Abstract
Chronic pressure overload to the heart leads to cardiac hypertrophy and failure through processes that involve reorganization of subcellular compartments and alteration of established signaling mechanisms. To identify proteins contributing to this process, we examined changes in nuclear-associated myofilament proteins as the murine heart undergoes progressive hypertrophy following pressure overload. Calsarcin-1, a negative regulator of calcineurin signaling in the heart, was found to be enriched in cardiac nuclei and displays increased abundance following pressure overload through a mechanism that is decoupled from transcriptional regulation. Using proteomics, we identified novel processing of this protein in the setting of cardiac injury and identified four residues subject to modification by phosphorylation. These studies are the first to determine mechanisms regulating calsarcin abundance during hypertrophy and failure and reveal the first evidence of post-translational modifications of calsarcin-1 in the myocardium. Overall, the findings expand the roles calsarcins to include nuclear tasks during cardiac growth.
Keywords: phosphorylation, signal transduction, proteomics, nucleus, cardiac hypertrophy
Introduction
Left ventricular cardiac hypertrophy is a result of stresses which induce growth on the cellular level. These stresses include exercise, pregnancy, neural/hormonal stimuli and disease (including myocardial infarction and atherosclerosis), all of which are characterized by cytosolic signaling processes that impinge on the nucleus to modulate gene expression. While the specific elements involved in the multiple forms of physiological and pathological hypertrophy are many, the fundamental need to execute transduction of a signal to the nucleus is conserved.
Among the cytoplasmic signaling molecules involved in hypertrophy, the serine/threonine phosphatase calcineurin has gained substantial attention for its conserved role as a positive regulator of hypertrophy induced by genetic models, aortic constriction, exercise, myocardial infarction and high salt diet (for review, see [1]). By directly binding to, dephosphorylating and inducing nuclear translocation of the transcription factor NFAT in response to increased intracellular calcium, calcineurin is responsible for up-regulation of genes that control growth. In addition to the well-characterized regulatory proteins involved in calcineurin signaling in non-muscle cells—such as calmodulin, which intimately regulates the calcium dependency of the protein [2]—a family of calcineurin binding proteins called calsarcins (also know as myozenins) were identified as muscle-specific calcineurin interacting proteins [3]. Calsarcin-1 and calsarcin-2 are expressed in all striated muscle during development, whereas only expression of calsarcin-1 persists in the adult heart. Initial studies demonstrated that calsarcins bind α-actinin and calcineurin directly and independently [3], and subsequent investigations have gone on to characterize in detail the role of calsarcins as negative regulators of calcinuerin signaling and thereby cardiac hypertrophy [4, 5].
Despite its known involvement in regulating calcineurin signaling in the diseased heart, several aspects of calsarcin function are unknown. In particular, its abundance during the development of pressure overload hypertrophy has never been established, along with mechanisms of endogenous modulation such as intracellular localization. Furthermore, how the protein is regulated post-translationally, including its endogenous protein species and residues modified by other proteins, have not been explored. Lastly, while alterations in myofilament biology are central to cardiac function and disease [6, 7], modulation of myofilament-nuclear interactions remains unclear. In the present study, we demonstrate nuclear localization of calsarcin-1 in the normal myocardium and report changes in protein abundance during the development of pressure overload hypertrophy in mice. In addition, we document phosphorylation and post-translational processing of calsarcin-1 in the heart. These observations are the first in depth analysis of calsarcin-1 post-translational modification in the heart and have potential importance for our understanding of established growth signaling mechanisms during disease.
Results
To explore the role of nuclear-cytoskeletal interactions in heart disease, we examined cytoskeletal and myofilament proteins enriched in nuclei isolated from mouse ventricular tissue. A series of pilot experiments were performed to select an isolation protocol to enrich for known nuclear proteins, such as histones, and reduce the presence of known residents of other organelles (including the adenine nucleotide transporter from mitochondria and the sodium/calcium exchanged from the plasma membrane) to low or undetectable levels. Electron microscopy was used to demonstrate that nuclei purified with this method have intact membranes (data not shown).
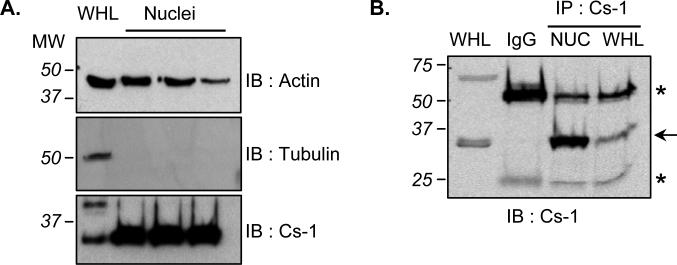
Compared with the total cardiac lysate, nuclei display little of the microtubule protein, tubulin, and although they contain actin, this protein is not enriched in this fraction as demonstrated by western blotting (Fig. 1A). In contrast, the myofilament associated protein calsarcin-1 was found to be associated with cardiac nuclei, indeed displaying enrichment in the organelle as compared with the whole heart (Fig. 1A). The abundant species of calsarcin in heart appears at ~30 kDa, and immunoprecipitation also confirmed enrichment in the nuclear compartment (Fig. 1B).
Figure 1. Enrichment of calsarcin-1 in cardiac nuclei.
A, Protein extracts from isolated cardiac nuclei were immunoblotted for the presence of actin, tubulin or calsarcin-1. As compared to whole heart lysate (WHL), only calsarcin-1 is enriched in nuclei (30μg loaded in each lane). B, To confirm calsarcin-1 presence in nuclei, the protein was immunoprecipitated (IP, with BD primary antibody) from this organelle as well as from the WHL. Arrow indicates calsarcin-1 band, substantially enriched in nuclei (IP was performed from 1mg total nuclear or WHL protein); asterisks indicate immunoglobulin bands.
