Abstract
Objectives
1) To determine allele frequencies of 3 LAMC1 single nucleotide polymorphisms (SNPs) in Caucasian and African-American (AA) women with > Stage II POP (cases) and in ethnicity-matched controls with < Stage 2 POP. 2) To determine if LAMC1 is associated with POP within ethnic groups.
Study design
Allelic discrimination was performed for LAMC1 SNPs rs10911193 (C/T), rs20563 (A/G), and rs20558 (T/C). SNP and haplotype-specific tests were used to examine associations between POP, ethnicity, and LAMC1.
Results
411 women were enrolled. Significant differences in allele and haplotype frequencies existed amongst AA and Caucasians: rs10911193 “T” (p=0.0014); rs20563 “G” (p<0.0001); rs20558 “C” (p<0.0001); rs20563, rs20558 “GC” (p<0.0001); and rs20563, rs20558 “AT” (p<0.0001). No significant associations between POP and LAMC1 SNPs or haplotypes were found within ethnicities.
Conclusions
While significant differences were identified between AA and Caucasian women, no associations were found between any LAMC1 gene variant and advanced POP.
Keywords: familial pelvic organ prolapsed, genetics, genetic predisposition, lamimin gamma 1, LAMC1, POP
Introduction
The pathophysiology of pelvic organ prolapse (POP) is multifactorial with risk factors that may be categorized as predisposing [1], inciting [2], promoting[3], or decompensating [4, 5]. Depending on the combination of these risk factors in an individual, prolapse may or may not develop over her lifetime. Childbirth in general is believed by many to be the major risk for pelvic organ prolapse yet the majority of parous women do not develop the condition [6–8] and young nulliparous women occasionally are affected [9]. An underlying heritable condition, therefore, may play a significant role in the development of POP.
Family-based studies and examination of monozygotic and dizygotic twins suggest that POP has a heritable component. Chiaffarino et al examined 108 women with pelvic floor disorders and found that the risk of developing POP was higher in women with a positive family history, including first-degree relatives only [1]. Genetic analysis of 10 families of women who developed POP at a young age found a dominant pattern of inheritance with incomplete penetrance[10] and a study of 101 pairs of nulliparous and parous sisters[9] found a high concordance of POP suggesting that familial predisposition plays a more important role in the development of this condition. Recent additional evidence for a predisposition gene for POP was revealed through a parametric linkage analysis of 70 affected women of European descent from 32 eligible families with at least 2 affected cases. Fifty-three percent of pedigrees had evidence of a predisposition gene on chromosome 9q21 with a HLOD score of 3.41 [11].
Epidemiological data also support this hypothesis. Ethnic and racial variations in the incidence of POP have been described with Caucasian and Hispanic women demonstrating a greater risk compared to women of African, Asian, and Native American ethnicity [12, 13]. Although this racial/ethnic disparity is not necessarily the result of genetic variation, it raises the possibility that the racial/ethnic differences are due to different frequencies of risk alleles for POP or, alternatively, alleles that confer a protective effect.
In trying to identify which factors are involved in cases of familial POP, several candidate genes involved in collagen and elastin biosynthesis and extracellular matrix metabolism have been evaluated. Nikilova identified one such gene, LAMC1 that codes for the laminin gamma 1 chain, a critical component of the extracellular matrix [14]. These authors found that the minor “T” allele of a single nucleotide polymorphism (SNP) (rs10911193) in the gene promoter that affects binding of the transcription factor NFIL3 was significantly more prevalent among probands (22% vs 6.7%) of a family who demonstrated an autosomal dominant mode of inheritance of POP than the general population. The authors, however, did not disclose the ethnicity of the family that was studied.
The goals of this study were to determine if polymorphisms in the LAMC1 gene are associated with advanced POP in different ethnic groups by 1) determining the allele frequencies of SNP rs10911193, in addition to 2 other functional SNPs that have been shown to change the amino acid sequence in the laminin gamma 1 chain protein, in Caucasian and AA women with advanced POP (cases) and in race-matched controls with normal pelvic support and by 2) determining if any of the allele frequencies are associated with POP within ethnic groups.
