SUMMARY
Histone H2B phosphorylation at Serine 14 (phosS14) has been proposed as an epigenetic marker of apoptotic cells whereas acetylation at the adjacent Lysine 15 (acK15) is a property of non-dying cells. We investigated the relationship and the potential regulatory mechanisms between these two epigenetic histone modifications and internucleosomal DNA degradation during apoptosis. Using rat primary thymocytes induced to undergo apoptosis with glucocorticoids we found that H2B phosphorylated at Ser14 was associated with soluble, cleaved DNA in apoptotic nuclei. In contrast acK15 was prevalent in non-apoptotic nuclei and scarce in apoptotic nuclei. This switch between K15 acetylation and S14 phosphorylation on H2B was also observed in apoptotic thymocytes from animals treated in vivo with glucocorticoids and in a rat hepatoma cell line (HTC) induced to die by UV-C or Fas ligand. Interestingly the combined use of a histone deacetylase inhibitor and glucocorticoid suppressed both S14 phosphorylation and internucleosomal DNA degradation without inhibiting apoptosis in thymocytes. Using synthetic peptides and a PKC phosphorylation assay system, we show that the deacetylation of K15 was necessary to allow the S14 phosphorylation. These findings suggest that selective chromatin post-translational modifications are associated with DNA degradation during apoptosis.
Keywords: chromatin structure, lymphocyte apoptosis, histone modification, glucocorticoid
INTRODUCTION
A fundamental unit of chromatin is the nucleosome which is composed of two sets of histones (H2A, H2B, H3 and H4) that are wrapped with 146 base pairs of DNA1. Histones have a tail in the N-terminal domain which is free from DNA and often has various covalent modifications2. In recent studies, it was shown that epigenetic histone modifications play an important role in chromatin function leading to the hypothesis that histone modifications act as a “histone code” possibly for chromatin remodeling3. In most apoptotic cells, internucleosomal degradation is considered a late biochemical hallmark of apoptosis4,5. Previously we demonstrated that histone H2B is phosphorylated in various mammalian apoptotic cells6,7 and can be phosphorylated by PKC both in vitro and in vivo6,8. Additionally, we determined that the site of the H2B phosphorylation is serine 14 (S14) and can involve the action of Mst-1 in vivo9. This S14 site can also be phosphorylated with PKC-delta in human B cells10, and S14 phosphorylation has been associated with apoptosis9 and with DNA double strand breaks11. However, the mechanisms regulating H2B phosphorylation at S14, particularly during apoptosis, are unknown.
