vaccari et al., 2008).
Interestingly, the ligands of Notch require endocytic internalization for ligand maturation in the signal-sending cell (Seugnet et al., 1997). Two RING E3-ubiquitin ligases, Neuralized and Mind bomb, have been implicated in this process (Itoh et al., 2003; Lai et al., 2005; Le Borgne et al., 2005; Pitsouli and Delidakis, 2005; Wang and Struhl, 2005). Although the mechanism is not entirely clear, it is believed that after endocytosis and endosomal maturation, the ligands are recycled back to the cell surface for Notch activation (Chitnis, 2006). An E3-ubiquitin ligase that targets the Notch ligands for lysosomal degradation is not known.
The Casitas B-lineage lymphoma (Cbl) family of E3-ubiquitin ligases negatively regulates receptor tyrosine kinases (RTKs) including EGFR, PDGF, CSF-1 and others (reviewed in Schmidt and Dikic, 2005; Thien and Langdon, 2005b). However, non-RTK receptors including mammalian Notch1 have also been shown to be regulated by Cbl proteins (Jehn et al., 2002). Cbl proteins are conserved both in structure and function. Mammals encode three Cbl proteins, c-Cbl, Cbl-b and Cbl-3 which restrict RTK signaling (Schmidt and Dikic, 2005; Thien and Langdon, 2005b). Sli-1, the Cbl ortholog in C. elegans, attenuates the activity of Let-23, the EGFR equivalent, in vulval development (Yoon et al., 1995). In Drosophila, D-Cbl restricts EGFR in oogenesis, guided migration of border cells, and cell fate specification in the eye (Jekely et al., 2005; Meisner et al., 1997; Pai et al., 2000; Pai et al., 2006; Wang et al., 2008b). D-Cbl also restricts PVR (PDGF/VEGF Receptor) signaling during border cell migration (Jekely et al., 2005).
All Cbl family members contain an N-terminal tyrosine kinase binding (TKB) domain followed by a RING E3-ubiquitin ligase domain (see Fig. 1A). c-Cbl and Cbl-b also contain a Pro-rich domain (SH3 binding) and a C-terminal ubiquitin-associated (UBA) domain (Schmidt and Dikic, 2005). Cbl-c and Sli-1 do not or have only a short Pro-rich domain and no UBA. Cbl proteins recognize and bind Tyr-phosphorylated EGFR through their TKB domain, enabling the E3 ligase activity of the RING domain to mono-ubiquitinate EGFR which promotes EGFR degradation through the endocytic pathway (Haglund et al., 2003; Joazeiro et al., 1999; Levkowitz et al., 1999; Waterman et al., 1999; Yokouchi et al., 1999). The Pro-rich domain promotes additional interactions with adaptor molecules such as the SH3-protein Grb-2 which stimulates increased EGFR endocytosis (Huang and Sorkin, 2005; Wang and Moran, 1996; Waterman et al., 2002).
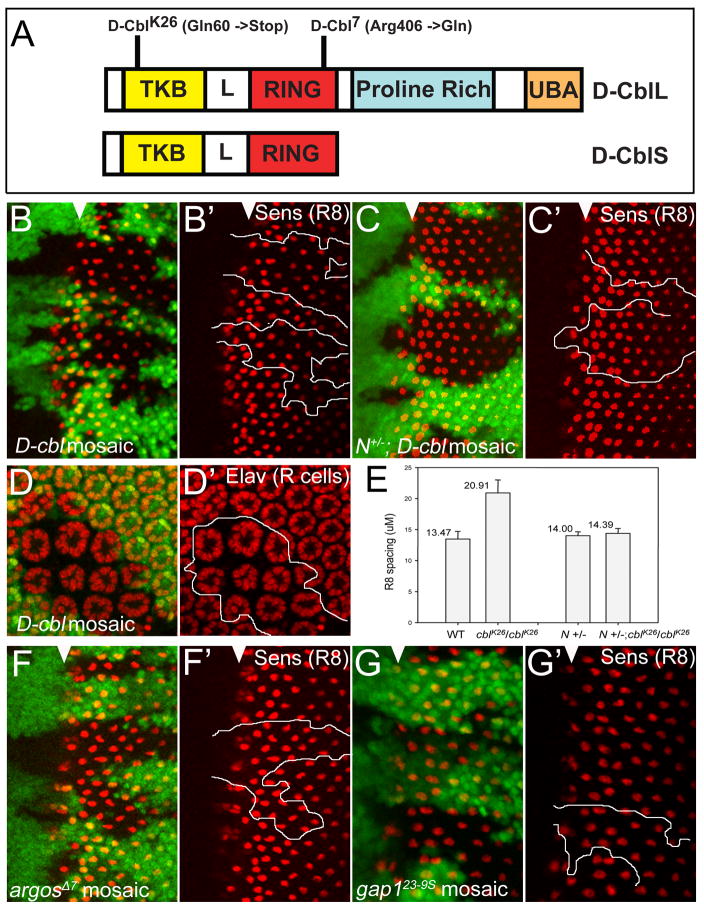
Figure 1. Increased R8 spacing in D-cbl clones is Notch-dependent.
(A) Domain structure of the long and short isoforms of D-Cbl. The relative locations of the non-sense mutation in D-cblK26 and the missense mutations in D-cbl7 are indicated. TKB - tyrosine kinase binding domain; L – Linker; RING - RING E3 ubiquitin ligase; UBA - ubiquitin-associated domain.
(B,B′) Increased spacing between R8 photoreceptor cells in D-cblK26 mutant clones. Anti-Senseless (Sens) labeling is shown in red to mark R8 cells. Anterior is to the left. Absence of GFP marks D-cbl mutant clones outlined by white lines in (B′). White triangles mark the position of the morphogenetic furrow. Genotype: ey-Flp; D-cblK26 FRT80/P[ubi-GFP] FRT80.
(C,C′) Heterozygosity of Notch (N) normalizes R8 spacing in D-cblK26 mutant clones. Mutant clones are outlined by white lines in (C′). White triangles mark the position of the morphogenetic furrow. Genotype: ey-Flp/Df(1)N8; D-cblK26 FRT80/P[ubi-GFP] FRT80. A similar result was obtained with N264-39 (data not shown).
(D,D′) Increased ommatidial spacing in D-cblK26 clones in 42 hours APF pupal eye discs labeled with Elav (marks photoreceptor neurons or R cells) in red. The D-cbl clone is marked by absence of GFP and outlined by a white line in (D′). Genotype as in (B).
(E) Summary of the analysis of R8 spacing. The distances between R8 cells in the MF and posterior to the MF in eye imaginal discs of the indicated genotypes are measured by Axiovision 6 program with the average length per R8 pairs in μM. A total of 100 R8 pairs from 10 different discs were measured for each data point.
(F,F′) argosΔ7 and (G,G′) gap123-9s mutant clones do not affect R8 spacing as determined by Sens labeling in red. Mutant clones are outlined by white lines in (F′,G′). White triangles mark the position of the morphogenetic furrow. Genotypes: ey-Flp; argosΔ7 or gap123-9s FRT80/P[ubi-GFP] FRT80.
D-cbl encodes two alternatively spliced isoforms, D-CblSHORT (D-CblS) and D-CblLONG (D-CblL), both of which contain the TBK and the RING E3-ubiquitin ligase domains, while D-CblL also has Pro-rich and UBA domains similar to c-Cbl and Cbl-b (Fig. 1A) (Hime et al., 2001; Hime et al., 1997; Meisner et al., 1997; Robertson et al., 2000). Although both D-cblL and D-cblS are expressed at all developmental stages, D-cblS is the prominent form of the D-cbl isoforms (Hime et al., 2001; Pai et al., 2006; Robertson et al., 2000). Overexpression of D-cblL results in phenotypes characteristic of reduced EGFR signaling (Pai et al., 2006). Overexpression of D-cblS has little, if any, effect on EGFR signaling (Meisner et al., 1997; Pai et al., 2006). Taken together, these data indicate that the two isoforms of D-cbl may play different roles in the developing organism. However, the precise function of D-cblS has not been resolved.
We have previously characterized the role of D-cbl for eye development (Wang et al., 2008b). Many of the D-cbl mutant phenotypes are characteristic of increased EGFR activity. However, some D-cbl phenotypes such as increased R8 spacing are independent of EGFR activity. Here, we provide genetic evidence that D-Cbl also restricts Notch signaling. Significantly, while the long isoform, D-CblL, is required for regulation of EGFR, the short isoform, D-CblS, preferentially restricts Notch activity. Specifically, our data suggest that D-CblS regulates the Notch ligand Delta. Thus, during Drosophila development D-cbl controls at least two pathways, EGFR and Notch/Delta, through production of two alternatively spliced isoforms.