Materials and Methods
Study Population
After obtaining institutional review board approval, Caucasian and African American women with > Stage II POP (cases) and a control population with < Stage II POP according to the Pelvic-Organ-Prolapse Quantification System (POP-Q) were prospectively recruited from the Urogynecology and benign gynecology clinics of a study author (CAM). Control subjects were recruited to match cases on the basis of age, race, menopausal status, smoking history, body-mass-index, and parity. Exclusion criteria for all study subjects included a personal history of a systemic collagen disorder such as Ehlers-Danlos or Marfan’s syndrome. Demographic data was obtained and recorded on data collection sheets at the time of written consent, including age, race, parity, menopausal status, smoking status, and BMI.
Genotyping
DNA samples were collected through buccal smears using Simhelix Buccal Swabs (Boca Scientific, Boca Raton, FL), the extraction of which was performed using ChargeSwitch gDNA Buccal Cell Kit ( Invitrogen, Carlsbad, CA). Three previously identified LAMC1 SNPs (rs10911193, rs20563, and/or rs20558) were evaluated in each subject. LAMC1 SNPs rs10911193, rs20563, and rs20558 were identified as NT_004487.18:g.33481002C>T, NM_002293.3:c.1372A>G, and NM_002293.3:c.2663T>C, respectively. PCR was performed on the samples using Taqman SNP Genotyping Assays (Applied BioSystems, Foster City,CA). Allelic PCR products were separated using the Applied Biosystems 7500 fast Sequence Detection System with SDS 1.3.1 software. Genotypes were auto-called by SDS 1.3.1 software with quality value set at 0.90.
To confirm rs10911193 C>T variant, we carried out PCR using a forward primer sequence of 5'- CACTGGCTGGTTACACTTTACCTCT-3’ and a reverse primer 5'-CCTTTTGAGTCCTAATGTCCAAGAC -3' yielding a 200 bp product. After initial denaturation at 95°C for 3 min, PCR was performed for 35 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 30 s and extension at 72°C for 30 s followed by a final 5 min elongation at 72°C. The PCR products were used for RFLP analysis with the restriction endonuclease MaeIII (Roche, Indianapolis, IN) for genotyping. The “C” allele yielded two fragments (141bp and 59bp) and the “T” allele yielded one fragment with a size of 200bp. To confirm the rs20563 A>G variant, we carried out PCR using a forward primer sequence of 5'-AGGGCTGACTTGAAGAGTGG -3’ and a reverse primer 5'- GCCTTCGACATTGTCTTTGC -3' yielding a 152 bp product. After initial denaturation at 94°C for 5 min, PCR was performed for 36 cycles of denaturation at 94°C for 30s, annealing at 52°C for 30 s and extension at 72°C for 30 s followed by a final 5 min elongation at 72°C. The rs20563 A>G variant was confirmed by RFLP analysis with the restriction endonuclease SspI (New England Biolabs, Ipswich,MA). The “A” allele yielded two fragments (110 bp and 42 bp) and the “G” yielded one fragment with a size of 152bp.
Statistical analysis
Descriptive statistics, including means and standard deviations, for demographic information were conducted using SAS v9.2 and assuming an alpha level of 0.05. Fisher’s exact tests implemented in the PLINK software [15] were used to test individual SNPs for allele frequency differences between races and for associations with POP within races. Inter-SNP linkage disequilibrium calculations for each race were performed in Haploview (version 4.0). Haplotype frequencies were also generated in PLINK based on the two SNPs found to be linked using Haploview and haplotype-specific tests were performed in PLINK to test for frequency differences between races and for associations with POP within races.
Results
A total of 411 subjects were available for analysis: 146 African Americans (63 cases and 83 controls) and 265 Caucasians (102 cases and 163 controls). The majority of the cases (82%) were Stage III POP. A comparison of demographic data of subjects by race and by case/control assignment is presented in Table 1. Significant differences in cases and controls included age (65.0 ± 10.8 vs. 59.8 ± 12.0, p<0.0001) and a greater percentage of post-menopausal women (90.3% vs. 78.9%, p=0.0013). When comparing races, AA women had a significantly higher body-mass-index (31.6 ± 6.3 vs. 27.9 ± 6.6, p<0.0001) and parity (3.5 ± 2.5 vs. 2.5 ± 1.2, p<0.0001) than Caucasian women.
Table 1.