Histone H2B has also been reported to be acetylated at the K5, K12, K15 and K20 sites12 which could potentially regulate the phosphorylation of S14 in the N-terminus of H2B. For example, it has been recently shown that phosphorylation of S10 in the yeast histone H2B, homologous to S14 in mammals, was regulated by K11 acetylation13, 14. To examine this question in mammalian cells we employed rat primary thymocytes induced by glucocorticoids to undergo apoptosis; a system used for both in vivo and in vitro analysis of apoptotic signal transduction15,16,17. We previously found that histone H2B was phosphorylated during the induction of apoptosis6. In this report, we examined the regulatory mechanism of the apoptosis-specific S14 H2B phosphorylation in relation to internucleosomal DNA degradation and deacetylation of the K15 lysine.
RESULTS
Glucocorticoids induce apoptosis in thymocytes
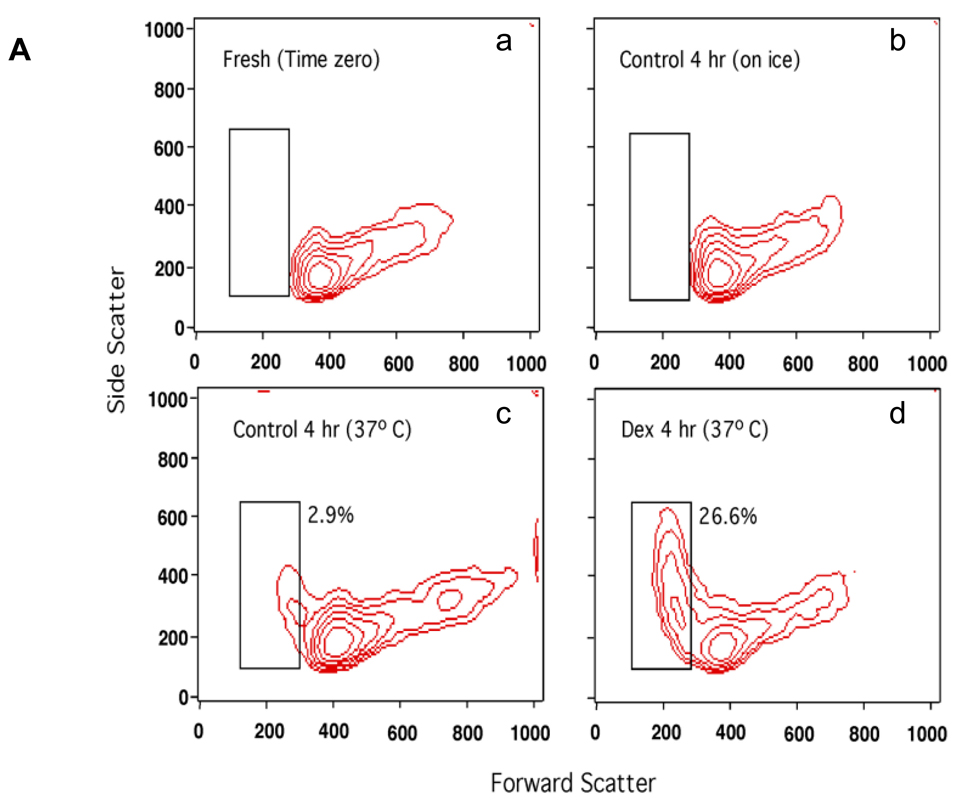
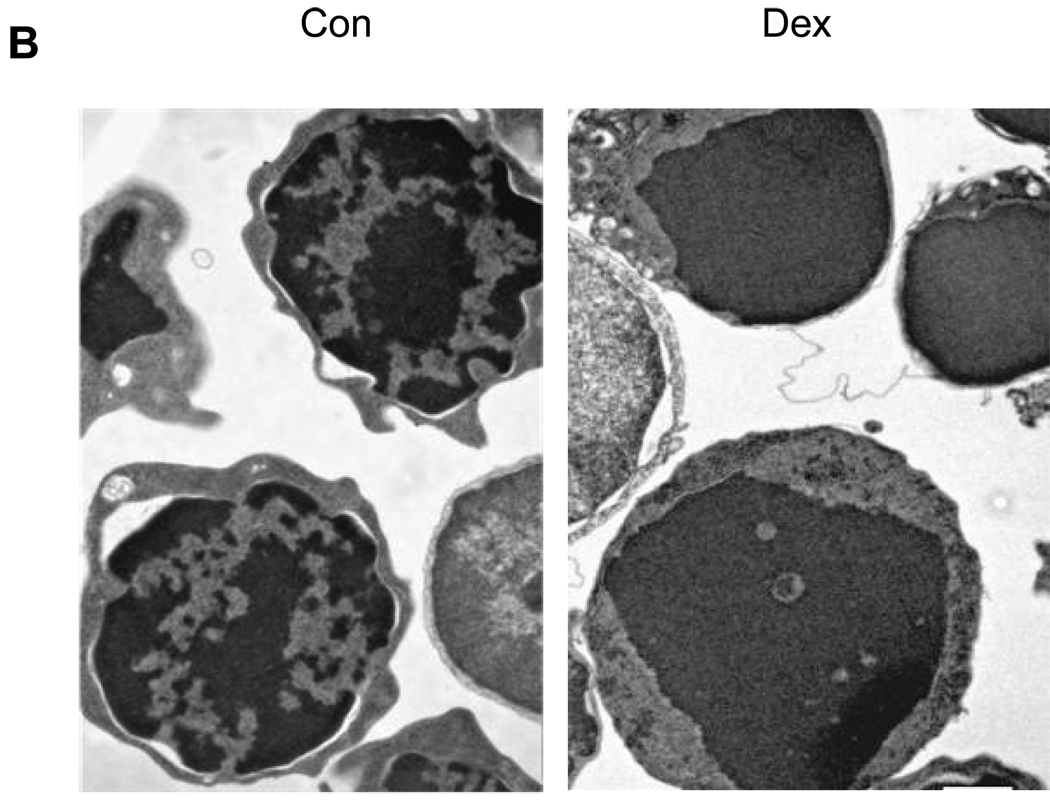
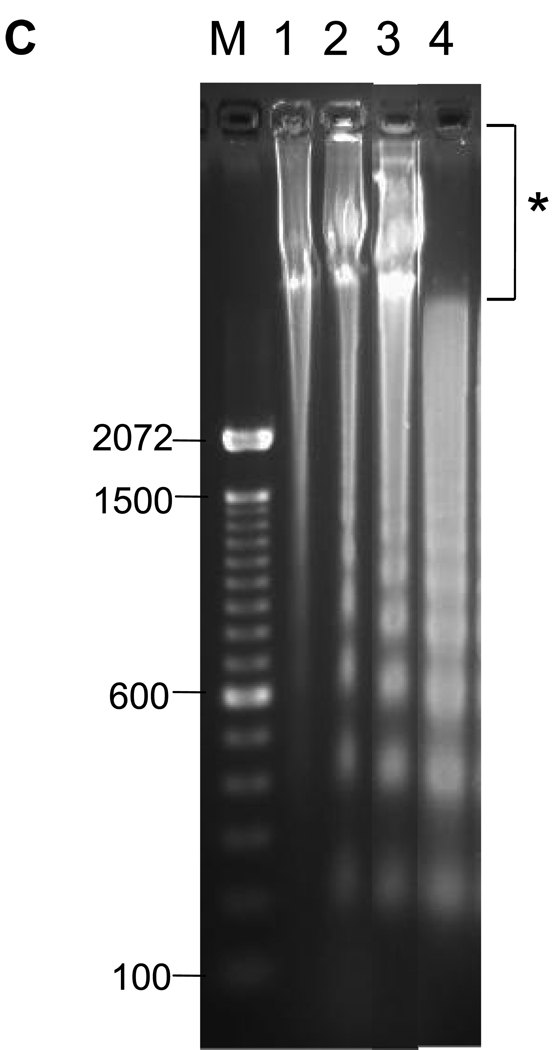
The induction of thymocyte apoptosis by dexamethasone was initially analyzed by flow cytometry (Fig. 1A). Thymocytes treated with 1 µM dexamethasone for 4 hrs contain a population of cells showing reduced forward scatter light but increased side scatter light. This occurrence of a shrunken, granular population of cells is characteristic of programmed cell death (Fig. 1A, d). In contrast, this cell population develops only to a small extent (2.9% vs. 26.6% in Dex treated) in the non-glucocorticoid treated control cells incubated at 37° C and is not detected in freshly isolated cells or cells incubated on ice for 4 hrs. This 2.9% of cells seen in our control sample likely reflects spontaneous apoptosis of these cells in culture (Fig. 1A, c). Ultrastructural analysis of thymocytes by electron microscopy shows distinct areas of euchromatin and heterochromatin (light and dark staining, respectively) in the nuclei of control thymocytes (Fig. 1B, left) in contrast to apoptotic cells where nuclear components appear degraded and showed homogeneous dark staining (Fig. 1B, right). Analysis of DNA by agarose gel electrophoresis (Fig. 1C) shows the characteristic pattern of internucleosomal DNA cleavage in nuclei from apoptotic thymocytes. After 4 hrs incubation of thymocytes with dexamethasone, only lower molecular weight DNA was observed in the soluble chromatin fraction (Fig. 1C, lane 4). Together these observations indicate that glucocorticoid treatment for 4 hrs is sufficient to induce classic apoptotic DNA fragments in rat thymocytes.