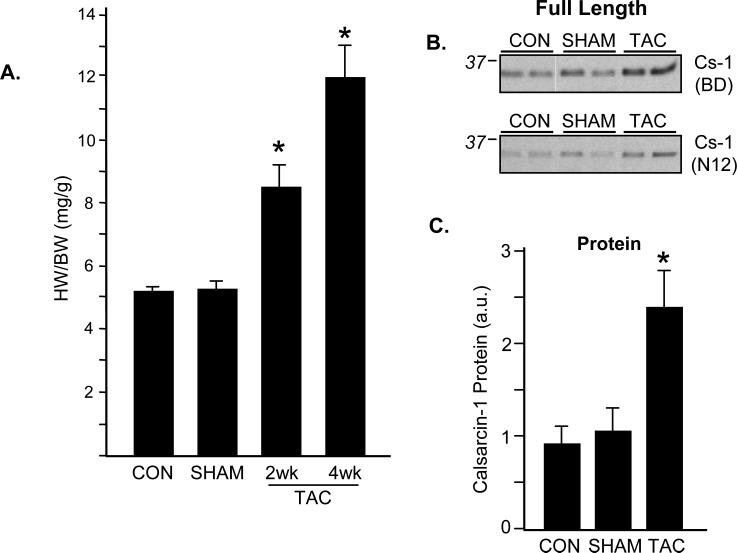
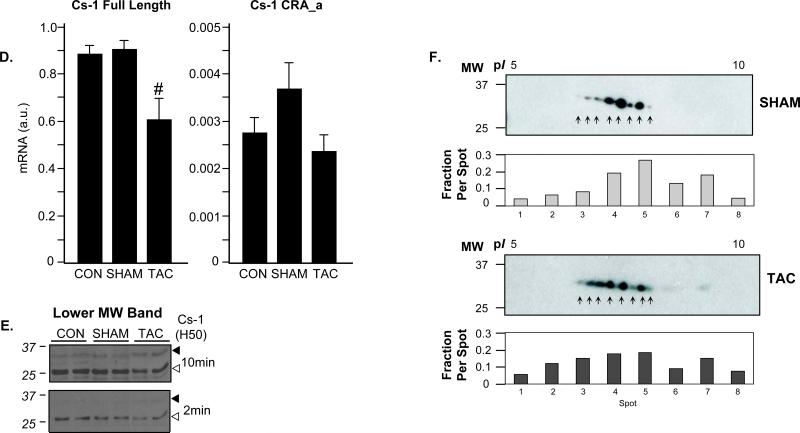
To examine changes in calsarcin in the diseased heart, we employed a model of transverse aortic constriction (TAC) to induce pressure overload. TAC-operated mice experienced a dramatic, time-dependent increase in heart weight to body weight ratio (Fig. 2A). This response is characterized by an initial improvement in cardiac function, usually from 0-2 weeks, followed by a transition to heart failure at ~4 weeks, which is accompanied by a significant decrease in ejection fraction and other indices of heart performance [8, 9]. Interestingly, cardiac hypertrophy was associated with increased calsarcin-1 abundance, as detected by western blotting, that peaked in intensity at 2 weeks (Fig. 2B and C) and did not further increase at 4 weeks. This increase in protein level was accompanied by a slight decrease in transcriptional activity of calsarcin-1 mRNA (Fig. 2D), suggesting regulation at both the post-translational and transcriptional levels. As mentioned above, the primary species of calsarcin-1 protein detected by western blotting was ~30 kDa which corresponds to the predicted length of the primary translation product as reported by other investigators [3]. However, several antibodies (including the antibody from BD Biosciences and the H50 reagent from Santa Cruz) against calsarcin-1 detected a lower molecular weight band, around ~25kDa that appeared with lower intensity (Fig. 2E) and displayed antithetic behavior to the full-length species (that is, the lower MW band decreased intensity with TAC whereas the full length increased). To our knowledge this protein species has not previously been reported.
Figure 2. Pressure overload hypertrophy alters expression of calsarcin-1.
A, Surgical constriction of the transverse aorta produces a progressive cardiac hypertrophy as demonstrated by increase in heart weight to body weight ratio. Data are mean±SEM, asterisk indicates p<0.0001 versus SHAM and N=4-6/group. B, Western blotting demonstrates a significant increase in calsarcin-1 protein levels 2 weeks after TAC (4 week displayed similar change, not shown) using two antibodies (BD and N12; N12 is a Santa Cruz antibody; BD is the company for the first antibody). C, Quantitation of full-length calsarcin-1 protein levels demonstrates a significant increase after TAC (* indicates p=0.03 vs. SHAM). D, RT-PCR analysis of calsarcin-1 was done for the full-length product (left panel) as well as using primers that recognize only the CRA-a isoform (right panel). Note the difference in relative abundance (y-axis), demonstrating the very low endogenous expression of the CRA_a product. There was a small but significant decrease in levels of the full-length product (# indicates p=0.007) following TAC. All protein and mRNA measurements are mean±SEM with N=4-5/group. E, Detection with a third antibody (H50) that targets the interior of the protein reveals a more complex pattern: the full-length calsarcin-1 (~30 kDa, filled arrow) behaves the same as with the BD and N12 antibodies, but a lower molecular weight species appears (~25 kDa, open arrow) that displays opposite behavior with the full length (it decreases with TAC). Two exposures of the same blot are shown to emphasize the different bands, further analyzed in Figure 3. F, Western blotting of 2D gels for calsarcin demonstrates distinct species of the protein, confirming mass spectrometry results. Inset quantification shows the per spot fraction of total calsarcin signal detected from the sum of all eight spots in the SHAM or TAC animals, respectively.