Material and methods
Drosophila genetics
Fly crosses were conducted using standard procedures at 25°C. Pupal developmental ages are expressed as hours after puparium formation (APF) with white pre-pupae defined as 0 hour APF. The following stocks were used: D-cblK26 and D-cbl7 (Wang et al., 2008b), egfrf2 (flbf2; Nusslein-Volhard et al., 1984), rl10a (Freeman, 1994), argosΔ7 (Freeman et al., 1992), gap123-9s (Bergmann et al., 1998), Df(1)N8 (Couso and Martinez Arias, 1994), N264-39 (obtained from Bloomington), UAS-D-cblL (line A18), UAS-D-cblS (lines A1 and 13-2) (Pai et al., 2006), UAS-EGFRDN (Freeman, 1996), UAS-Delta and, UAS-Ser (provided by Hugo Bellen), ey-Flp; P[ubi-GFP] FRT80 (provided by Georg Halder), hs-Flp actin-Gal4 UAS-GFP; tub-Gal80 FRT80 (provided by Hugo Bellen), hs-Flp; tub<GFP<Gal4 (provided by Hyung Don Ryoo).
Generation of mutant clones and expression of transgenes
To generate D-cbl mutant clones, D-cblK26 FRT80 and D-cbl7 FRT80 flies were crossed to ey-FLP; P[ubi-GFP] FRT80 and hs-FLP; P[ubi-GFP] FRT80. Clones are marked by loss of GFP. For induction of MARCM clones (Lee and Luo, 2001) in Fig. 3, D-cblK26 FRT80 and D-cbl7 FRT80 flies were crossed to hs-Flp actin-Gal4 UAS-GFP; tub-Gal80 FRT80. In this case, mutant clones are positively marked by GFP. For induction of D-cblL- and D-cblS-expressing clones, the UAS-based D-cbl transgenes were crossed to hs-Flp; tub<GFP<Gal4 (< = FRT). Clones were induced by heat shock at 37°C for 1 hour in a water bath between 24 and 48 hours after egg laying, and are marked by the absence of GFP. In the rescue experiment (Fig. 7), we used the MARCM system to express D-cblL or D-cblS in D-cblK26 mutant clones, positively marked by GFP. For expression along the anterioposterior (AP) boundary of wing imaginal discs, UAS-D-cblL, UAS-D-cblS, UAS-EGFRDN, UAS-Delta and UAS-Ser transgenes were crossed to dpp-Gal4 UAS-GFP.
Figure 3. D-Cbl antagonizes Notch signaling.
(A,B) Adult wing of a Notch heterozygote (A) and a trans-heterozygote of Notch and D-cblK26 (B). The Notch allele used is Df(1)N8.
(C-C″) Anti-Cut (red) labeling of D-cblK26 mosaics in wing imaginal discs induced by the MARCM system. Homozygous mutant clones are marked by GFP (C,C′). The white line in (C″) marks the clonal boundary. The arrow points to non-autonomous increase of Cut labeling. Genotype: hs-Flp actin-Gal4 UAS-GFP; tub-Gal80 FRT80/D-cblK26 FRT80.
(D-D″) Anti-Wingless (Wg) (red) labeling of D-cblK26 mosaics in wing imaginal discs induced by the MARCM system. Homozygous mutant clones are marked by GFP (D,D′). The white line in (D″) marks the clonal boundary. Note the increased Wg immunolabeling in D-cbl mutant tissue (arrow in D″). Genotype in (C) and (D): hs-Flp actin-Gal4 UAS-GFP; tub-Gal80 FRT80/D-cblK26 FRT80.
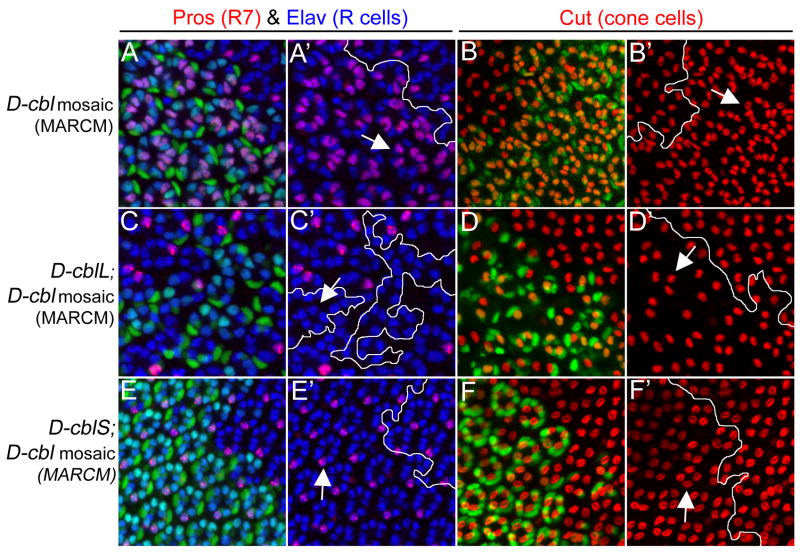
Figure 7. Distinct rescue of the D-cbl over-recruitment phenotype by expression of the D-cbl isoforms.
Shown are pupal (42h APF) eye discs labeled with anti-Pros/Elav antibodies (red/blue) for R7/photoreceptor (R) cells and anti-Cut antibody (red) for cone cells. D-cbl mutant clones are positively labeled by GFP due to the MARCM technique (outlined by white lines) and express either no transgene (A,A′,B,B′), D-cblL (C,C′,D,D′) or D-cblS (E,E′F,F′). GFP-negative cells are non-mutant (wild-type or heterozygous) and do not express any transgene. Arrows point to representative examples of the phenotypes in the mutant clones as explained in the text. The null allele D-cblK26, and the transgenes UAS-D-cblL (line A18) and UAS-D-cblS (line A1) transgenes were used.
Genotypes:
(A,B) hs-Flp actin-Gal4 UAS-GFP; D-cblK26 FRT80/tub-Gal80 FRT80;
(C,D) hs-Flp actin-Gal4 UAS-GFP; UAS-D-cblL/+; D-cblK26 FRT80/tub-Gal80 FRT80;
(E,F) hs-Flp actin-Gal4 UAS-GFP; UAS-D-cblS/+; D-cblK26 FRT80/tub-Gal80 FRT80.
Immunohistochemistry
Eye imaginal discs from the indicated larval or pupal stages were dissected and immunohistochemical labeling was performed as described (Herz et al., 2006). The following antibodies were used: mouse anti-Pros (1:50), rat anti-Elav (1:60), mouse anti-Cut (1:200), mouse anti-Wingless (1:20) and Mouse anti-Delta (1:100) obtained from the DSHB, University of Iowa; Rabbit anti-dpERK (1:2,000; Sigma), Guinea pig anti-Senseless (1:500) was kindly provided by Hugo Bellen, and rabbit anti-phosphotyrosine (1:500) by Georg Halder. Cy3- and Cy-5 fluorescently-conjugated secondary antibodies are obtained from Jackson ImmunoResearch and were used at dilutions of 1:400. Images were captured using an Olympus Optical FV500 confocal microscope. The distances between R8 cells in the MF and posterior to the MF in eye imaginal discs of the indicated genotypes in Fig. 1E were measured by Axiovision 6 program with the average length per R8 pairs in μM. A total of 100 R8 pairs from 10 different discs were measured for each data point.
Results
Increased R8 spacing in D-cbl clones is Notch-dependent
Previously, it has been reported that D-Cbl negatively regulates EGFR signaling (Hime et al., 1997; Jekely et al., 2005; Meisner et al., 1997; Pai et al., 2000; Pai et al., 2006; Robertson et al., 2000; Wang et al., 2008b). We showed that during eye development the null allele D-cblK26 which contains a nonsense mutation at amino acid residue 60 (Fig. 1A), displays inhibition of apoptosis and specification defects (Wang et al., 2008b). These phenotypes are similar to the phenotypes described for other negative regulators of EGFR such as argos, gap1 and sprouty (Casci et al., 1999; Freeman et al., 1992; Gaul et al., 1992; Kramer et al., 1999; Kretzschmar et al., 1992; Okano et al., 1992; Schweitzer et al., 1995). However, at least two phenotypes appear to be specific for D-cbl. D-cbl mutant heads and imaginal discs are overgrown (Wang et al., 2008b). In addition, we noted that the spacing between individual ommatidia is wider in D-cbl clones compared to wild-type. This is particularly striking during pupal stages (Fig. 1D,D′). To determine the origin of the increased ommatidial spacing, we labeled 3rd instar eye imaginal discs with an antibody against Senseless (Sens), a R8 cell marker (Nolo et al., 2000). R8 cells are the founder cells of the ommatidia, and are thus a convenient marker for ommatidial spacing. By Sens labeling, we found that the spacing of R8 cells in D-cbl clones is increased approximately 1½ fold compared to wild-type (Fig. 1B,B′; quantified in Fig. 1E). Importantly, increased R8 and ommatidial spacing originates at the MF (Fig. 1B,B′) and is therefore not the result of over-recruitment of several cell types into the ommatidia later in development.