Demographic characteristics of study subjects
| By Race | African American N=146 |
Caucasian N=265 |
P-value |
|---|---|---|---|
| Age (years) | 62.7 ± 11.8 | 61.4 ± 11.8 | 0.2725 |
| BMI (kg/m2) | 31.6 ± 6.3 | 27.9 ± 6.6 | <0.0001 |
| Parity | 3.5 ± 2.5 | 2.5 ± 1.2 | <0.0001 |
| Smoking (%) | 12.3% | 15.1% | 0.4639 |
| Post-menopausal (%) | 84.9% | 82.6% | 0.8129 |
| By POP | Controls N=246 |
Cases N=165 |
|
| Age (years) | 59.8 ± 12.0 | 65.0 ± 10.8 | <0.0001 |
| BMI (kg/m2) | 29.2 ± 7.4 | 29.2 ± 5.5 | 0.9455 |
| Parity | 2.8 ± 1.8 | 3.0 ± 1.8 | 0.2783 |
| Smoking (%) | 15.0% | 12.7% | 0.5647 |
| Post-menopausal (%) | 78.9% | 90.3% | 0.0013 |
The allele frequencies of rs10911193, rs20563, and rs20558 by ethnicity are presented in Table 2. Significant differences in the allele frequencies for all three SNPs existed amongst AA and Caucasians: rs10911193 “T” (4.8% vs. 11.5%, odds ratio (OR) 0.39, 95% CI (0.21,0.71), p=0.0014), the promoter SNP; rs20563 “G” (39.0% vs. 53.4%, OR 0.56, 95% CI (0.42, 0.75), p<0.0001), which produces a non-synonymous amino acid substitution, Ile458Val; and rs20558 “C” (38.4% vs. 53.6%, OR 0.54, 95% CI (0.40, 0.72), p<0.0001), which also results in non-synonymous amino acid substitution Leu888Pro.
Table 2.
Allele frequencies and haplotypes among populations
| LAMC1 | Allele | African American (frequency) |
Caucasian (frequency) |
OR (95% CI) | P-value |
|---|---|---|---|---|---|
| SNP | |||||
| rs10911193 | T | 0.048 | 0.115 | 0.39 (0.21, 0.71) | 0.0014 |
| rs20563 | G | 0.390 | 0.534 | 0.56 (0.42, 0.75) | <0.0001 |
| rs20558 | C | 0.384 | 0.536 | 0.54 (0.40, 0.72) | <0.0001 |
| Haplotypes | ChiSq | P-value | |||
| GC | 0.386 | 0.535 | 16.62 | <0.0001 | |
| AT | 0.614 | 0.465 |
Haplotype SNP order: rs20563, rs20558. OR, odds ratio; CI, confidence interval; ChiSq, chi-squared.
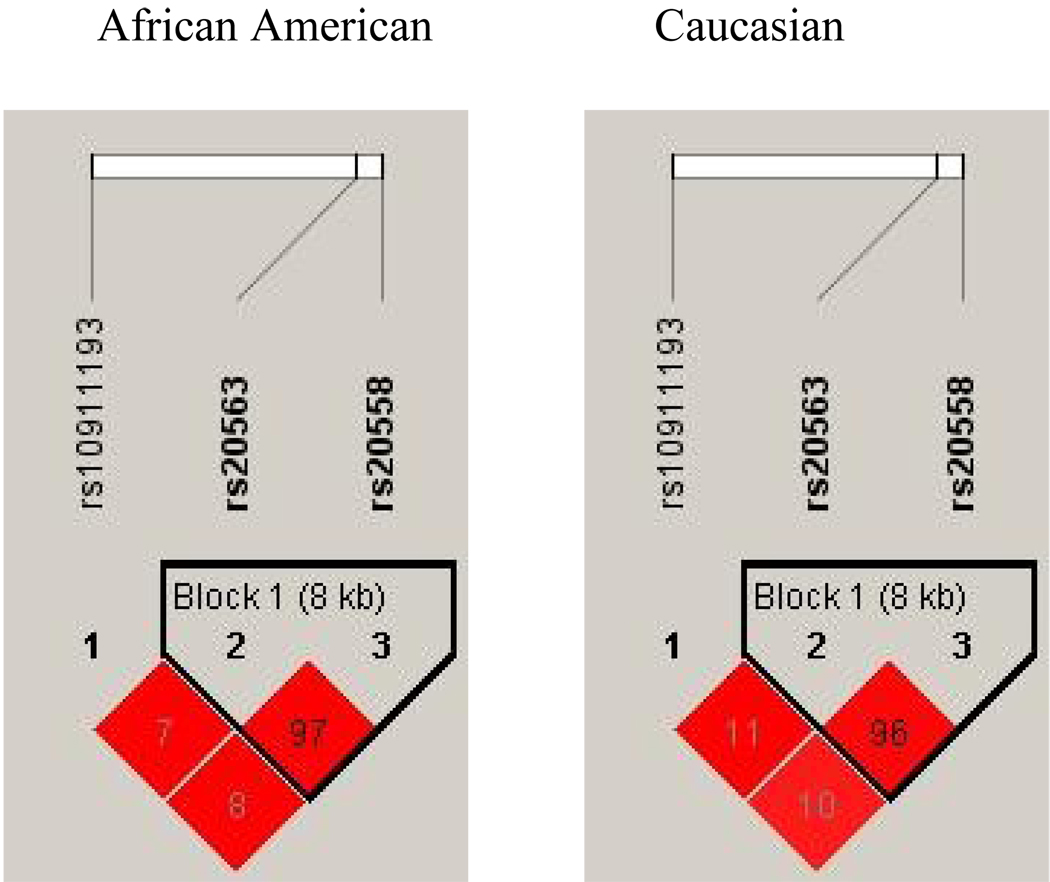