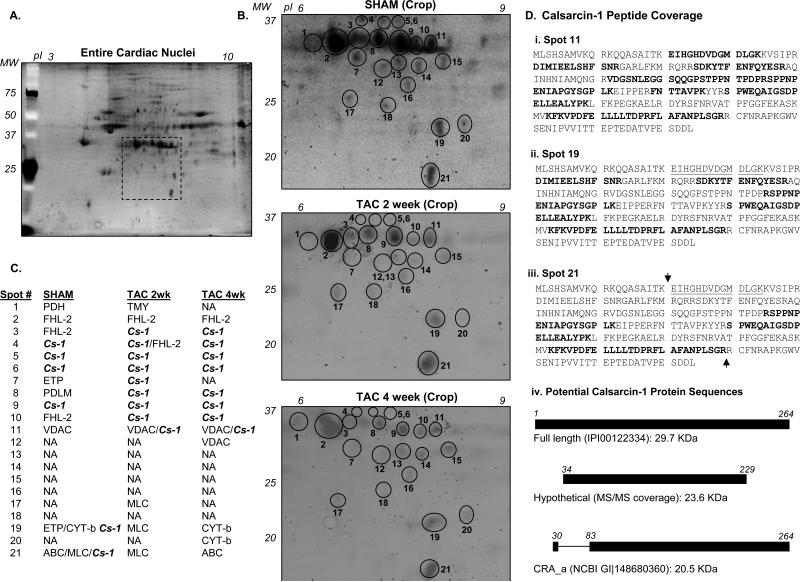
To further investigate changes in calsarcin-1 modification during hypertrophy, we employed 2D electrophoresis to separate nuclear proteins (Fig. 3A), focusing first on the region of separation corresponding to full-length calsarcin-1 (Fig. 3B). We observe calsarcin-1 present in several distinct spots around 30 kDa, stretching across a wide isoelectric range. Several of these species are immunodecorated with calsarcin-1 antibody (Fig. 2F). As shown in Figure 3C, calsarcin-1 was the primary protein (as judged by having the highest combined Xcorr and the most peptides identified) that appeared in 6-9 different spots around 30 kDa, as well as 2 spots (#19 and #21) at lower molecular weight on the gel. Peptides used for identification were sufficient to distinguish calsarcin-1 from the other isoforms of this protein, calsarcin-2 and calsarcin-3 (see also Fig. 5). During pressure overload, the spot pattern for calsarcin-1 changed, most notable for the disappearance of the protein from spots #19 and #21 (Fig. 3B and C). Proteins listed in Figure 1C were identified with 4 or more peptides from LC/MS/MS experiments with the exception of calsarcin-1 from spots #19 and #21 (here calsarcin was detected with 3 peptides); furthermore, we lowered the threshold for search criteria to 1 peptide for both the spots #19 and #21 from the 2 week and 4 week TAC samples to insure that calsarcin-1 was absent even at this low threshold. It is important to note, however, that SYPRO's staining sensitivity is lower than what can be detected with a mass spectrometer such as the Orbitrap used in this study (in other words, the mass spectrometer can detect proteins not visible on the gel). As such, it is possible that more that one protein may reside in a spot and/or that protein may be detected by mass spectrometry that did not resolve as an independent spot in the electrophoresis stage but was nevertheless excised from the gel along with a visible spot. Analysis of the localization of the identified peptides on calsarcin-1 from different representative spots indicates that a tryptic peptide towards the amino terminus that was detected in the spots corresponding to the full-length calsarcin-1 protein was conspicuously absent from every identification of calsarcin-1 in the lower molecular weight spots (Fig. 3D). These spots correspond roughly in molecular weight to the lower mass bands detected by western blotting, and could arise from protein cleavage or a proposed alternative transcriptional start site ([10];Fig. 3D, panel iv), as further addressed below.
Figure 3. Proteomic analysis of nuclear calsarcin-1 during cardiac hypertrophy.
A, To examine proteins in cardiac nuclei, 2D electrophoresis was used to separate and visualize the most abundant proteins associated with this organelle. B, Cropped region of 2D display of cardiac nuclei corresponding to the region where calsarcin-1 was identified. Spots examined near where calsarcin-1 was identified are labeled in gels from SHAM, 2 week TAC and 4 week TAC. Proteins identified by LC/MS/MS analyses of the spots yielded are listed in C, which revealed multiple species of calsarcin-1 in the basal state (column 2, SHAM), that are significantly modified following TAC (NA indicates no proteins identified with 4 or more peptides). In particular, the lower MW species of calsarcin-1 in the basal condition (spots 19 and 21) are gone following TAC. Full names and accession numbers of proteins identified in these spots are in Supplemental Table 1. D, Calsarcin-1 peptide coverage (determined from MS/MS data) for spots 11, 19 and 21 from a SHAM experiment with identified peptides in bold. Mass spectrometric analysis of lower molecular weight species (spots 19 and 20) never yielded the peptide underlined in i. and iii., whereas this was consistently identified in spot 11 (i.) and others spots containing the full-length protein (not shown). Panel iv. is a diagram of the full length calsarcin-1 as compared with a version that would result from hypothetical cleavage of sequence around the arrows indicated in iii.; also shown is the predicted CRA_a isoform of calsarcin-1, produced by an internal start site.