We tested whether the increased R8 spacing in D-cbl clones is caused by increased EGFR signaling. However, halving the gene dose of EGFR has no detectable effect on R8 spacing in D-cbl clones (data not shown). To exclude the possibility that EGFR is not dosage sensitive for increased R8 spacing in D-cbl, we tested whether increasing the signaling strength of EGFR by inducing mutant clones of argos and gap1, two negative regulators of EGFR signaling, can increase R8 spacing. However, that was found not to be the case (Fig. 1F,F′,G,G′) consistent with a previous report (Baonza et al., 2001).
Notch signaling controls R8 spacing by lateral inhibition (Baker and Zitron, 1995; Cagan and Ready, 1989; Li and Baker, 2001). Thus, we tested whether the increased R8 spacing in D-cbl clones may be caused by increased Notch signaling. Consistently, removing one copy of Notch significantly suppressed the increased R8 spacing in D-cbl clones (Fig. 1C,C′; quantified in Fig. 1E). Thus, the increased R8 and subsequently ommatidial spacing in D-cbl mutant clones is largely mediated through increased Notch signaling. These observations suggest that D-Cbl restricts Notch signaling for R8 spacing in eye development.
The specification defects in D-cbl clones are dependent on both EGFR and Notch
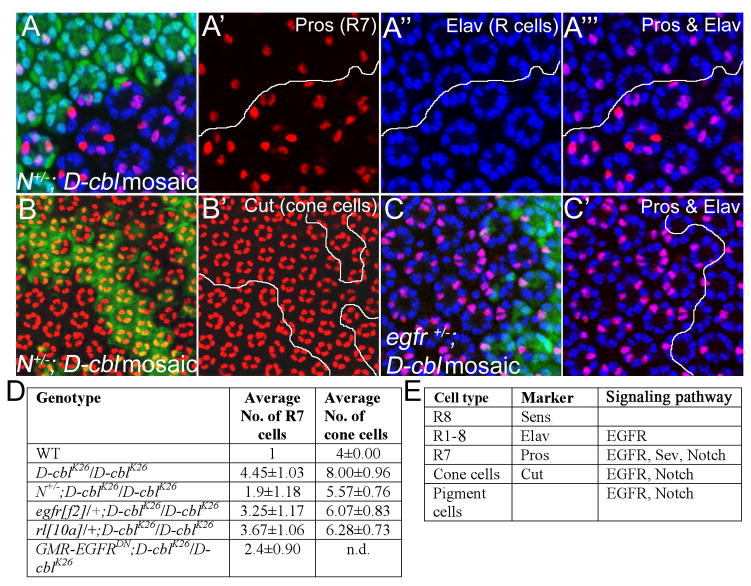
In previous work, we have shown that D-cbl mutant ommatidia display severe specification defects in the developing eye (Wang et al., 2008b). Specifically, in D-cbl mutant ommatidia we found increased numbers of R7 neurons (4.45 ± 1.03 versus 1 in wild-type), cone cells (8 ± 0.96 versus 4 in wild-type) and pigment cells (see Figs. 2D and 7A,B as examples). Although these over-recruitment phenotypes are typical for increased EGFR activity, it is noteworthy that the affected cell types (R7, cone cells) also require Notch for specification (Fig. 2E) (Cooper and Bray, 2000; Nagaraj and Banerjee, 2007; Tomlinson and Struhl, 2001; Tsuda et al., 2002). Thus, we tested whether inappropriate activation of Notch also contributes to the over-recruitment of these cell types in D-cbl mutants. A table indicating the cell type-specific markers used is shown in Fig. 2E. Indeed, heterozygosity of Notch significantly suppresses the over-recruitment of R7 cells (1.9 ± 1.18 per ommatidium, n=35) (Fig. 2A-A‴, quantified in Fig. 2D) and cone cells (5.6 ± 0.76 per ommatidium, n=35) in D-cbl mutant clones (Fig. 2B,B′,D). Surprisingly, the suppression of the specification defect in D-cbl clones is much more pronounced in heterozygous Notch background compared to heterozygous egfr or rl (rolled, encoding the Drosophila MAPK homolog) (Fig. 2C,C′,D). Even expression of a dominant negative EGFR (EGFRDN) construct does not suppress the R7 over-recruitment phenotype of D-cbl mutants as well as heterozygous Notch (Fig. 2D). Thus, these observations provide further evidence to support the notion that D-Cbl restricts Notch signaling during eye development.
Figure 2. Partial rescue of R7 and cone cell defects in egfr+/− and N+/− background.
Analysis of R7, photoreceptors (R cells) (A-A‴) and cone cells (B,B′) in D-cblK26 clones in heterozygous Notch (N) background by Pros/Elav (red/blue) double labelings (A-A‴) and Cut (red) labelings (B,B′) in 42 hrs APF pupal eye imaginal discs. Mutant clones are outlined by white lines in A′-A‴ and B′. Compared to D-cblK26 clones alone (Fig. 7A,B), R7 and cone cells are significantly reduced in N/+;D-cbl mutant ommatidia (see quantification in D). Genotype: ey-Flp/Df(1)N8; D-cblK26 FRT80/P[ubi-GFP] FRT80.
(C,C′) Anti-Pros (red) and anti-Elav (blue) double labeling of D-cblK26 mosaics heterozygous for egfr in 42 hours APF eye disc. Mutant clones are outlined by white lines in C′. Genotype: ey-Flp; egfrf2/+; D-cblK26 FRT80/P[ubi-GFP] FRT80.
(D) Summary of the total number of R7 and cone cells in 42 hrs APF pupal eye imaginal discs of the indicated genotypes as determined by Anti-Pros and Elav staining. A total of 35 ommatidia were counted for each genotype.
(E) List of cell type-specific markers and the signaling pathways involved in specification of the cell types listed.
D-Cbl negatively regulates Notch signaling during wing development
In the previous two sections, we demonstrated that Notch can dominantly suppress D-cbl mutant phenotypes. We also tested the reciprocal interaction, whether D-cbl mutants affect Notch-dependent phenotypes in the Drosophila wing. Heterozygous Notch flies are characterized by a notched wing margin phenotype (Fig. 3A). This Notch phenotype is dominantly suppressed by removing one copy of D-cbl (Fig. 3B), suggesting that D-cbl antagonizes Notch in the wing.
To further characterize this genetic interaction, we generated D-cbl mutant clones in wing imaginal discs of 3rd instar larvae using the MARCM system (Lee and Luo, 2001). Expression of Cut is a well characterized marker of Notch activity. Cut is cell-autonomously induced along the presumptive wing margin of 3rd instar larval wings by high levels of Notch signaling (de Celis et al., 1996; Micchelli et al., 1997). We found that in D-cbl mutant clones (marked by GFP) Cut labeling is increased indicating elevated Notch signaling (Fig. 3C-C″). There are two interesting observations in this experiment. First, loss of D-cbl does not induce ectopic expression of Cut (Fig. 3C′,C″), i.e. Notch activity. Instead, the signaling strength appears to be increased. Second, a non-autonomous increase of Cut-labeling is detectable immediately adjacent to D-cbl clones (arrow in Fig. 3C″). Because D-Cbl protein is expected to act autonomously in the cell, it may affect a protein which acts in a non-autonomous manner. Because the Notch signaling system acts in a juxtacrine manner and because the non-autonomous effect extents approximately one cell outside of the D-cbl clone (Fig. 3C′,C″), D-Cbl targets potentially – directly or indirectly - a ligand of Notch. We also found that the levels of Wingless (Wg) protein, a second marker of Notch activity along the wing margin (Couso et al., 1994; Rulifson and Blair, 1995) are increased in D-cblK26 mutant tissue (Fig. 3D-D″). However, because of the secreted nature of Wg, statements about autonomy/non-autonomy cannot be made. Taken together, we conclude that D-Cbl negatively regulates the Notch signaling pathway during development.