Haplotype analysis showed rs20563 and rs20558 to be in high LD for both African American and Caucasians (R2=0.972 and R2=0.962 respectively) (Figure 1). Two haplotypes of these SNPs were identified: GC and AT (SNP order: rs20563, rs20558). Haplotype frequencies by ethnicity are presented in Table 2. Significant differences in the haplotype frequencies existed amongst AA and Caucasians (GC (38.6% vs. 53.5%), AT (61.4% vs. 46.5%), chi-squared (ChiSq) 16.62, p<0.0001).
Figure 1.
Linkage disequilibrium (LD) plots with inter-single nucleotide polymorphism (SNP) R2 for LAMC1 SNPs rs10911193, rs20563, and rs20558 within the African American and Caucasian cohorts.
No associations between POP and allele frequencies were found within ethnic groups: AA and Caucasian rs10911193 “T” (p=0.408 and p=0.780 respectively); AA and Caucasian rs20563 “G” (p=0.334 and p=0.421 respectively); AA and Caucasian rs20558 “C” (p=0.332 and p=0.371 respectively) (Table 3). Additionally, no associations between POP and haplotype frequencies were found for either African Americans or Caucasians (p=0.298 and p=0.352 respectively) (Table 3).
Table 3.
Case-control study of LAMC1 SNPs and haplotypes and POP.
| LAMC1 | Allele | African American | Caucasian | ||||||
|---|---|---|---|---|---|---|---|---|---|
| SNP | POP OR |
Control OR |
P-value | POP OR |
Control OR |
P-value | |||
| rs10911193 | T | 0.063 | 0.036 | 0.408 | 0.108 | 0.120 | 0.780 | ||
| rs20563 | G | 0.357 | 0.416 | 0.334 | .0490 | 0.451 | 0.421 | ||
| rs20558 | C | 0.349 | 0.410 | 0.332 | 0.490 | 0.448 | 0.371 | ||
| Haplotype | ChiSq | P-value | ChiSq | P-value | |||||
| GC | 0.352 | 0.412 | 1.084 | 0.298 | 0.510 | 0.551 | 0.868 | 0.352 | |
| AT | 0.648 | 0.588 | 0.490 | 0.449 | |||||
Haplotype SNP order: rs20563, rs20558. OR, odds ratio; ChiSq, chi-squared.
Given the significant difference in SNP allele and haplotype frequencies between ethnic groups and the lack of any observed association between POP and either allele frequency or haplotype frequency within these groups, attempting to search for an association between cases and gene expression across the combined cohort would introduce the potential for population stratification. Therefore, this analysis was not included.