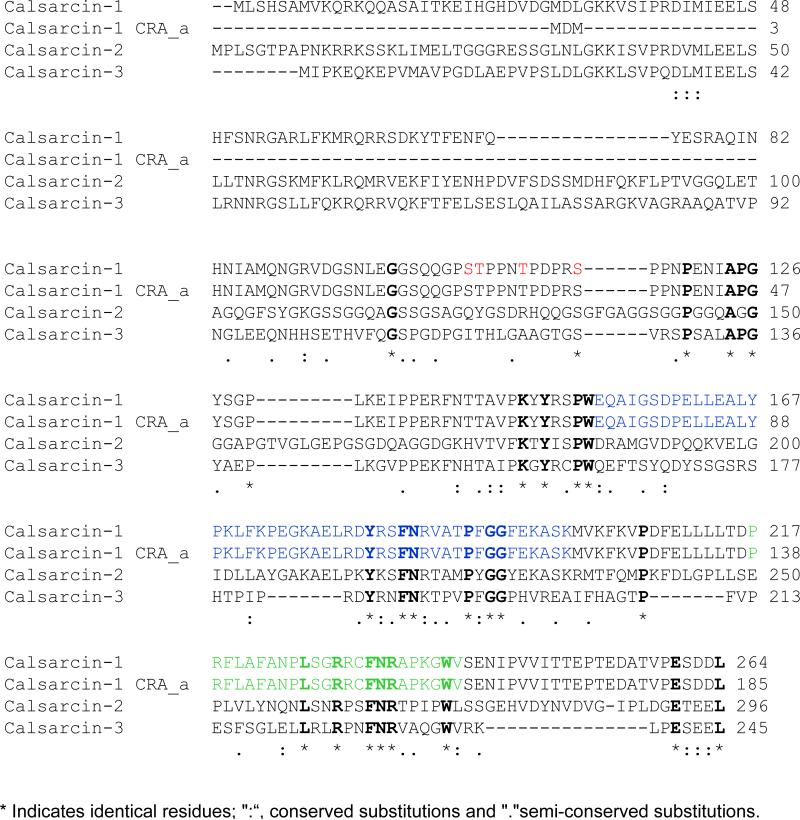
Figure 5. Calsarcin-1 phosphorylation and sequence alignment.
Alignment of the three murine calsarcin genes, calsarcin-1 (myozenin-2, UniProt Q9JJW5), 2 (myozenin-1, Q9JK37) and 3 (myozenin-3, Q8R4E4) with a truncated version of calsarcin-1 identified in a large-scale mouse genome analysis (NCBI GI|148680360) called CRA_a. Regions of sequence identity between all four protein sequences are bold; red amino acids indicate the residues shown to be phosphorylated in the present manuscript; blue and green residues indicated α-actinin-2 and calcineurin binding regions[3], respectively.
Multiple calsarcin-1 protein species suggest the presence of multiple transcriptional products and/or post-translational modifications. A previous shotgun genome analysis [10] identified a potential internal start site that would produce a shorter version of calsarcin-1 at the mRNA level (Fig. 3D.iv). To examine abundance of this transcript, we performed quantitative realtime-PCR analysis of the myozenin-2 gene (encoding calsarcin-1) using a second set of primers that would detect only this shorter version (termed CRA_a in the NCBI database). As shown in Figure 2D, while a small amount of this transcript was detectable, it was present at a level ~260-fold less than the full-length product and was not regulated during hypertrophy. To explore potential phosphorylation of the protein, we searched our protein identification data from collision-induced dissociation experiments on the Orbi-trap for phosphorylated peptides (Fig. 4A and B and Supplemental Fig. 2A-D), several of which were observed and localized to four different residues on the protein (Fig. 5). To confirm these observations, we performed electron-transfer dissociation fragmentation experiments on an LTQ mass spectrometer (Fig. 4C and Supplemental Fig. 2E) and observed phosphorylation on all the same residues of calsarcin-1 from cardiac nuclei. In addition, we detected occurrences of single peptides with two phosphate groups intact (Fig. 4B and Supplemental Fig. 2D), but these were rare. All spectra reported in this study are from the calsarcin-1 protein from the spot pattern around 37 KDa. In addition, we detected phosphorylation of calsarcin-1 in both the 2 and 4 week time points after TAC, as well as in the SHAM hearts. Unfortunately, the approach used in this study does not allow us to confidently quantify differences in the abundance of distinct phosphorylated species (i.e. with single residue resolution) between these phenotypic states.
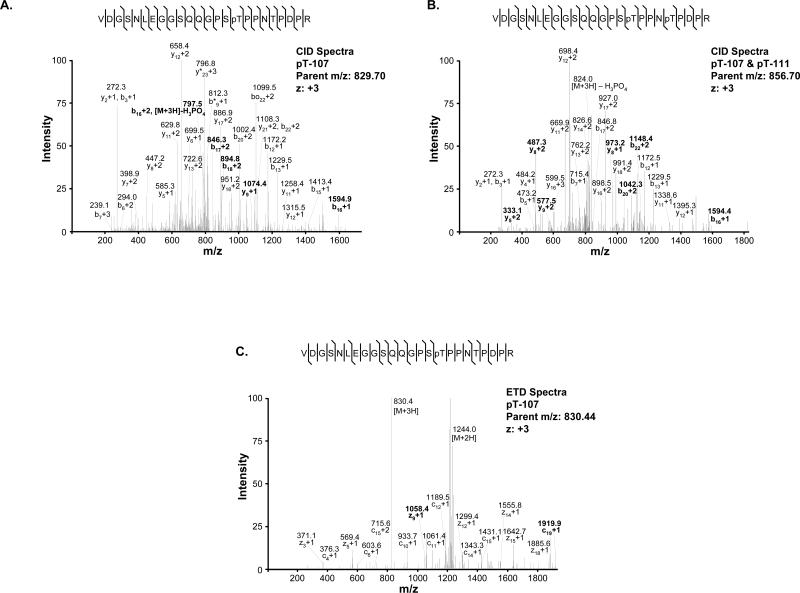
Figure 4. Mass spectrometric detection of phosphorylation on calsarcin-1.