The short isoform D-CblS preferentially antagonizes Notch signaling
Next, we determined which of the two alternatively spliced isoforms of D-cbl (Fig. 1A) accounts for the control of Notch signaling. However, the seven described D-cbl alleles affect both the long and the short isoforms (Pai et al., 2000; Pai et al., 2006; Wang et al., 2008b) precluding us from using allele-specific mutants to address the individual functions of the isoforms. Nevertheless, we used UAS-based transgenes of the isoforms to address this question. These transgenes express the isoforms several fold over endogenous levels (Pai et al., 2006). Using these transgenes, it has been shown that the long isoform, D-CblL, inhibits EGFR activity (Pai et al., 2006). We tested which of the D-Cbl isoforms regulates Notch activity by assaying Cut expression along the wing margin of the disc. Cut expression was unaffected when D-cblL was expressed either in heat-shock-induced clones (Fig. 4A,A′), or along the anterioposterior (AP) boundary perpendicular to Cut using dpp-Gal4 (Fig. 4D,D′). Significantly, however, expression of the short isoform, D-cblS, inhibited Cut expression both in heat-shock-induced clones (Fig. 4B,B′) and in the dpp expression domain (Fig. 4E,E′). Interestingly, expression of D-cblS also appears to have a non-autonomous effect on Cut expression (Fig. 4B,B′,E,E′). In addition, Wg protein levels are also reduced by expression of the short isoform (Suppl. Fig. S1). As a control, to rule out that expression of D-cblS reduces EGFR activity which then in turn may suppress Cut expression, we directly reduced EGFR activity by expressing EGFRDN in heat shock-induced clones or along the AP boundary by dpp-Gal4. However, Cut expression is unaltered in EGFRDN-expressing areas (Fig. 4C,C′,F,F′) supporting the notion that the Cut-suppressing activity of D-CblS is not an indirect effect of reduced EGFR activity. Instead, it supports our observation that the short isoform, D-CblS, can regulate Notch activity.
Figure 4. D-CblS suppresses the Notch target gene Cut.
Wing imaginal discs were labeled for Cut as Notch activity marker in red. Clones over-expressing D-cblL (A,A′), D-cblS (B,B′) and EGFRDN (C,C′) were obtained by crossing the corresponding UAS lines to hs-Flp; tub<GFP<Gal4 flies, and are negatively marked by the absence of GFP. The top panels of each experiment are the combined GFP and Cut channels, the (′) panels are the Cut labelings only. Expression of D-cblS reduced expression of Cut (B,B′, arrow). Induction of D-cblL (A,A′) and EGFRDN (C,C′) did not affect Cut expression. White lines mark the clone boundaries.
Expression of D-CblS (E,E′) specifically along the AP boundary of wing discs by dpp-Gal4, positively marked by GFP, strongly suppresses Cut expression (arrow in E′). Expression of D-cblL (D,D′) and EGFRDN (F,F′) have no effect in this assay. The UAS-D-cblL (line A18) and UAS-D-cblS (line A1) transgenes were used.
D-Cbl regulates RTK activity via the long isoform D-CblL
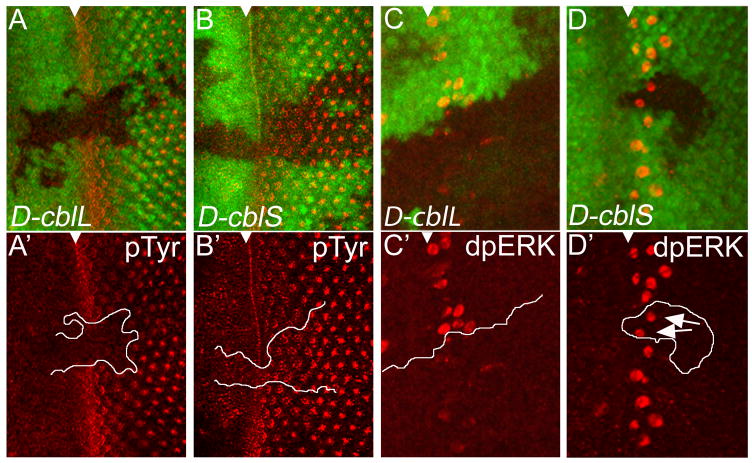
It has previously been shown that the D-cblL and D-cblS transgenes are expressed several fold over endogenous levels (Pai et al., 2006). However, to rule out the possibility that the D-cblL transgene does not produce functional D-CblL protein (which may explain the failure of D-CblL to affect Notch markers), we tested the ability of D-CblL and D-CblS to regulate RTK activity markers. First, we used anti-phosphotyrosine (pTyr) antibody labelings as molecular marker of RTK activity in third instar larval eye discs. Expression of D-cblL significantly reduces pTyr labeling (Fig. 5A,A′), whereas expression of D-cblS does not affect pTyr (Fig. 5B,B′), clearly visible in clones crossing the MF and posterior to the MF. Non-autonomous effects of D-CblL regulation on RTK activity were not detected. Similarly, overexpression of D-cblL, but not D-cblS, results in loss of dpERK labeling, a marker of activated MAPK downstream of RTK signaling (Fig. 5C,C′,D,D′). These results show that the D-cblL transgene produces functional D-CblL protein and are consistent with previous reports that in the Drosophila ovary expression of D-cblL, but not D-cblS, suppresses expression of EGFR markers such as pipe and argos, and induces endocytosis of the EGFR ligand Gurken (Chang et al., 2008; Pai et al., 2006; Wang et al., 2008a). In summary, the results presented in Figures 4 and 5 strongly suggest that the two isoforms of D-Cbl have distinct preferences for EGFR and Notch signaling.
Figure 5. D-CblL negatively regulates EGFR activity.
Heat shock-induced expression of D-cblL (marked by absence of GFP) reduces pTyr labeling (in red) (A,A′) and dpERK protein (red) levels (C,C′) in 3rd instar eye discs. The top panels of each experiment are the combined GFP and pTyr/dpERK channels, the (′) panels are the pTyr/dpERK labelings only. Heat shock-induced clones expressing D-cblS do not affect pTyr (B,B′) and dpERK levels (D,D′). The white triangle marks the position of the MF. White lines in (A′,B′,C′,D′) mark the clone boundaries. The arrows in (D′) point to dpERK positive cells in D-cblS expressing clones. The UAS-D-cblL (line A18) and UAS-D-cblS (line A1) transgenes were used.
D-CblS regulates Delta
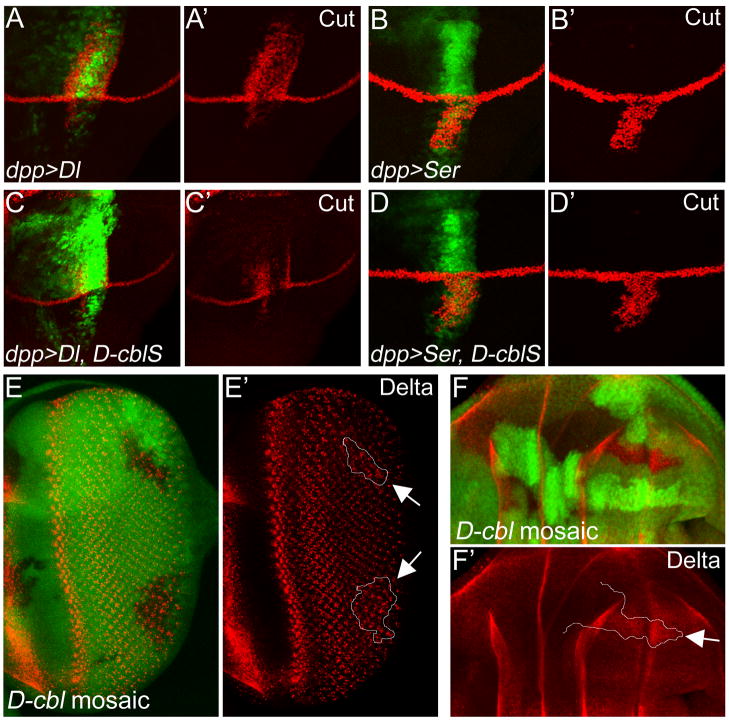
Because of the non-autonomous effects of D-cbl mutant clones (Fig. 3C′,C″) and D-cblS overexpression in the wing (Fig. 4B,B′,E,E′), we considered the possibility that D-cblS regulates Delta and/or Serrate (Ser), the two Notch ligands. To address this possibility, we expressed Delta and Ser with or without D-cblS using dpp-Gal4 and scored for Cut expression. As previously reported, dpp-driven expression of Delta induces Cut expression in the dorsal half of the dpp expression domain in wing discs (Fig. 6A,A′), whereas dpp-driven Ser induces Cut expression in the ventral half of the disc (Fig. 6B,B′) (Doherty et al., 1996; Fleming et al., 1997; Klein et al., 1997; Panin et al., 1997). Significantly, co-expression of D-cblS strongly suppresses Delta-induced Cut expression (Fig. 6C,C′), but has little effect on Ser-induced Cut expression (Fig. 6D,D′). This finding strongly implies that D-CblS preferentially regulates Delta, but not Ser.
Figure 6. D-CblS negatively regulates Delta.
(A,B) Overexpression of Delta (Dl) (A,A′) and Serrate (Ser) (B,B′) along the AP boundary using dpp-Gal4 induces ectopic Cut expression in the dorsal or ventral part of the dpp-expression domain. GFP marks the dpp expression domain.
(C,D) Co-expression with D-cblS suppresses Delta-induced Cut expression (C,C′), but has little effect on Ser-induced Cut expression (D,D′). GFP marks the dpp expression domain.