Comment
While significant differences in allele frequencies existed between races for all three SNPs and the two haplotypes, we found no association between any LAMC1 gene variant and advanced POP within each race. Nikolova et al previously reported that the minor “T” allele LAMC1 variant (rs10911193) in the gene promoter was strongly linked to familial POP and suggested that it might be a useful marker for the development of the disease [14]. Based on our reported results, the allele frequencies described in the report of Nikolova et al. (22% among probands vs. 6.7% for a general “European” population) raise concerns regarding the definition of the proband and general population ancestries. In our study, we demonstrated a significantly higher minor “T” allele frequency in the Caucasian cohort (11.5%) whereas the minor allele frequency of 4.8% in our African American subjects was in far closer approximation to their reported results. Furthermore, the minor alleles for rs20563 and rs20558 identified in our study differed between the two cohorts (“G” and “C” respectively for African Americans and “A” and “T” respectively for Caucasians). These observations of wide differences in allele frequencies highlight the potential for population stratification to confound association studies of the LAMC1 gene and POP and we challenge the conclusions drawn from the previous results.
After determining the allele frequencies of each SNP in each population, a power analysis was performed to determine the number of subjects needed to demonstrate an association between the minor allele and POP with an Odds Ratio of 2.0, based on a power of 0.80 and alpha of 0.05. Given our sample sizes, we can exclude a role for the LAMC1 SNPs in contributing to an odds ratio for POP > 3.3 in Caucasians, but cannot exclude lesser contributions.
Ethnic variation in POP prevalence has been described yet no comparative studies have yet provided evidence for a significant association between particular risk alleles and disease prevalence. We recently reported on the lack of any association between a SNP in the lysyl oxidase gene and POP amongst a cohort of Caucasian and African American women with and without POP [16]. A predisposition gene for pelvic floor disorders on chromosome 9 based on a linkage analysis of women of European descent has been identified [11]. Which particular genes are involved and whether there is significant ethnic variation in allele frequencies is currently unknown and presents several opportunities for further study. While we did observe an approximately 2.5-fold higher chance of our Caucasian cohort expressing the minor “T” risk allele for rs109111193 than the African Americans, an ethnic group with a lower prevalence of POP, the lack of any significant association between the risk allele and disease within each group decreases the potential impact of this gene variant.
Several limitations in this study exist, the principal of which is the sample size. A significantly larger numbers of subjects would be needed in each group to demonstrate the potentially small impact (odds ratio between 1.5 and 2) that a particular risk allele may confer. Our sample size is big enough, however, to show that none of the studied LAMC1 polymorphisms has a strong association with POP in either our African American or Caucasian cohort. In addition to a limited sample size, our cases and controls did have significant differences in age and number of women who were menopausal, both of which are known risk factors for disease prevalence. We could have included women in our control group who carry the risk allele and have not yet reached the age threshold for developing POP. The differences in demographic variables, however, do not alter the observed differences in ethnic allele frequencies, the principal outcome of this study.
A final limitation is that we did not confine our study to women with a strong potential for a genetic predisposition for POP. Evaluation of a genetic basis for POP is problematic given the multitude of factors that have been shown to affect disease prevalence. The Lifespan Model for Pelvic Floor Disorders presented by DeLancey et al includes three phases of which genetic constitution is categorized under Phase I: predisposing factors [17]. It is certainly plausible that a significant genetic predisposition exists for some, but not the majority of women with POP. Perhaps similar to breast cancer which occurs at a higher frequency and a younger age in women with genetic mutations such as BRCA1, POP that is related to heritable causes may too present at a younger age and more advanced stage. Performing genetic studies on women in the general population who may reflect the cumulative effect of exposure to other known risk factors (Phase II, inciting factors and Phase III, intervening factors [17]) could potentially hide the impact of genetic factors that are observed in more specific populations. The inclusion of predominantly post-menopausal women from the general population may explain why we found no association between any risk allele and the presence/absence of POP.
In conclusion, several LAMC1 single nucleotide polymorphisms exist with significant ethnic differences in allele and haplotype frequencies yet no LAMC1 gene variant was associated with POP in either group. In the general population, LAMC1 is not a candidate gene that confers risk or protection for the development of the disease. Any future studies regarding the LAMC1 gene need to account for the observed differences in ethnic allele frequencies to avoid the confounding factor of population stratification.
Acknowledgments
This research was supported by National Institutes of Health grant P60 MD002256. LDH is supported by the VCU Physician-Scientist Training Program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accepted for oral presentation at the 30th Annual American Urogynecology Society Meeting, Hollywood Florida.
Reprints will not be available
Significant ethnic differences in allele and haplotype frequencies for 3 LAMC1 gene single nucleotide polymorphisms exist, but none was associated with advanced pelvic organ prolapse.