FIG. 1. Dexamethasone induces apoptosis in thymocytes.
(A) Rat primary thymocytes induced to undergo apoptosis by glucocorticoid treatment. Thymocytes were incubated with 1µM dexamethasone in RPMI1640 medium for 4 hrs at 37°C, and analyzed by flow cytometry. X-axis, forward scatter indicating cell size. Y-axis, side scatter indicating cell granularity. Apoptotic cells show a decrease in cell size (forward scatter light) with a concomitant increase in granularity (side scatter light). Dexamethasone treatment increases the percentage of apoptotic cells (box). a. freshly isolated thymocytes, b. untreated thymocytes incubated on ice for 4 hrs, c. untreated thymocytes incubated at 37°C for 4 hrs, d. dexamethasone treated thymocytes at 37°C for 4 hrs.
(B) Electron microscopic observation of apoptotic thymocytes. Thymocytes treated with dexamethasone were fixed as described in the Methods. Con, Control thymocytes. Dex, Dexamethasone. Nuclear materials of apoptotic cells are degraded. Magnification: 9, 900 ×.
(C) Analysis of DNA fragments from whole or soluble chromatin. Thymocytes treated with dexamethasone were harvested after 4 hrs. The DNA (1 µg) was run on 1.8% agarose gel and stained with ethidium bromide. Lane 1: Freshly isolated DNA from nuclei, lane 2: Control nuclei incubated 4 hrs, lane 3: nuclei from 1 µM dexamethasone treated nuclei for 4 hrs, lane 4: DNA from soluble chromatin from nuclei of the dexamethasone treated cells. Marker DNA; 100 bp DNA ladder. Note: DNA of soluble chromatin (lane 4) does not have HMW DNA as indicated by *.
Phosphorylated histone H2B at S14 is abundant in the soluble chromatin
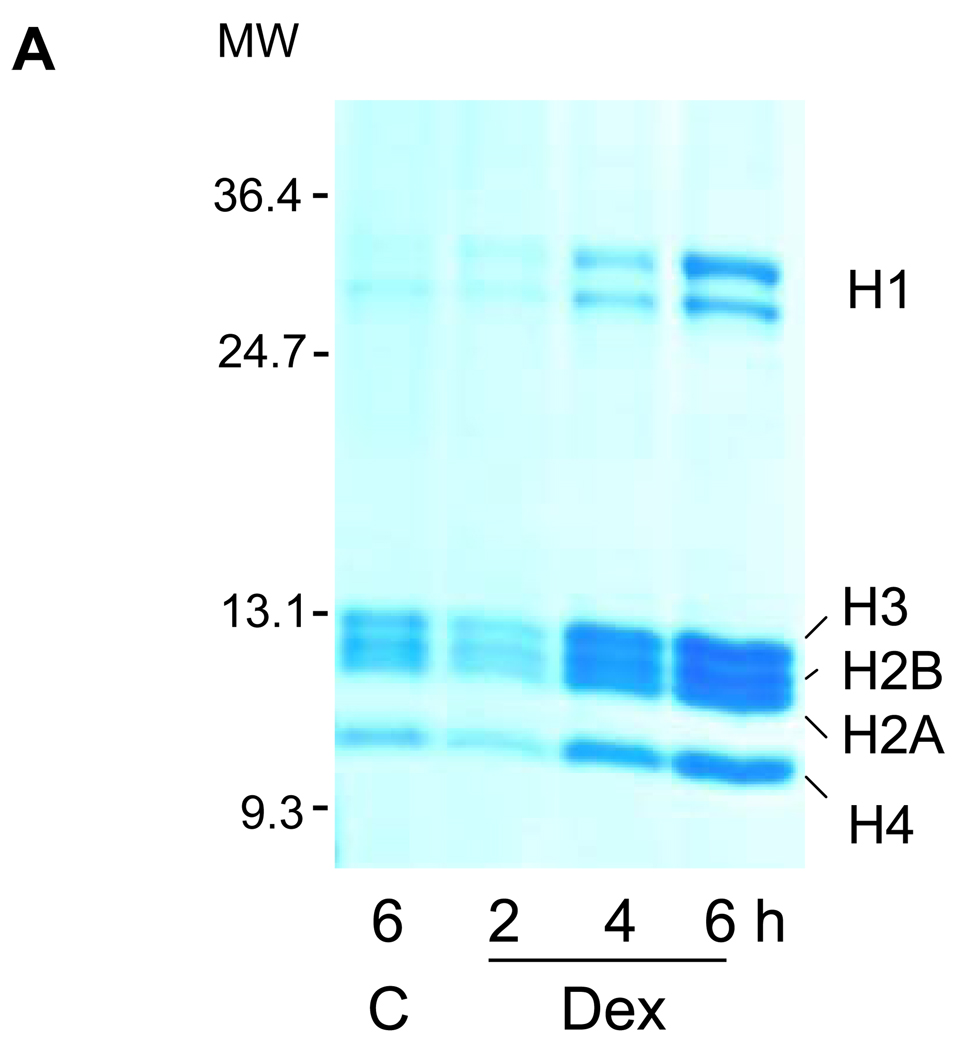
Degraded DNA fragments with their associated histones are separable from whole genomic DNA based on their solubility in nonionic detergent. Soluble chromatin from thymocytes exposed to 4 hrs of 1 µM dexamethasone was analyzed by SDS gel electrophoresis (Fig. 2A). The relative amount of histones H1, H2A, H2B, H3 and H4 present in the soluble