(E,F) Accumulation of Delta protein in D-cbl7 mutant clones posterior to the MF (E,E′) and in the wing disc (E,F). D-cbl7 affects the RING E3-ubiquitin ligase domain of D-Cbl (Fig. 1A). Delta protein does not accumulate in D-cbl clones in regions where Delta is normally not present. Clones are marked by the absence of GFP and outlined by white lines in E′ and F′ (see arrows). Genotypes: (E,E′) ey-Flp; D-cbl7 FRT80/P[ubi-GFP] FRT80. (F,F′) hs-Flp; D-cbl7 FRT80/P[ubi-GFP] FRT80.
To further characterize this genetic interaction, we analyzed protein levels of Delta in D-cbl mutant clones in wing and eye imaginal discs. Consistently, in clones of the RING domain mutant D-cbl7, a subtle but detectable accumulation of Delta protein posterior to the MF (Fig. 6E,E′) and in wing discs is detectable (Fig. 6F,F′) suggesting that lack of ubiquitylation and potentially proteasomal degradation accounts for the accumulation of Delta. Clones of the null allele, D-cblK26, gave similar results (data not shown). Loss of D-cbl does not induce ectopic accumulation of Delta protein in regions where it is normally not expressed. Protein levels of Ser and Notch are not detectably altered in D-cbl clones (data not shown). In summary, this analysis demonstrates that D-Cbl regulates the Notch-ligand Delta.
D-cblS rescues the D-cbl over-recruitment phenotype in the developing eye
Next, we performed rescue experiments of the D-cbl mutant phenotype in the developing eye by expressing the individual D-cbl isoforms. It was previously shown that both isoforms partially or completely rescue the lethality of D-cbl mutant animals (Jekely et al., 2005; Pai et al., 2006). However, the relative contribution of each isoform in the absence of the other for eye development has not been determined yet. In this set of experiments, we expressed either D-cblL (Fig. 7C,D) or D-cblS (Fig. 7E,F) in D-cblK26 null mutant clones using the MARCM system (Lee and Luo, 2001). D-cbl mutant clones expressing the D-cbl isoforms are positively labeled by GFP. GFP-negative cells are non-mutant (wild-type or heterozygous) and do not express any transgene. D-cbl mutant ommatidia contain 4.45 ± 1.03 R7 cells compared to one in wild-type, and 8.00 ± 0.96 cone cells compared to 4 in wild-type (arrows in Fig. 7A,B; Fig. 2D). Individual expression of D-cblL or D-cblS reduces the number of R7 and cone cells in D-cbl clones, indicating rescue of the D-cbl over-recruitment phenotype. However, the rescue is qualitatively and quantitatively different for the isoforms. Over-expression of D-cblL in D-cblK26 mutant ommatidia not only suppresses the over-recruitment phenotype, but even transforms it into a under-recruitment phenotype relative to wild-type, characterized by loss of R7 and other types of photoreceptor neurons as well as cone cells (arrows in Fig. 7C′,D′). This under-recruitment phenotype is reminiscent of reduced EGFR signaling. Thus, consistent with previous reports (Pai et al., 2006), D-CblL preferentially restricts EGFR signaling.
Strikingly, expression of D-cblS rescues the D-cbl null mutant phenotype to almost normal (Fig. 7E,E′,F,F′). The rescued ommatidia contain eight R cells with one R7 cell, and usually four cone cells (arrows in Fig. 7E′,F′). The increased ommatidial spacing in D-cbl mutants is restored upon expression of D-cblS. The differences of the rescue by D-cblL and D-cblS are not due to low level expression of the transgenes, as both transgenes are expressed several fold over the levels of the endogenous isoforms (Pai et al., 2006). Therefore, this analysis suggests that the long isoform is dispensable for eye differentiation which was also concluded for ovary development (Pai et al., 2006) and may explain why D-cblL-specific alleles were not isolated in several mutagenesis screens (Pai et al., 2000; Pai et al., 2006; Wang et al., 2008b).
Discussion
In this study we provide genetic evidence that D-Cbl not only restricts EGFR signaling, but also Notch signaling (summarized in Fig. 8). It had previously been shown that the long isoform, D-CblL, accounts for the control of EGFR (Pai et al., 2006) which we also confirm in this study. However, a specific function of the short isoform, D-CblS, had not been assigned previously. Here, we provide genetic evidence that D-CblS preferentially restricts Notch activity in vivo through regulation of Delta (Fig. 8).
Figure 8. Summary of D-Cbl activities.
Shown are the characterized activities of D-CblL and D-CblS in this report and by (Wang et al., 2008b) during eye and wing development. The dotted line indicates a potential minor activity of D-CblS towards EGFR.
Importantly, we found that the regulation of EGFR and Notch occurs through distinct isoforms of D-cbl. Expression of the short isoform, D-cblS, affects Notch-dependent Cut and Wg expression, but does not alter EGFR markers. In contrast, expression of the long isoform, D-cblL, does not influence Notch activity markers, but does affect EGFR activity. These findings suggest that D-CblL restricts EGFR, while D-CblS negatively regulates Notch signaling.
We have previously reported that loss of D-cbl causes an over-recruitment phenotype which affects R7 photoreceptor neurons and cone cells (Wang et al., 2008b) (see also Fig. 7A,B and Fig. 2D). We performed rescue experiments of the over-recruitment phenotype with the two D-cbl isoforms. D-CblL expression transforms the over-recruitment phenotype into an under-recruitment phenotype characterized by loss of R7, other photoreceptor neurons and cone cells (Fig. 7C,D). This phenotype is reminiscent of a hypomorphic EGFR condition, consistent with similar D-cblL-induced phenotypes in ovaries (Pai et al., 2006). In contrast, expression of D-cblS rescues the over-recruitment phenotype of D-cbl clones back to wild-type (Fig. 7E,F). This result suggests that the long isoform is dispensable for eye specification. In this context, it is also noteworthy that D-cblS is the prominent isoform during all developmental stages (Hime et al., 2001; Pai et al., 2006; Robertson et al., 2000).
There are two possibilities to explain the wild-type rescue of the over-recruitment phenotype of D-cbl mutants by D-CblS. First, the over-recruitment phenotype is the result of increased Notch, but not EGFR, activity. This possibility may explain why reducing EGFR and MAPK activity only weakly suppresses the over-recruitment phenotype, while Notch heterozygosity strongly suppresses it (Fig. 2D). Second, D-CblS does not only control Notch, but also EGFR activity. Consistent with this possibility, it was shown that D-CblS can physically interact with EGFR in vitro (Hime et al., 1997; Meisner et al., 1997) and that D-cblS can control EGFR in the ovary (Pai et al., 2006). However, this possibility does not explain why overexpression of D-cblS fails to affect EGFR markers (Fig. 5). In any case, our genetic data suggest that D-CblS preferentially regulates Notch, and may have a minor activity towards EGFR (Fig. 8).
There has been one biochemical study which links mammalian c-Cbl with Notch1 regulation (Jehn et al., 2002). However, while we cannot exclude the possibility that D-CblS also regulates Notch in Drosophila, we have made several observations which suggest that D-CblS restricts Notch signaling through regulation of Delta. First, the non-autonomous effects on Notch activity in D-cbl mutant clones (Fig. 3C′,C″) and in response to D-cblS overexpression (Fig. 4B′,E′) argue in favor of a Notch ligand as D-CblS target. Second, the ectopic induction of Cut by Delta can be suppressed by co-expression of D-cblS (Fig. 6C). Third, Delta protein accumulates in D-cbl mutant clones (Fig. 6E,F). In contrast, we did not observe an accumulation of Notch protein in D-cbl clones (data not shown).
It is unclear how D-CblS regulates Delta. However, the fact that in clones of the RING mutant D-cbl7, which affects the E3-ubiquitin ligase domain (Wang et al., 2008b), Delta protein accumulates suggests that D-CblS regulates Delta by ubiquitination and potentially lysosomal degradation. Two RING E3-ubiquitin ligases, Neuralized (Neur) and Mind bomb (Mib), have previously been shown to target Delta for ubiquitylation (Itoh et al., 2003; Lai et al., 2005; Le Borgne et al., 2005; Pitsouli and Delidakis, 2005; Wang and Struhl, 2005). However, Neur and Mib are required for ligand maturation and Notch activation. In contrast, the genetic behavior of D-cbl mutants and D-cblS expression indicates a requirement of D-Cbl for inactivation of Delta activity. Therefore, similar to the regulation of Notch by several E3-ubiquitin ligases (Dx versus Su(dx) and Nedd4), the regulation of Delta also involves a system of E3 ligases with opposing outcome on Delta activity. How ubiquitylation of Delta by these ligases promotes a different outcome is currently unknown. Alternatively, it is also possible that D-CblS regulates the activity of other proteins required for Delta activity such as Neur and Mib, or other accessory and adaptor proteins.