Bibliography
- 1.Chiaffarino F, et al. Reproductive factors, family history, occupation and risk of urogenital prolapse. Eur J Obstet Gynecol Reprod Biol. 1999;82(1):63–67. doi: 10.1016/s0301-2115(98)00175-4. [DOI] [PubMed] [Google Scholar]
- 2.Mant J, Painter R, Vessey M. Epidemiology of genital prolapse: observations from the Oxford Family Planning Association Study. Br J Obstet Gynaecol. 1997;104(5):579–585. doi: 10.1111/j.1471-0528.1997.tb11536.x. [DOI] [PubMed] [Google Scholar]
- 3.Weber AM, Richter HE. Pelvic organ prolapse. Obstet Gynecol. 2005;106(3):615–634. doi: 10.1097/01.AOG.0000175832.13266.bb. [DOI] [PubMed] [Google Scholar]
- 4.Bump RC, Norton PA. Epidemiology and natural history of pelvic floor dysfunction. Obstet Gynecol Clin North Am. 1998;25(4):723–746. doi: 10.1016/s0889-8545(05)70039-5. [DOI] [PubMed] [Google Scholar]
- 5.Nygaard I, Bradley C, Brandt D. Pelvic organ prolapse in older women: prevalence and risk factors. Obstet Gynecol. 2004;104(3):489–497. doi: 10.1097/01.AOG.0000136100.10818.d8. [DOI] [PubMed] [Google Scholar]
- 6.Dannecker C, Anthuber C. The effects of childbirth on the pelvic-floor. J Perinat Med. 2000;28(3):175–184. doi: 10.1515/JPM.2000.025. [DOI] [PubMed] [Google Scholar]
- 7.Gill EJ, Hurt WG. Pathophysiology of pelvic organ prolapse. Obstet Gynecol Clin North Am. 1998;25(4):757–769. doi: 10.1016/s0889-8545(05)70041-3. [DOI] [PubMed] [Google Scholar]
- 8.Hendrix SL, et al. Pelvic organ prolapse in the Women's Health Initiative: gravity and gravidity. Am J Obstet Gynecol. 2002;186(6):1160–1166. doi: 10.1067/mob.2002.123819. [DOI] [PubMed] [Google Scholar]
- 9.Buchsbaum GM, et al. Pelvic organ prolapse in nulliparous women and their parous sisters. Obstet Gynecol. 2006;108(6):1388–1393. doi: 10.1097/01.AOG.0000245784.31082.ed. [DOI] [PubMed] [Google Scholar]
- 10.Jack GS, et al. Familial transmission of genitovaginal prolapse. Int Urogynecol J Pelvic Floor Dysfunct. 2006;17(5):498–501. doi: 10.1007/s00192-005-0054-x. [DOI] [PubMed] [Google Scholar]
- 11.Allen-Brady K, et al. Significant linkage evidence for a predisposition gene for pelvic floor disorders on chromosome 9q21. Am J Hum Genet. 2009;84(5):678–682. doi: 10.1016/j.ajhg.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Swift S, et al. Pelvic Organ Support Study (POSST): the distribution, clinical definition, and epidemiologic condition of pelvic organ support defects. Am J Obstet Gynecol. 2005;192(3):795–806. doi: 10.1016/j.ajog.2004.10.602. [DOI] [PubMed] [Google Scholar]
- 13.Sewell CA, Chang E, Sultana CJ. Prevalence of genital prolapse in 3 ethnic groups. J Reprod Med. 2007;52(9):769–773. [PubMed] [Google Scholar]
- 14.Nikolova G, et al. Sequence variant in the laminin gamma1 (LAMC1) gene associated with familial pelvic organ prolapse. Hum Genet. 2007;120(6):847–856. doi: 10.1007/s00439-006-0267-1. [DOI] [PubMed] [Google Scholar]
- 15.Purcell S, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferrell G, et al. A Single nucleotide polymorphism in the promoter of the LOXL1 gene and its relationship to pelvic organ prolapse and preterm premature rupture of membranes. Reprod Sci. 2009;16(5):438–446. doi: 10.1177/1933719108330567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delancey JO, et al. Graphic integration of causal factors of pelvic floor disorders: an integrated life span model. Am J Obstet Gynecol. 2008;199(6):610 e1–610 e5. doi: 10.1016/j.ajog.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]