chromatin fraction increased in a time dependent manner upon incubation with dexamethasone, indicating the release of histones from chromatin during apoptosis. In the control cells incubated for 6 h without dexamethasone, only small amounts of histones were also released which likely reflects spontaneous apoptosis of a small percentage of our cultured cells18.
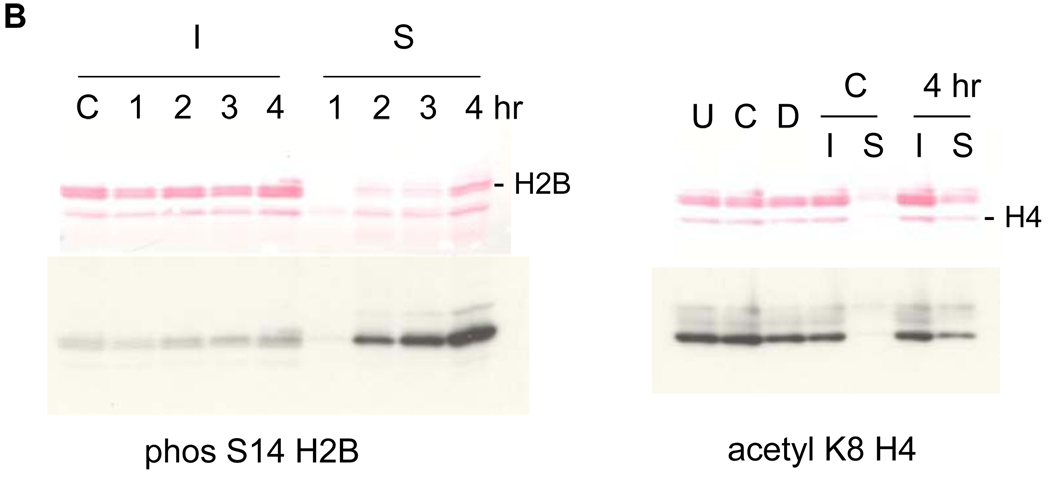
FIG. 2. Soluble chromatin contains S14 phosphorylated H2B.
(A) Histones were released during apoptotic internucleosomal degradation. Nuclei were isolated from control and dexamethasone treated cells and incubated with a lysis buffer containing 0.5% Triton X-100 at 4°C. Proteins contained in the lysis buffer were analyzed on SDS polyacrylamide gel electrophoresis and stained with 0.1% Coomassie Brilliant Blue.
(B) Thymocytes were incubated at 37°C for 1, 2, 3 and 4 hrs with 1 µM dexamethasone. Soluble (S) and insoluble (I) chromatin were isolated from nuclei. Histones were extracted from the chromatin and analyzed on SDS PAGE. left; Western blots were conducted with an anti-phosS14. C; untreated thymocytes incubated at 37°C. Proteins were stained with Ponceau-S. Right; Western blots were conducted with an acK8H4 antibody. U; untreated thymocytes on ice, C; control thymocytes, D; dexamethasone treated thymocytes at 37°C.
To analyze the amount of apoptosis-specific H2B phosphorylation at Ser14 in the soluble chromatin, a western blot against anti-phosSer14 H2B was conducted (Fig. 2B). Four hours following dexamethasone treatment, the amount of phosS14 is significantly increased in the soluble fraction, compared to the insoluble fraction of chromatin (Fig. 2B, left). In contrast, other histone modifications such as H4 acetylation at K8 are present in both the soluble and insoluble fraction in roughly equivalent amounts (Fig. 2B, right). These data suggest that histone H2B phosphorylation is selectively associated with histone release and internucleosomal DNA degradation during apoptosis.