Such a system of opposing E3 ubiquitin ligases may also explain why in D-cbl clones Notch activity is only mildly increased, and loss of D-cbl does not ectopically induce Notch signaling in the paradigms we have looked at. Loss of D-cbl alone is not sufficient for ectopic activation of Notch/Delta signaling. The other ubiquitin ligases may be needed for full Notch activation. Even the EGFR-associated phenotypes of D-cbl in the ovary are very modest (Pai et al., 2000) and the protein levels of the EGFR are not detectably altered in D-cbl mutant clones (Jekely et al., 2005). Therefore, only in regions of the imaginal discs where Notch is normally activated, can the accumulation of Delta in D-cbl clones further increase the signaling strength of the Notch pathway.
In summary, our data show that distinct isoforms of D-Cbl restrict EGFR and Notch signaling. While the long isoform restricts EGFR signaling, the short isoform preferentially controls Notch activity through regulation of Delta. Future genetic and biochemical work is needed to dissect the mechanism of Delta regulation by D-CblS.
Implications for mammalian Cbl proteins and oncogenesis
The proto-oncogene Casitas B-lineage lymphoma (cbl) is the cellular homologue of the retroviral transforming gene, v-cbl, that induces pre-B cell lymphoma and myeloid leukemia (Langdon et al., 1989). Mammals contain three cbl genes, c-cbl, cbl-b, and cbl-c (Schmidt and Dikic, 2005). Best characterized are c-cbl and cbl-b, both of which have a domain structure very similar to D-CblL and both control EGFR activity (Thien and Langdon, 2005a). However, cbl-c encodes a shorter isoform lacking the UBA domain and most of the Pro-rich domain (Keane et al., 1999; Kim et al., 1999), thus exhibiting similarity to D-CblS. Not much is known about its in vivo function, except that it is mainly expressed in epithelial cells (Griffiths et al., 2003). However, based on our studies we would predict that Cbl-c may regulate Notch activity in mammals. It is also possible that c-cbl or cbl-b are alternatively spliced and give rise to shorter isoforms similar to D-CblS.
Pathological changes in cbl signaling pathways are associated with several human diseases. Traditionally, most work on Cbl function has focused on its role as EGFR regulator. Excessive EGFR signaling released from cbl negative regulation may cause tumor growth, metastasis and angiogenesis (Peschard and Park, 2003). However, our work emphasizes an important role of Cbl for Notch regulation. Because the available D-cbl alleles affect both isoforms, the resulting phenotype is the combined result of increased EGFR and Notch activity. This may explain the overgrowth phenotype observed in eyes and heads predominantly mutant for D-cbl (Wang et al., 2008b) which has not been observed for mutants affecting negative regulators of EGFR signaling such as gap1, argos and sprouty. Furthermore, many human malignancies including lymphomas are caused by constitutive activation of Notch (Sjolund et al., 2005). Thus, genetic studies in model organisms may contribute to our understanding of oncogenic processes in mammals.
Supplementary Material
(A,B,C) Heat shock-induced clones expressing D-cblS (line A1) (marked by absence of GFP and arrow) suppresses Wg expression along the dorsoventral boundary in 3rd instar wing disc.
Acknowledgments
We would like to thank Hugo Bellen, Georg Halder, Shigeo Hayashi, Li-Mei Pai, Hyung Don Ryoo, Trudi Schüpbach, the Bloomington Stock Center and the Developmental Studies Hybridoma Bank for fly stocks and reagents; G. Halder and members of the Bergmann lab for discussions. This project was supported by grants R01GM068016, R01GM074977 and R01GM081543 from the National Institute of General Medical Sciences. The content is solely the responsibility of the authors and does not represent the official views of the NIGMS or NIH. We gratefully acknowledge support by the Robert A. Welch Foundation (G-1496). We would also like to thank an anonymous donor for a generous gift.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baker NE, Zitron AE. Drosophila eye development: Notch and Delta amplify a neurogenic pattern conferred on the morphogenetic furrow by scabrous. Mech Dev. 1995;49:173–189. doi: 10.1016/0925-4773(94)00314-d. [DOI] [PubMed] [Google Scholar]
- Baonza A, Casci T, Freeman M. A primary role for the epidermal growth factor receptor in ommatidial spacing in the Drosophila eye. Curr Biol. 2001;11:396–404. doi: 10.1016/s0960-9822(01)00125-7. [DOI] [PubMed] [Google Scholar]
- Bergmann A, Agapite J, McCall K, Steller H. The Drosophila gene hid is a direct molecular target of Ras-dependent survival signaling. Cell. 1998;95:331–341. doi: 10.1016/s0092-8674(00)81765-1. [DOI] [PubMed] [Google Scholar]
- Cagan RL, Ready DF. The emergence of order in the Drosophila pupal retina. Dev Biol. 1989;136:346–362. doi: 10.1016/0012-1606(89)90261-3. [DOI] [PubMed] [Google Scholar]
- Casci T, Vinos J, Freeman M. Sprouty, an intracellular inhibitor of Ras signaling. Cell. 1999;96:655–665. doi: 10.1016/s0092-8674(00)80576-0. [DOI] [PubMed] [Google Scholar]
- Chang WL, Liou W, Pen HC, Chou HY, Chang YW, Li WH, Chiang W, Pai LM. The gradient of Gurken, a long-range morphogen, is directly regulated by Cbl-mediated endocytosis. Development. 2008;135:1923–1933. doi: 10.1242/dev.017103. [DOI] [PubMed] [Google Scholar]
- Chitnis A. Why is delta endocytosis required for effective activation of notch? Dev Dyn. 2006;235:886–894. doi: 10.1002/dvdy.20683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper MT, Bray SJ. R7 photoreceptor specification requires Notch activity. Curr Biol. 2000;10:1507–1510. doi: 10.1016/s0960-9822(00)00826-5. [DOI] [PubMed] [Google Scholar]
- Couso JP, Bishop SA, Martinez Arias A. The wingless signalling pathway and the patterning of the wing margin in Drosophila. Development. 1994;120:621–636. doi: 10.1242/dev.120.3.621. [DOI] [PubMed] [Google Scholar]
- Couso JP, Martinez Arias A. Notch is required for wingless signaling in the epidermis of Drosophila. Cell. 1994;79:259–272. doi: 10.1016/0092-8674(94)90195-3. [DOI] [PubMed] [Google Scholar]
- de Celis JF, Garcia-Bellido A, Bray SJ. Activation and function of Notch at the dorsal-ventral boundary of the wing imaginal disc. Development. 1996;122:359–369. doi: 10.1242/dev.122.1.359. [DOI] [PubMed] [Google Scholar]
- Doherty D, Feger G, Younger-Shepherd S, Jan LY, Jan YN. Delta is a ventral to dorsal signal complementary to Serrate, another Notch ligand, in Drosophila wing formation. Genes Dev. 1996;10:421–434. doi: 10.1101/gad.10.4.421. [DOI] [PubMed] [Google Scholar]
- Fleming RJ, Gu Y, Hukriede NA. Serrate-mediated activation of Notch is specifically blocked by the product of the gene fringe in the dorsal compartment of the Drosophila wing imaginal disc. Development. 1997;124:2973–2981. doi: 10.1242/dev.124.15.2973. [DOI] [PubMed] [Google Scholar]
- Fostier M, Evans DA, Artavanis-Tsakonas S, Baron M. Genetic characterization of the Drosophila melanogaster Suppressor of deltex gene: A regulator of notch signaling. Genetics. 1998;150:1477–1485. doi: 10.1093/genetics/150.4.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman M. The spitz gene is required for photoreceptor determination in the Drosophila eye where it interacts with the EGF receptor. Mech Dev. 1994;48:25–33. doi: 10.1016/0925-4773(94)90003-5. [DOI] [PubMed] [Google Scholar]
- Freeman M. Reiterative use of the EGF receptor triggers differentiation of all cell types in the Drosophila eye. Cell. 1996;87:651–660. doi: 10.1016/s0092-8674(00)81385-9. [DOI] [PubMed] [Google Scholar]