Spots excised from 2D gels were subjected to tryptic digestion and analyzed by LC/MS/MS on an Orbitrap (using CID) and an LTQ (using ETD). A, CID spectra of calsarcin-1 peptide indicating phosphorylation at T107, resulting from parent m/z=829.70. Abundant ions are labeled in the spectra (0 indicates H2O loss, * indicated NH3 loss). B, CID spectra of the peptide phosphorylated at T107 and T111. C, ETD spectra of calsarcin-1 peptide indicating phosphorylation at T107, displaying characteristic domination of daughter ion spectra by unfragmented parents (+3 and +2). Softer fragmentation with ETD produced longer series of c or z ions with the modification intact. In all spectra shown, the daughter ion pattern allowed accurate assignment of the modification to the specified residue on the peptide. Bold labels indicate features used to resolve modified site. Additional spectra for the other phosphorylation sites are available in Supplemental Figure 1.
Discussion
Regulation of cardiac growth fundamentally involves changes in gene expression, which in turn requires a means for signaling processes to impinge on the nucleus. While it is clear that kinases and phosphatases can physically interact with this organelle, as well as indirectly influence it via activation of transcription factors in the cytoplasm, the proteins responsible for docking of signaling molecules at the nucleus are poorly studied. The present investigation demonstrates that calsarcin-1, an established z-disc myofilament protein, localizes to cardiac nuclei, is upregulated during TAC-induced hypertrophy and undergoes extensive post-translational modifications in the normal heart and following pressure overload. These observations have important implications for our understanding of signal transduction to the nucleus during hypertrophy, as well as in the understanding of how nuclei are regulated by components of the contractile apparatus.
Calsarcins were originally identified by a yeast two-hybrid screen for calcineurin-binding proteins [3]. Of the three isoforms characterized to date, calsarcin-1 mRNA is abundantly expressed in striated muscle—especially the heart—throughout life whereas calsarcin-2 is restricted to skeletal muscle in the adult following transient expression in the embryonic heart. Our studies confirm this observation at the protein level based on unequivocal mass spectrometry data: peptides identified from calsarcins map only to the calsarcin-1 isoform. Early studies documented association of calsarcin-1 with calcineurin and α-actinin, displaying localization of the protein along z-discs in cardiac myocytes [3]. More recently it was shown that loss of calsarcin-1 does not alter basal heart weight to body weight ratio, but accentuates the hypertrophic response of the heart following pressure overload or calcineurin activation [4]. Together, these observations support calsarcin-1 as a negative regulator of the hypertrophic phosphatase, calcineurin. By examining genomic regions associated with early onset of cardiomyopathy in a patient population, a separate study independently identified the myozenin-2 gene as a novel inducer of human hypertrophic cardiomyopathy [11]. What remains unclear are the endogenous sites of action for calsarcin and the means by which it is regulated post-transcriptionally.
Because we observed enrichment of calsarcin-1 in cardiac nuclei, we sought to examine endogenous heterogeneity at the protein level using proteomics. Two dimensional electrophoresis and LC/MS/MS revealed several species of calsarcin-1 protein in cardiac nuclei, including extensively phosphorylated protein. One previous high-throughput study reported phosphorylation of calsarcin-1 on T-107 and T-111 from mouse liver [12] (in a tissue where, incidentally, there is extremely low expression of the protein [3]). We detected these modifications in the heart for the first time, along with two other phosphorylation events on nearby residues. It is noteworthy that all phosphorylation events reported in this study were detected with both CID and ETD, passed the criteria for not being identified in a reversed database search and came from manually evaluated spectra, decreasing the likelihood of false positive identifications. The surrounding sequence of calsarcin-1 near the sites of modification was not amenable to dissection with other enzymes that may have added to the understanding of how this region was regulated by phosphorylation. Likewise, the size of one of the two tryptic peptides in this region was quite large (24 amino acids), a fortuitous occurrence that made the peptide likely to carry more charge and therefore amenable to ETD fragmentation. It remains unclear whether these modifications serve as markers for disease as has been demonstrated for other sarcomeric proteins such as troponin I [13]. Although we detected single peptides with two phosphate groups intact, it was not surprising that this was quite infrequent, given the labile nature of the modification. We never detected a peptide with more than two phosphate groups intact by either CID or ETD, although this should not be interpreted as nonexistence, because multiply phosphorylated peptides are notoriously difficult to detect due to altered retention on RP columns, lower ionization efficiency and ion suppression by more abundant, less modified co-eluting counterparts[14].
The close physical proximity of these modifications suggests that this region of the cardiac calsarcin-1 protein is a site of regulatory importance, independent of the regions already established to be involved in calcineurin and α-actinin binding (Fig. 5). One site previously suggested to be involved in calcineurin docking (PxIxIT; [15]) is not observed in calsarcin-1, nor is the sequence of a peptide implicated as a calcineurin substrate (FDRRVSVAAE; [16]), so it remains unclear whether the phosphorylation events described in the current study are subject to direct dephosphorylation by calcineurin. Site directed mutagenesis will be required to sort out the relative importance of these different residues in calsarcin-1 function and cardiac disease.
Another intriguing observation in the present investigation is the presence of lower molecular weight species of calsarcin-1, the abundance of which decreases (while overall levels of full-length calsarcin-1 increase) during hypertrophy. There are two possible explanations for this form of calsarcin: first, it could be the result of altered transcriptional activity, possibly representing the first protein-level detection of the CRA_a isoform of calsarcin-1, resulting from an internal transcriptional start site, which to date has only been predicted from shotgun genome analyses[10] (Figs. 3D and 5); second, this lower molecular weight species could be the result of degradation/processing at the protein level. While our RT-PCR data detected the CRA_a transcript, it was extremely low abundance in comparison to the full-length calsarcin-1 species. For this lower molecular weight species to result from this alternative start site, the ratio of full length to CRA_a species would have to be significantly lower at the protein level versus what we observe at the transcript level to make it technically feasible for us to detect by mass spectrometry. This would require a heretofore undetermined mechanism to dramatically decouple calsarcin-1 protein and mRNA abundance. Our peptide identifications for calsarcin-1, while extensive (Fig. 3D), could not distinguish between (a) absence of detection of amino terminal peptides in the full-length species and (b) presence of the CRA_a species (which would necessitate detection of the extreme amino terminus of CRA_a, the peptide MDMHNIAMQNGR, to distinguish from the full length calsarcin-1, see also Fig. 5). Parallel studies to examine calsarcin-1 associated proteins by immunoprecipitation and mass spectrometry identified the serine protease HTRA2 (data not shown) which is known to be involved in degradation and caspase activation in the heart following stress [17, 18]. It is interesting to speculate that this or some other stress-activated protease may modulate calsarin-1 during stress, thereby influencing hypertrophic signaling.