Intracellular distribution of S14 phosphorylation in apoptotic cells
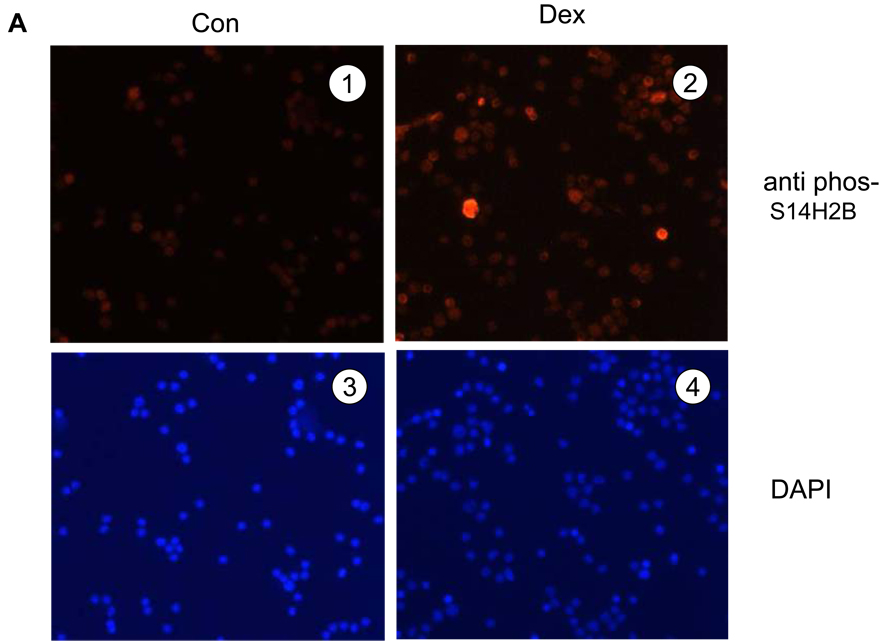
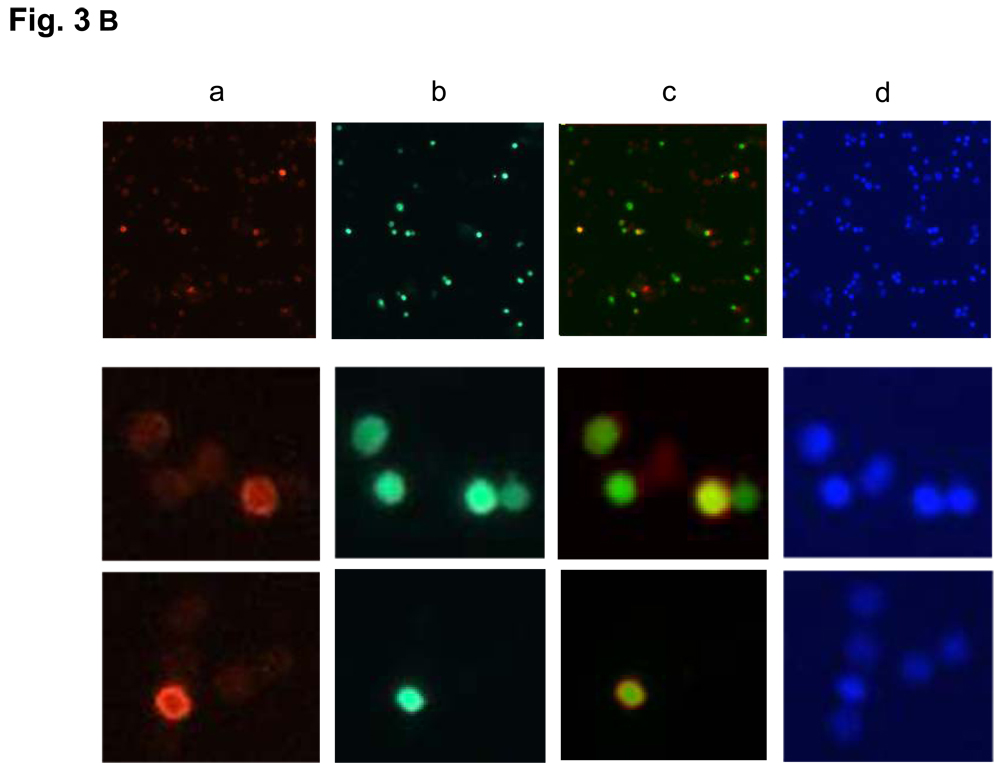
Next, we evaluated the distribution of the phosS14 in thymocytes by immunofluorescence (Fig. 3A). Thymocytes treated with 1 µM dexamethasone for 4 hrs or left untreated were stained with anti-phosS14 antibody (red) and DAPI (blue). There was an increase in cells positive for phosS14 in dexamethasone treated cells (Fig. 3A, 1 and 2). To determine if the H2B phosphorylation is associated with degraded DNA, thymocytes were examined by immunofluorescence of phosS14 (Fig. 3B, a), TUNEL staining (Fig. 3B, b), merge of the both (Fig. 3B, c) and DAPI (Fig. 3B, d). As shown in Fig. 3B, cells positive for phosS14 are also TUNEL positive. Interestingly, not all TUNEL positive cells stain with phosS14 because the TUNEL reaction occurs in both high molecular weight (HMW) DNA fragments and internucleosomal DNA fragments, whereas phosphorylation of S14H2B occurs primarily in internucleosomal DNA fragments.
FIG. 3. Immunofluorescence of S14 phosphorylation and TUNEL staining of dexamethasone treated thymocytes.