- Freeman M, Klambt C, Goodman CS, Rubin GM. The argos gene encodes a diffusible factor that regulates cell fate decisions in the Drosophila eye. Cell. 1992;69:963–975. doi: 10.1016/0092-8674(92)90615-j. [DOI] [PubMed] [Google Scholar]
- Gaul U, Mardon G, Rubin GM. A putative Ras GTPase activating protein acts as a negative regulator of signaling by the Sevenless receptor tyrosine kinase. Cell. 1992;68:1007–1019. doi: 10.1016/0092-8674(92)90073-l. [DOI] [PubMed] [Google Scholar]
- Griffiths EK, Sanchez O, Mill P, Krawczyk C, Hojilla CV, Rubin E, Nau MM, Khokha R, Lipkowitz S, Hui CC, Penninger JM. Cbl-3-deficient mice exhibit normal epithelial development. Mol Cell Biol. 2003;23:7708–7718. doi: 10.1128/MCB.23.21.7708-7718.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haglund K, Sigismund S, Polo S, Szymkiewicz I, Di Fiore PP, Dikic I. Multiple monoubiquitination of RTKs is sufficient for their endocytosis and degradation. Nat Cell Biol. 2003;5:461–466. doi: 10.1038/ncb983. [DOI] [PubMed] [Google Scholar]
- Herz HM, Chen Z, Scherr H, Lackey M, Bolduc C, Bergmann A. vps25 mosaics display non-autonomous cell survival and overgrowth, and autonomous apoptosis. Development. 2006;133:1871–1880. doi: 10.1242/dev.02356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hime GR, Abud HE, Garner B, Harris KL, Robertson H. Dynamic expression of alternate splice forms of D-cbl during embryogenesis. Mech Dev. 2001;102:235–238. doi: 10.1016/s0925-4773(01)00292-1. [DOI] [PubMed] [Google Scholar]
- Hime GR, Dhungat MP, Ng A, Bowtell DD. D-Cbl, the Drosophila homologue of the c-Cbl proto-oncogene, interacts with the Drosophila EGF receptor in vivo, despite lacking C-terminal adaptor binding sites. Oncogene. 1997;14:2709–2719. doi: 10.1038/sj.onc.1201223. [DOI] [PubMed] [Google Scholar]
- Hori K, Fostier M, Ito M, Fuwa TJ, Go MJ, Okano H, Baron M, Matsuno K. Drosophila deltex mediates suppressor of Hairless-independent and late-endosomal activation of Notch signaling. Development. 2004;131:5527–5537. doi: 10.1242/dev.01448. [DOI] [PubMed] [Google Scholar]
- Huang F, Sorkin A. Growth factor receptor binding protein 2-mediated recruitment of the RING domain of Cbl to the epidermal growth factor receptor is essential and sufficient to support receptor endocytosis. Mol Biol Cell. 2005;16:1268–1281. doi: 10.1091/mbc.E04-09-0832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh M, Kim CH, Palardy G, Oda T, Jiang YJ, Maust D, Yeo SY, Lorick K, Wright GJ, Ariza-McNaughton L, Weissman AM, Lewis J, Chandrasekharappa SC, Chitnis AB. Mind bomb is a ubiquitin ligase that is essential for efficient activation of Notch signaling by Delta. Dev Cell. 2003;4:67–82. doi: 10.1016/s1534-5807(02)00409-4. [DOI] [PubMed] [Google Scholar]
- Jehn BM, Dittert I, Beyer S, von der Mark K, Bielke W. c-Cbl binding and ubiquitin-dependent lysosomal degradation of membrane-associated Notch1. J Biol Chem. 2002;277:8033–8040. doi: 10.1074/jbc.M108552200. [DOI] [PubMed] [Google Scholar]
- Jekely G, Sung HH, Luque CM, Rorth P. Regulators of endocytosis maintain localized receptor tyrosine kinase signaling in guided migration. Dev Cell. 2005;9:197–207. doi: 10.1016/j.devcel.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Joazeiro CA, Wing SS, Huang H, Leverson JD, Hunter T, Liu YC. The tyrosine kinase negative regulator c-Cbl as a RING-type, E2-dependent ubiquitin-protein ligase. Science. 1999;286:309–312. doi: 10.1126/science.286.5438.309. [DOI] [PubMed] [Google Scholar]
- Keane MM, Ettenberg SA, Nau MM, Banerjee P, Cuello M, Penninger J, Lipkowitz S. cbl-3: a new mammalian cbl family protein. Oncogene. 1999;18:3365–3375. doi: 10.1038/sj.onc.1202753. [DOI] [PubMed] [Google Scholar]
- Kim M, Tezuka T, Suziki Y, Sugano S, Hirai M, Yamamoto T. Molecular cloning and characterization of a novel cbl-family gene, cbl-c. Gene. 1999;239:145–154. doi: 10.1016/s0378-1119(99)00356-x. [DOI] [PubMed] [Google Scholar]
- Klein T, Brennan K, Arias AM. An intrinsic dominant negative activity of serrate that is modulated during wing development in Drosophila. Dev Biol. 1997;189:123–134. doi: 10.1006/dbio.1997.8564. [DOI] [PubMed] [Google Scholar]
- Kramer S, Okabe M, Hacohen N, Krasnow MA, Hiromi Y. Sprouty: a common antagonist of FGF and EGF signaling pathways in Drosophila. Development. 1999;126:2515–2525. doi: 10.1242/dev.126.11.2515. [DOI] [PubMed] [Google Scholar]
- Kretzschmar D, Brunner A, Wiersdorff V, Pflugfelder GO, Heisenberg M, Schneuwly S. Giant lens, a gene involved in cell determination and axon guidance in the visual system of Drosophila melanogaster. Embo J. 1992;11:2531–2539. doi: 10.1002/j.1460-2075.1992.tb05318.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar JP, Tio M, Hsiung F, Akopyan S, Gabay L, Seger R, Shilo BZ, Moses K. Dissecting the roles of the Drosophila EGF receptor in eye development and MAP kinase activation. Development. 1998;125:3875–3885. doi: 10.1242/dev.125.19.3875. [DOI] [PubMed] [Google Scholar]
- Lai EC, Roegiers F, Qin X, Jan YN, Rubin GM. The ubiquitin ligase Drosophila Mind bomb promotes Notch signaling by regulating the localization and activity of Serrate and Delta. Development. 2005;132:2319–2332. doi: 10.1242/dev.01825. [DOI] [PubMed] [Google Scholar]
- Langdon WY, Hartley JW, Klinken SP, Ruscetti SK, Morse HC., 3rd v-cbl, an oncogene from a dual-recombinant murine retrovirus that induces early B-lineage lymphomas. Proc Natl Acad Sci U S A. 1989;86:1168–1172. doi: 10.1073/pnas.86.4.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Borgne R. Regulation of Notch signalling by endocytosis and endosomal sorting. Curr Opin Cell Biol. 2006;18:213–222. doi: 10.1016/j.ceb.2006.02.011. [DOI] [PubMed] [Google Scholar]
- Le Borgne R, Remaud S, Hamel S, Schweisguth F. Two distinct E3 ubiquitin ligases have complementary functions in the regulation of delta and serrate signaling in Drosophila. PLoS Biol. 2005;3:e96. doi: 10.1371/journal.pbio.0030096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 2001;24:251–254. doi: 10.1016/s0166-2236(00)01791-4. [DOI] [PubMed] [Google Scholar]
- Levkowitz G, Waterman H, Ettenberg SA, Katz M, Tsygankov AY, Alroy I, Lavi S, Iwai K, Reiss Y, Ciechanover A, Lipkowitz S, Yarden Y. Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-Cbl/Sli-1. Mol Cell. 1999;4:1029–1040. doi: 10.1016/s1097-2765(00)80231-2. [DOI] [PubMed] [Google Scholar]
- Li Y, Baker NE. Proneural enhancement by Notch overcomes Suppressor-of-Hairless repressor function in the developing Drosophila eye. Curr Biol. 2001;11:330–338. doi: 10.1016/s0960-9822(01)00093-8. [DOI] [PubMed] [Google Scholar]
- Matsuno K, Diederich RJ, Go MJ, Blaumueller CM, Artavanis-Tsakonas S. Deltex acts as a positive regulator of Notch signaling through interactions with the Notch ankyrin repeats. Development. 1995;121:2633–2644. doi: 10.1242/dev.121.8.2633. [DOI] [PubMed] [Google Scholar]
- Meisner H, Daga A, Buxton J, Fernandez B, Chawla A, Banerjee U, Czech MP. Interactions of Drosophila Cbl with epidermal growth factor receptors and role of Cbl in R7 photoreceptor cell development. Mol Cell Biol. 1997;17:2217–2225. doi: 10.1128/mcb.17.4.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micchelli CA, Rulifson EJ, Blair SS. The function and regulation of cut expression on the wing margin of Drosophila: Notch, Wingless and a dominant negative role for Delta and Serrate. Development. 1997;124:1485–1495. doi: 10.1242/dev.124.8.1485. [DOI] [PubMed] [Google Scholar]
- Nagaraj R, Banerjee U. Combinatorial signaling in the specification of primary pigment cells in the Drosophila eye. Development. 2007;134:825–831. doi: 10.1242/dev.02788. [DOI] [PubMed] [Google Scholar]