How might altered calsarcin-1 species impact the development of hypertrophy? Because it has previously been shown to inhibit calcineurin, an increase in calsarcin-1 protein during pressure overload may contribute to the compensatory hypertrophic response to increased demand for work on the heart. As such, it would be intriguing to investigate how calsarcin-1 abundance and modifications change when the heart progresses further into failure, beyond the 4 week time point examined in these studies. Furthermore, while we observed substantial enrichment in calsarcin-1 in cardiac nuclei, it seems unlikely that this protein is located physically within the nucleus, but rather is nuclear-associated. Future quantitative microscopic analyses could provide insight into intracellular localization changes in calsarcins during disease, however these approaches would not capture post-translational modification/processing events and subspecies of the protein. In addition, it is interesting to speculate on the role of calsarcin in this subcellular location, including potential scaffolding and docking roles for other signaling molecules. The signaling functions that result from distinct populations—including nuclear and non-nuclear—as well as different species of calsarcins in vivo, remain to be determined.
Materials & Methods
All experiments conform to the NIH guide for the Care and Use of Laboratory Animals and were approved by the UCLA Animal Research Committee.
Antibodies, Chemicals and Reagents
Primary antibodies: calsarcin-1 (N12) #46175, calsarcin-1 (C12) #46172, calsarcin-1 (H50) #66990, α-actin #1616 (from Santa Cruz Biotechnology); calsarcin-1 #612579 (from BD Biosciences); β-tubulin #ab21057 (from Abcam Inc). Secondary Antibodies: HRP-conjugated chicken anti-rabbit sc2963, goat anti-mouse sc2055, bovine anti-goat sc2384, donkey anti-rabbit sc 2077, donkey anti-goat sc2056.
Mice
All experiments were performed on adult (8-12 week old) male balb/c mice from Charles River Laboratories.
Transverse aortic constriction surgery
The murine model of transverse aortic banding to induce pressure overload hypertrophy was performed as described previously [8, 9]. Briefly, animals were anesthetized with 3.0% isoflurane, intubated, and ventilated with 2.0% isoflurane in 98% O2 / 2% CO2. The chest was entered from the left side via the third intercostal space, the aorta identified at the T8 region and a venous vascular clamp placed around the vessel. The internal diameter of the resulting modified clamp was that of a 27 gauge needle. The chest was then closed using 6-0 proline suture, during which negative pressure was reestablished in the thorax by removing air with a PE-50 chest tube attached to a syringe. SHAM operated mice underwent the same procedure without placement of aortic clamp. Pressure overload was confirmed by measuring the gradient in carotid artery pressure by echocardiography and the development of hypertrophy confirmed by post-mortem analysis of heart weight to body weight or tibia length measurements. At least three mice in each group were used for proteomic analyses, with at least two gels run from each heart.
Isolation of Cardiac Nuclei
Hearts were excised and homogenized in 2 ml homogenization buffer containing (in mmol/L) 250 sucrose, 10 Tris-HCl, pH 7.4, 1 EDTA, protease inhibitor cocktail (Roche Diagnostics), 1 PMSF, 1 Na3VO4, 1 NaF. The initial homogenate was centrifuged at 1000 × g for 10 min at 4° C to pellet the nuclear fraction. The nuclear pellet was resuspended in 0.5 ml of homogenization buffer, overlaid on 1.5 ml of 1 M sucrose cushion and centrifuged at 1600 × g for 5 min at 4° C. The sucrose cushion centrifugation was performed twice after which the pellet was washed in cold PBS and pelleted at 1600 × g for 5 min at 4° C.
Protein Sample Preparation
The nuclear pellet was resuspended in cell lysis buffer and the protein concentration was taken using a Bradford Assay (BioRad). Samples for analysis by 2D electrophoresis were then acetone precipitated by adding at least 6 volumes of acidic acetone (1 drop of 6N HCl in 10 ml of cold acetone) to the sample, incubating at - 80° C for 30 min, then centrifuging at 13,000 × g for 20 min at 4 ° C to pellet the precipitated protein. The pellet was then allowed to air dry. Resulting pellets were re-suspended in 2D buffer containing protease and phosphatase inhibitors (7M urea, 2M thiourea, 33mM CHAPS, 45mM ASB-14, 0.5% (v/v) Triton x-100, Protease inhibitor cocktail-Roche, 1 mM PMSF, 1 mM Na3VO4, 1 mM NaF). For each experiment, 300 μg of protein were incubated for 30 min in 120 μl of 2D buffer containing 15 mM DTT and 1% (v/v) IPG buffer pH 3-10. Each sample was passively rehydrated overnight at room temperature into 7 mm IPG strips pH 3-10. Strips were isoelectrically focused on a Protean IEF Cell (Biorad) using a programmed voltage gradient consisting of 200 V held for 20 min, linearly increasing to 3500 V over 2.8 kVh and focused at 3500 V for 4.8 kVh. Proteins were separated by molecular weight using SDS-PAGE (10% Tris-glycine gel). Resulting gels were stained with Sypro Ruby (Bio-Rad) and technical replicates were performed for all gels.