(A) Thymocytes were fixed with 4% paraformaldehyde and plated on cover slips and the cells were treated with anti-phosS14 and a secondary antibody (Cy3) at 37°C for 1 h. left (1 and 3) is control, and right (2 and 4) are dexamethsone treated cells. 1 and 2 show immunofluorescence of phosS14, 3 and 4; DAPI.
(B) Thymocytes were treated as above. a; Immunofluorescence of phosS14, b; TUNEL staining, c; merged, d; DAPI. Magnification is 100 × in the four pictures of the top panel, and 800 × in the eight pictures of the middle and the bottom panels.
Alterations in histone acetylation at K15 within Histone H2B
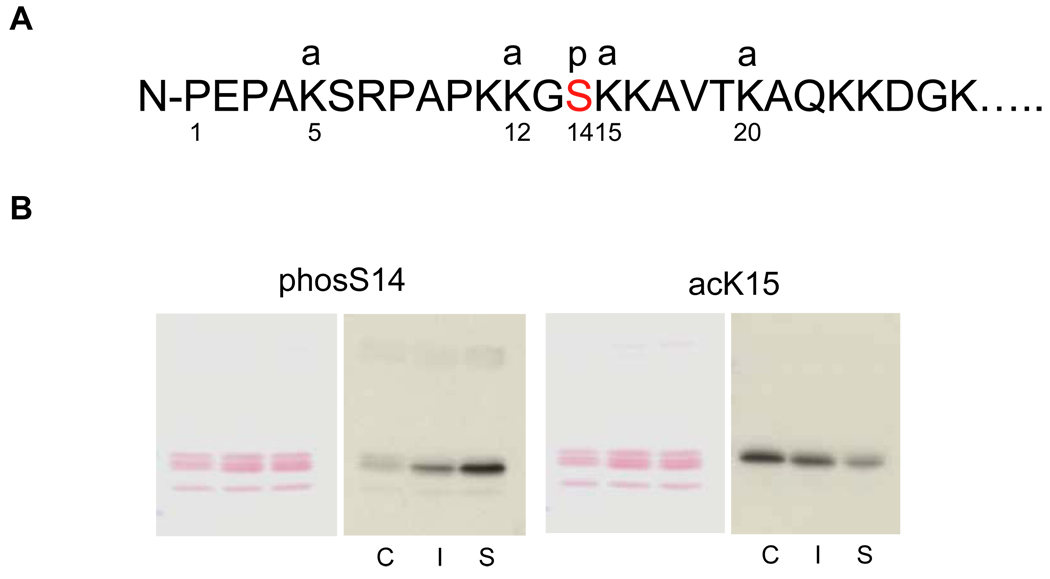
Although no significant difference in the acetylation of histone H4 at K8 was observed between soluble and insoluble chromatin (Fig. 2B), we were interested in possible changes in the level of acetylation of other sites in histone H2B. Figure 4A shows the amino acid sequence and known sites of modifications of H2B at the N-terminal tail. There are four major acetylation sites in H2B (K5, K12, K15 and K20)12.
FIG. 4. Changes of site-specific histone modifications in the soluble and insoluble chromatin in apoptotic thymocytes.
(A) The sites of H2B modifications at the N-terminal tail. Four sites are known to be acetylated at 5, 12, 15 and 20th residues of lysine, whereas a phosphorylation occurs at the 14th serine in the middle of an acetylation group. a: acetylation, p: phosphorylation.
(B) Acetylation and phosphorylation of H2B in the soluble and insoluble chromatin. Soluble and insoluble chromatin was prepared from dexamethasone treated thymocytes and histones were extracted. The histones were blotted against antibodies for site-specific modifications (phosS14 and acetyl K15) (right panel). Proteins were stained with Ponceau S (left panel). C. Control chromatin, I: Insoluble chromatin from dexamethasone treated thymocytes. S: Soluble chromatin from dexamethasone treated thymocytes. The data shown is representative of an N=4.
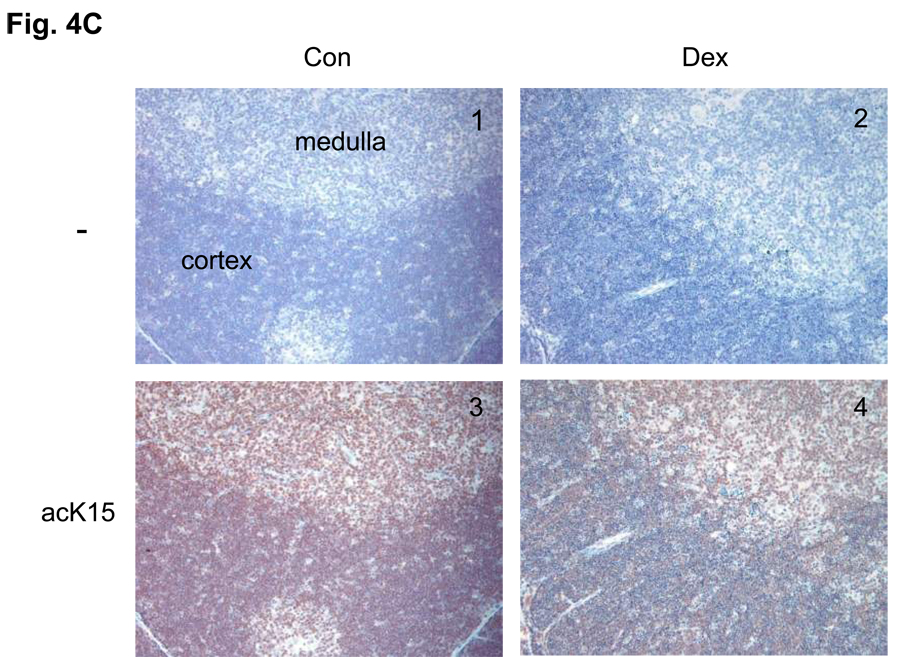
(C) Immunohistochemical analysis of phosS14 and acK15 in thymus tissues. Rat thymus was dissected and the tissues were treated with or without antibodies for histone modifications and stained with horse radish peroxidase. Brown color was observed as a positive reaction of a specific antibody. Pictures from 1 to 4 are a series of immunohistochemical analysis against acK15 and from 5 to 8 are for the analysis of phosS14. 1 and 5: Thymus section from the control animal treated without antibody. 2 and 6: Thymus section from the dexamethasone treated animal and treated without antibody. 3 and 7: Control section with the antibody. 4 and 8: Dexamethasone section with antibody.