- Nolo R, Abbott LA, Bellen HJ. Senseless, a Zn finger transcription factor, is necessary and sufficient for sensory organ development in Drosophila. Cell. 2000;102:349–362. doi: 10.1016/s0092-8674(00)00040-4. [DOI] [PubMed] [Google Scholar]
- Nusslein-Volhard C, Wieschaus E, Kluding H. Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster. I. Zygotic loci on the second chromosome. Roux's Arch. Dev. Biol. 1984;193:267–282. doi: 10.1007/BF00848156. [DOI] [PubMed] [Google Scholar]
- Okano H, Hayashi S, Tanimura T, Sawamoto K, Yoshikawa S, Watanabe J, Iwasaki M, Hirose S, Mikoshiba K, Montell C. Regulation of Drosophila neural development by a putative secreted protein. Differentiation. 1992;52:1–11. doi: 10.1111/j.1432-0436.1992.tb00494.x. [DOI] [PubMed] [Google Scholar]
- Pai LM, Barcelo G, Schupbach T. D-cbl, a negative regulator of the Egfr pathway, is required for dorsoventral patterning in Drosophila oogenesis. Cell. 2000;103:51–61. doi: 10.1016/s0092-8674(00)00104-5. [DOI] [PubMed] [Google Scholar]
- Pai LM, Wang PY, Chen SR, Barcelo G, Chang WL, Nilson L, Schupbach T. Differential effects of Cbl isoforms on Egfr signaling in Drosophila. Mech Dev. 2006;123:450–462. doi: 10.1016/j.mod.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Panin VM, Papayannopoulos V, Wilson R, Irvine KD. Fringe modulates Notch-ligand interactions. Nature. 1997;387:908–912. doi: 10.1038/43191. [DOI] [PubMed] [Google Scholar]
- Parks AL, Turner FR, Muskavitch MA. Relationships between complex Delta expression and the specification of retinal cell fates during Drosophila eye development. Mech Dev. 1995;50:201–216. doi: 10.1016/0925-4773(94)00336-l. [DOI] [PubMed] [Google Scholar]
- Peschard P, Park M. Escape from Cbl-mediated downregulation: a recurrent theme for oncogenic deregulation of receptor tyrosine kinases. Cancer Cell. 2003;3:519–523. doi: 10.1016/s1535-6108(03)00136-3. [DOI] [PubMed] [Google Scholar]
- Pitsouli C, Delidakis C. The interplay between DSL proteins and ubiquitin ligases in Notch signaling. Development. 2005;132:4041–4050. doi: 10.1242/dev.01979. [DOI] [PubMed] [Google Scholar]
- Ready DF, Hanson TE, Benzer S. Development of the Drosophila retina, a neurocrystalline lattice. Dev Biol. 1976;53:217–240. doi: 10.1016/0012-1606(76)90225-6. [DOI] [PubMed] [Google Scholar]
- Robertson H, Hime GR, Lada H, Bowtell DD. A Drosophila analogue of v-Cbl is a dominant-negative oncoprotein in vivo. Oncogene. 2000;19:3299–3308. doi: 10.1038/sj.onc.1203624. [DOI] [PubMed] [Google Scholar]
- Rulifson EJ, Blair SS. Notch regulates wingless expression and is not required for reception of the paracrine wingless signal during wing margin neurogenesis in Drosophila. Development. 1995;121:2813–2824. doi: 10.1242/dev.121.9.2813. [DOI] [PubMed] [Google Scholar]
- Sakata T, Sakaguchi H, Tsuda L, Higashitani A, Aigaki T, Matsuno K, Hayashi S. Drosophila Nedd4 regulates endocytosis of notch and suppresses its ligand-independent activation. Curr Biol. 2004;14:2228–2236. doi: 10.1016/j.cub.2004.12.028. [DOI] [PubMed] [Google Scholar]
- Schmidt MH, Dikic I. The Cbl interactome and its functions. Nat Rev Mol Cell Biol. 2005;6:907–919. doi: 10.1038/nrm1762. [DOI] [PubMed] [Google Scholar]
- Schweitzer R, Howes R, Smith R, Shilo BZ, Freeman M. Inhibition of Drosophila EGF receptor activation by the secreted protein Argos. Nature. 1995;376:699–702. doi: 10.1038/376699a0. [DOI] [PubMed] [Google Scholar]
- Seugnet L, Simpson P, Haenlin M. Requirement for dynamin during Notch signaling in Drosophila neurogenesis. Dev Biol. 1997;192:585–598. doi: 10.1006/dbio.1997.8723. [DOI] [PubMed] [Google Scholar]
- Sjolund J, Manetopoulos C, Stockhausen MT, Axelson H. The Notch pathway in cancer: differentiation gone awry. Eur J Cancer. 2005;41:2620–2629. doi: 10.1016/j.ejca.2005.06.025. [DOI] [PubMed] [Google Scholar]
- Thien CB, Langdon WY. c-Cbl and Cbl-b ubiquitin ligases: substrate diversity and the negative regulation of signalling responses. Biochem J. 2005a;391:153–166. doi: 10.1042/BJ20050892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thien CB, Langdon WY. Negative regulation of PTK signalling by Cbl proteins. Growth Factors. 2005b;23:161–167. doi: 10.1080/08977190500153763. [DOI] [PubMed] [Google Scholar]
- Tomlinson A, Struhl G. Delta/Notch and Boss/Sevenless signals act combinatorially to specify the Drosophila R7 photoreceptor. Mol Cell. 2001;7:487–495. doi: 10.1016/s1097-2765(01)00196-4. [DOI] [PubMed] [Google Scholar]
- Tsuda L, Nagaraj R, Zipursky SL, Banerjee U. An EGFR/Ebi/Sno pathway promotes delta expression by inactivating Su(H)/SMRTER repression during inductive notch signaling. Cell. 2002;110:625–637. doi: 10.1016/s0092-8674(02)00875-9. [DOI] [PubMed] [Google Scholar]
- Vaccari T, Lu H, Kanwar R, Fortini ME, Bilder D. Endosomal entry regulates Notch receptor activation in Drosophila melanogaster. J Cell Biol. 2008;180:755–762. doi: 10.1083/jcb.200708127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voas MG, Rebay I. Signal integration during development: insights from the Drosophila eye. Dev Dyn. 2004;229:162–175. doi: 10.1002/dvdy.10449. [DOI] [PubMed] [Google Scholar]
- Wang PY, Chang WL, Pai LM. Smiling Gurken gradient: An expansion of the Gurken gradient. Fly (Austin) 2008a;2 doi: 10.4161/fly.6526. [DOI] [PubMed] [Google Scholar]
- Wang W, Struhl G. Distinct roles for Mind bomb, Neuralized and Epsin in mediating DSL endocytosis and signaling in Drosophila. Development. 2005;132:2883–2894. doi: 10.1242/dev.01860. [DOI] [PubMed] [Google Scholar]
- Wang Y, Werz C, Xu D, Chen Z, Li Y, Hafen E, Bergmann A. Drosophila cbl is essential for control of cell death and cell differentiation during eye development. PLoS ONE. 2008b;3:e1447. doi: 10.1371/journal.pone.0001447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Moran MF. Requirement for the adapter protein GRB2 in EGF receptor endocytosis. Science. 1996;272:1935–1939. doi: 10.1126/science.272.5270.1935. [DOI] [PubMed] [Google Scholar]
- Waterman H, Katz M, Rubin C, Shtiegman K, Lavi S, Elson A, Jovin T, Yarden Y. A mutant EGF-receptor defective in ubiquitylation and endocytosis unveils a role for Grb2 in negative signaling. Embo J. 2002;21:303–313. doi: 10.1093/emboj/21.3.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman H, Levkowitz G, Alroy I, Yarden Y. The RING finger of c-Cbl mediates desensitization of the epidermal growth factor receptor. J Biol Chem. 1999;274:22151–22154. doi: 10.1074/jbc.274.32.22151. [DOI] [PubMed] [Google Scholar]
- Wilkin MB, Carbery AM, Fostier M, Aslam H, Mazaleyrat SL, Higgs J, Myat A, Evans DA, Cornell M, Baron M. Regulation of notch endosomal sorting and signaling by Drosophila Nedd4 family proteins. Curr Biol. 2004;14:2237–2244. doi: 10.1016/j.cub.2004.11.030. [DOI] [PubMed] [Google Scholar]
- Yokouchi M, Kondo T, Houghton A, Bartkiewicz M, Horne WC, Zhang H, Yoshimura A, Baron R. Ligand-induced ubiquitination of the epidermal growth factor receptor involves the interaction of the c-Cbl RING finger and UbcH7. J Biol Chem. 1999;274:31707–31712. doi: 10.1074/jbc.274.44.31707. [DOI] [PubMed] [Google Scholar]
- Yoon CH, Lee J, Jongeward GD, Sternberg PW. Similarity of sli-1, a regulator of vulval development in C. elegans, to the mammalian proto-oncogene c-cbl. Science. 1995;269:1102–1105. doi: 10.1126/science.7652556. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A,B,C) Heat shock-induced clones expressing D-cblS (line A1) (marked by absence of GFP and arrow) suppresses Wg expression along the dorsoventral boundary in 3rd instar wing disc.