Western immunoblotting and immunoprecipitations
Ventricular proteins were isolated in buffer containing: (in mmol/L) 150 NaCl, 20 Tris- HCl, 1 EDTA, 1 EGTA, 2.5 sodium pyrophosphate, 1 β-glycerophosphate, 1 Na3VO4, 1 NaF, 1 PMSF phosphatase inhibitors, 1% NP-40 and protease inhibitor cocktail (Roche Diagnostics). After Bradford protein assay, proteins were separated by standard SDS-PAGE on 10% Tris-glycine gels, transferred to nitrocellulose and blotted with corresponding antibodies. For immunoprecipitation, 1mg of starting protein was precleared with 100μl A/G beads and then incubated with 40 μl beads and 5 μl antibody for 4 hours at 4°C with end over end mixing. The beads were washed three times and proteins eluted with Laemmli buffer followed by electrophoresis and western blotting. Protein loading was confirmed by Ponceau staining of membranes. All blots are representative of at least three technical replicates.
Protein Identification by Mass Spectrometry
Protein bands or spots were in-gel digested with trypsin as described [19] with minor modifications. Briefly, the gel plug is dehydrated in acetonitrile (ACN) and dried completely in a Speedvac. Samples are reduced and alkylated with 10 mM dithiotreitol and 10mM TCEP solution in 50 mM NH4HCO3 (1 h at 37°C) and 100 mM iodoacetamide (45 min at 56°C in dark), respectively. Gels are washed with 50 mM NH4HCO3, dehydrated with ACN, and dried down in a Speedvac. Gel pieces are then swollen in digestion buffer containing 50mM NH4HCO3, and 20.0 ng/μL of trypsin (37°C, overnight). Peptides are extracted by 0.1%TFA in 50%ACN solution, dried down and resuspended in LC buffer A. Ten μl of extracted peptides were analyzed by nano-flow LC/MS/MS on a Thermo Orbitrap with with dedicated Eksigent nanopump using a reversed phase column (75 μm i.d. 10 cm, BioBasic C18 5μm particle size, New Objective). The flow rate is 200 nL/min for separation: mobile phase A is 0.1% formic acid, 2% ACN in water, and mobile phase B is 0.1% formic acid, 20% water in ACN. A shallow gradient is used for analyses: linear gradient from 5% B to 40% B over 75 min, then to 100% B over 19 min, and finally keeping constant 100% B for 10 min. Spectra were acquired in data-dependent mode with dynamic exclusion where the instrument selects the top six most abundant ions in the parent spectra for fragmentation and data were searched against the mouse IPI database (version 3.46) using the SEQUEST algorithm in the Bioworks software program. Any proteins identified in a reversed database search were removed, producing an effective false positive rate of zero at the database searching level. All spectra used for identification had deltaCN>0.1 and met the following Xcorr criteria: >2 (+1), >3 (+2), >4 (+3), and >5 (+4). In addition to these criteria, all spectra used for identification of phosphorylated peptides were manually examined to ensure correct mass of parent ion, presence or absence of phosphate in daughter ion spectra, and neutral loss peaks. An additional 10 μl of extracted peptides were analyzed by electron transfer dissociation on a Thermo LTQ instrument. ETD experiments were performed using a 100 ms reaction time and a 2e5 anion target for the fluoranthene reagent. Supplemental activation (resonance CID) was utilized to improve fragmentation of precursor ions. At least two technical replicates were done on all mass spectrometry experiments.
RNA Purification and Quantitative Real Time PCR Analysis
Total RNA was isolated from murine hearts using TRIzol (Invitrogen) according to the manufacturer's protocol and RT-PCR performed as described [9, 20]. Expression of full length and truncated (CRA_a) calsarcin-1 was determined using the following sets of primers: FL_CS1_F1: 5′-GGTGAAGCAAAGGAAACAGC-3′, CRA_CS1_F2: 5′-ATGGACATGCACAATATCGC-3′, FL_CS1_R1: 5’- TGCATGGCGATATTGTGATT-3’ and CRA_CS1_R2: 5′-TTTGGGTACAAAGCCTCCAG-3′. GAPDH expression was determined using the following primers: GAPDH_F1: 5’-CCCACTAACATCAAATGGGG-3’ and GAPDH_R1: 5’-CCTTCCACAATGCCAAAGTT-3’. Expression levels were analyzed using the iQ5 Optical Systems software v2.0 and normalized against GAPDH by subtracting the mean cycle number for each experimental group from the mean cycle number for GAPDH from the same group. The normalized means were then applied to the formula ((2(cycle#))-1) to calculate fold change.
Statistical analyses
All data are expressed ±SEM with p≤0.05 considered to be statistically significant. Comparisons were made using student's t-test for equal variances.
Supplementary Material
Supplemental Figure 1. Additional spectra of phosphorylated peptides. CID spectra used for detection of single phosphorylation at T-111 (A), S-106 (B) and S-116 (C), as well as for dual phosphorylation at S-106 and T-111 (D) are shown. ETD spectra for S-116 (E) is also provided. Bold labels indicate features used to resolve modified site.