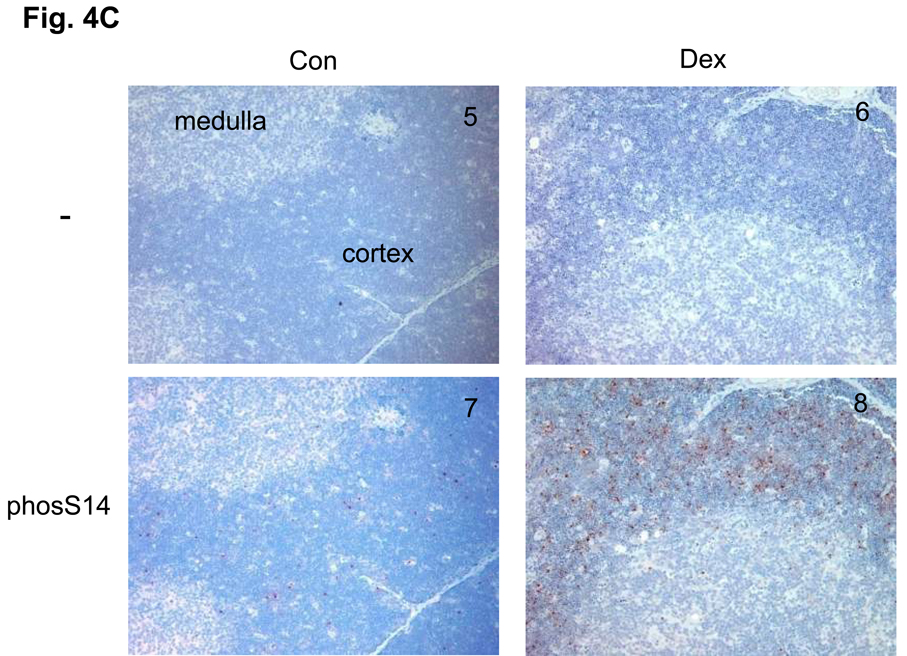
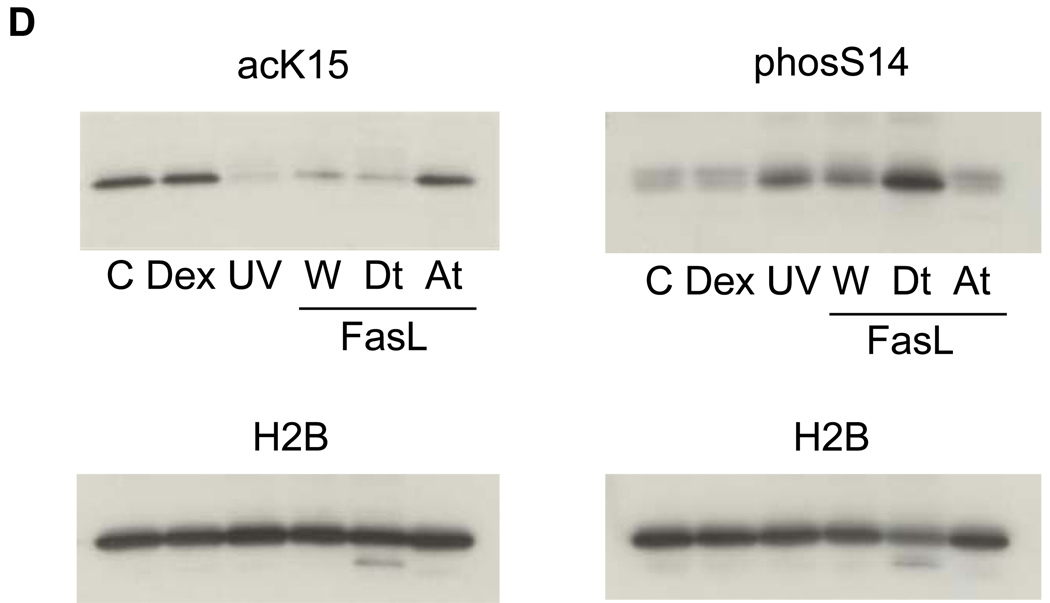
(D) A switch of histone modifications between acK15 and phosS14 in H2B of apoptotic HTC cells. Apoptosis was induced in HTC cells by UV-C or Fas ligand. Attached and detached cells were separated by a gentle shaking from glass substratum. Histones were extracted and analyzed by a western blotting against anti-phosS14. C, Control; Dex, dexamethasone; UV, whole cells treated with UV-C; W, whole cells treated with Fas ligand, At and Dt; attached and detached whole cells, respectively from Fas ligand treated cells. Amount of each sample was normalized by H2B detected with an anti-H2B antibody. The data shown is representative of an N=3.