Acknowledgements
This study was supported by NIH/NHLBI grants to TMV and YW. SF is the recipient of an NIH Ruth Kirschstein Fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
LITERATURE CITED
- 1.Wilkins BJ, Molkentin JD. Calcium-calcineurin signaling in the regulation of cardiac hypertrophy. Biochem Biophys Res Commun. 2004 Oct 1;322(4):1178–91. doi: 10.1016/j.bbrc.2004.07.121. [DOI] [PubMed] [Google Scholar]
- 2.Klee CB, Crouch TH, Krinks MH. Calcineurin: a calcium- and calmodulin-binding protein of the nervous system. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6270–3. doi: 10.1073/pnas.76.12.6270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frey N, Richardson JA, Olson EN. Calsarcins, a novel family of sarcomeric calcineurin-binding proteins. Proc Natl Acad Sci U S A. 2000 Dec 19;97(26):14632–7. doi: 10.1073/pnas.260501097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frey N, Barrientos T, Shelton JM, Frank D, Rutten H, Gehring D, et al. Mice lacking calsarcin-1 are sensitized to calcineurin signaling and show accelerated cardiomyopathy in response to pathological biomechanical stress. Nat Med. 2004 Dec;10(12):1336–43. doi: 10.1038/nm1132. [DOI] [PubMed] [Google Scholar]
- 5.Frey N, Frank D, Lippl S, Kuhn C, Kogler H, Barrientos T, et al. Calsarcin-2 deficiency increases exercise capacity in mice through calcineurin/NFAT activation. J Clin Invest. 2008 Nov;118(11):3598–608. doi: 10.1172/JCI36277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kostin S, Hein S, Arnon E, Scholz D, Schaper J. The cytoskeleton and related proteins in the human failing heart. Heart Fail Rev. 2000 Oct;5(3):271–80. doi: 10.1023/A:1009813621103. [DOI] [PubMed] [Google Scholar]
- 7.Pyle WG, Solaro RJ. At the crossroads of myocardial signaling: the role of Z-discs in intracellular signaling and cardiac function. Circ Res. 2004 Feb 20;94(3):296–305. doi: 10.1161/01.RES.0000116143.74830.A9. [DOI] [PubMed] [Google Scholar]
- 8.Takimoto E, Champion HC, Li M, Ren S, Rodriguez ER, Tavazzi B, et al. Oxidant stress from nitric oxide synthase-3 uncoupling stimulates cardiac pathologic remodeling from chronic pressure load. J Clin Invest. 2005 May;115(5):1221–31. doi: 10.1172/JCI21968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mitchell-Jordan SA, Holopainen T, Ren S, Wang S, Warburton S, Zhang MJ, et al. Loss of Bmx nonreceptor tyrosine kinase prevents pressure overload-induced cardiac hypertrophy. Circ Res. 2008 Dec 5;103(12):1359–62. doi: 10.1161/CIRCRESAHA.108.186577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mural RJ, Adams MD, Myers EW, Smith HO, Miklos GL, Wides R, et al. A comparison of whole-genome shotgun-derived mouse chromosome 16 and the human genome. Science. 2002 May 31;296(5573):1661–71. doi: 10.1126/science.1069193. [DOI] [PubMed] [Google Scholar]
- 11.Osio A, Tan L, Chen SN, Lombardi R, Nagueh SF, Shete S, et al. Myozenin 2 is a novel gene for human hypertrophic cardiomyopathy. Circ Res. 2007 Mar 30;100(6):766–8. doi: 10.1161/01.RES.0000263008.66799.aa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Villen J, Beausoleil SA, Gerber SA, Gygi SP. Large-scale phosphorylation analysis of mouse liver. Proc Natl Acad Sci U S A. 2007 Jan 30;104(5):1488–93. doi: 10.1073/pnas.0609836104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McDonough JL, Van Eyk JE. Developing the next generation of cardiac markers: disease-induced modifications of troponin I. Prog Cardiovasc Dis. 2004 Nov-Dec;47(3):207–16. doi: 10.1016/j.pcad.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 14.Steen H, Jebanathirajah JA, Rush J, Morrice N, Kirschner MW. Phosphorylation analysis by mass spectrometry: myths, facts, and the consequences for qualitative and quantitative measurements. Mol Cell Proteomics. 2006 Jan;5(1):172–81. doi: 10.1074/mcp.M500135-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Li H, Zhang L, Rao A, Harrison SC, Hogan PG. Structure of calcineurin in complex with PVIVIT peptide: portrait of a low-affinity signalling interaction. J Mol Biol. 2007 Jun 22;369(5):1296–306. doi: 10.1016/j.jmb.2007.04.032. [DOI] [PubMed] [Google Scholar]
- 16.Donella-Deana A, Krinks MH, Ruzzene M, Klee C, Pinna LA. Dephosphorylation of phosphopeptides by calcineurin (protein phosphatase 2B). Eur J Biochem. 1994 Jan 15;219(1-2):109–17. doi: 10.1111/j.1432-1033.1994.tb19920.x. [DOI] [PubMed] [Google Scholar]
- 17.Bhuiyan MS, Fukunaga K. Activation of HtrA2, a mitochondrial serine protease mediates apoptosis: current knowledge on HtrA2 mediated myocardial ischemia/reperfusion injury. Cardiovasc Ther. 2008;26(3):224–32. doi: 10.1111/j.1755-5922.2008.00052.x. Fall. [DOI] [PubMed] [Google Scholar]
- 18.Liu HR, Gao E, Hu A, Tao L, Qu Y, Most P, et al. Role of Omi/HtrA2 in apoptotic cell death after myocardial ischemia and reperfusion. Circulation. 2005 Jan 4;111(1):90–6. doi: 10.1161/01.CIR.0000151613.90994.17. [DOI] [PubMed] [Google Scholar]
- 19.Edmondson RD, Vondriska TM, Biederman KJ, Jun Z, Jones RC, Zheng YT, et al. Protein kinase C epsilon signaling complexes include metabolism- and transcription/translation-related proteins - Complementary separation techniques with LC/MS/MS. Molecular & Cellular Proteomics. 2002 Jun;1(6):421–33. doi: 10.1074/mcp.m100036-mcp200. [DOI] [PubMed] [Google Scholar]
- 20.Mitchell S, Ota A, Foster W, Zhang B, Fang Z, Patel S, et al. Distinct gene expression profiles in adult mouse heart following targeted MAP kinase activation. Physiol Genomics. 2006 Mar 13;25(1):50–9. doi: 10.1152/physiolgenomics.00224.2005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Additional spectra of phosphorylated peptides. CID spectra used for detection of single phosphorylation at T-111 (A), S-106 (B) and S-116 (C), as well as for dual phosphorylation at S-106 and T-111 (D) are shown. ETD spectra for S-116 (E) is also provided. Bold labels indicate features used to resolve modified site.