We focused our studies on lysine 15 because of its proximity to S14. Thus, we examined the level of lysine 15 acetylation and S14 phosphorylation in both soluble and insoluble chromatin fractions of apoptotic nuclei from glucocorticoid treated thymocytes. Histones were isolated from soluble and insoluble chromatin obtained from dexamethasone treated thymocytes. Total chromatin was obtained from untreated (control) cells. Figure 4B shows western blots for the acetylation site at K15 (acK15) as well as the phosphorylation of S14 (phosS14). PhosS14 is more abundant in the soluble than the insoluble fraction from 4 hrs dexamethasone treated thymocytes, and is much greater than the amount in the control (Fig. 4B, c). In contrast, acK15 was abundant in the control and insoluble fraction but was reduced in the soluble fraction of dexamethasone treated thymocytes. Therefore, K15 appeared to be highly acetylated in the H2B histones in chromatin from control cells, but partially deacetylated following the induction of apoptosis by dexamethasone. Using synthetic peptide substrates, each specific antibody was evaluated for its ability to recognize its respective epitope (acetylated-lysine K15 or phosphorylated S14). Adjacent acetylation and/or phosphorylation had no significant influence on the ability of the antibodies to recognize their appropriate substrates (see supplemental data S1).
To evaluate whether this reciprocal change occurs between deacetylation of K15 and phosphorylation at S14 during apoptosis of thymocytes in vivo, we injected rats with dexamethasone and after 18 h the thymus was dissected, sectioned and examined by immunohistochemistry. The control thymus tissue was strongly stained with anti-acK15 (Fig. 4C, 3), whereas staining was less pronounced in dexamethasone treated tissue (Fig. 4C, 4), indicating H2B histones are deacetylated at K15 following treatment. In contrast, staining for phosS14 was minimal in the tissue from controls (Fig. 4C, 7), but abundant in the thymus of the dexamethasone treated animals. This staining was concentrated in the thymic cortex area that harbors immature T-cells which are particularly sensitive to glucocorticoid induced apoptosis (Fig. 4C, 8). Together these data suggest that the switch between K15 deacetylation and S14 phosphorylation also occurs after glucocorticoid-treatment in vivo.
An important question that arises from our studies is whether this epigenetic switch also occurs during apoptosis in other cell types. To address this issue we used HTC cells as a model system. This cell line undergoes apoptosis upon UV-C irradiation or Fas ligand treatment19 but not with glucocorticoid treatment, despite the presence of an intact glucocorticoid receptor signaling system19. After the induction of apoptosis in HTC cells, some cells remained attached to a glass substratum (At), and approximately 30 % of the cells are detached and are floating in the medium (Dt). We observed an increase in the phosphorylation of S14 in the detached UV-C or FasL treated apoptotic cells (Fig. 4D, Dt) compared to the attached cells (Fig. 4D, At). In contrast K15 acetylation was reduced upon induction of apoptosis and release from the substratum which is a reflection of their state of apoptosis. Similar results were also observed when studying apoptosis of HL-60, a human promyelocytic leukemic cell line, induced by etoposide (data not shown). Importantly, HTC cells treated with glucocorticoids (Fig. 4D, Dex) show no significant difference compared to control cells in their levels of K15 acetylation and S14 phosphorylation. The fact that we detected no increase in phosS14 in dexamethasone treated HTC cells is strong evidence that this epigenetic event is specifically related to apoptosis and not simply a result of glucocorticoid administration, since these cells are known to be glucocorticoid responsive19. Together these data summarized in Table 1 indicate that the switch between deacetylation of K15 and phosphorylation of S14 in H2B occurs during apoptosis of primary cells as well as established cell lines. Furthermore, this switch appears to be independent of the type of apoptotic stimulus used to initiate the cell death process.
TABLE 1. Summary of Results from Fig. 4 (B, C and D) and HL-60 cells.
Summary of results from Fig. 4 (B, C and D). Histones were prepared from nuclei or chromatin fractions from cells treated with apoptosis inducing agents as described in the Materials and Methods and analyzed by western blot against either anti-phosS14 or anti-acK15. Con; Control, Dex; dexamethasone, W; whole chromatin or nuclei, Sol and Ins; soluble and insoluble chromatin respectively from thymocyte nuclei. Dt and At; nuclei from detached and attached cells respectively isolated from the FasL treated HTC cells. The intensity of the signal of the western blot was represented as −; negligible, +; positive, and ++; intensive.
| Cells | Agents | Fractions | phosS14 | acK15 |
|---|---|---|---|---|
| Thymocytes | Con | − | + | |
| Dex | W | + | − | |
| Dex | Sol | ++ | − | |
| Dex | Ins | − | ++ | |
| HTC cells | Con | − | + | |
| Dex | − | + | ||
| UV-C | + | − | ||
| FasL | W | + | − | |
| FasL | Dt | ++ | − | |
| FasL | At | − | ++ | |
| HL-60 | Con | − | + | |
| VP-16 | + | − |
Effect of TSA on the S14 phosphorylation in thymocytes
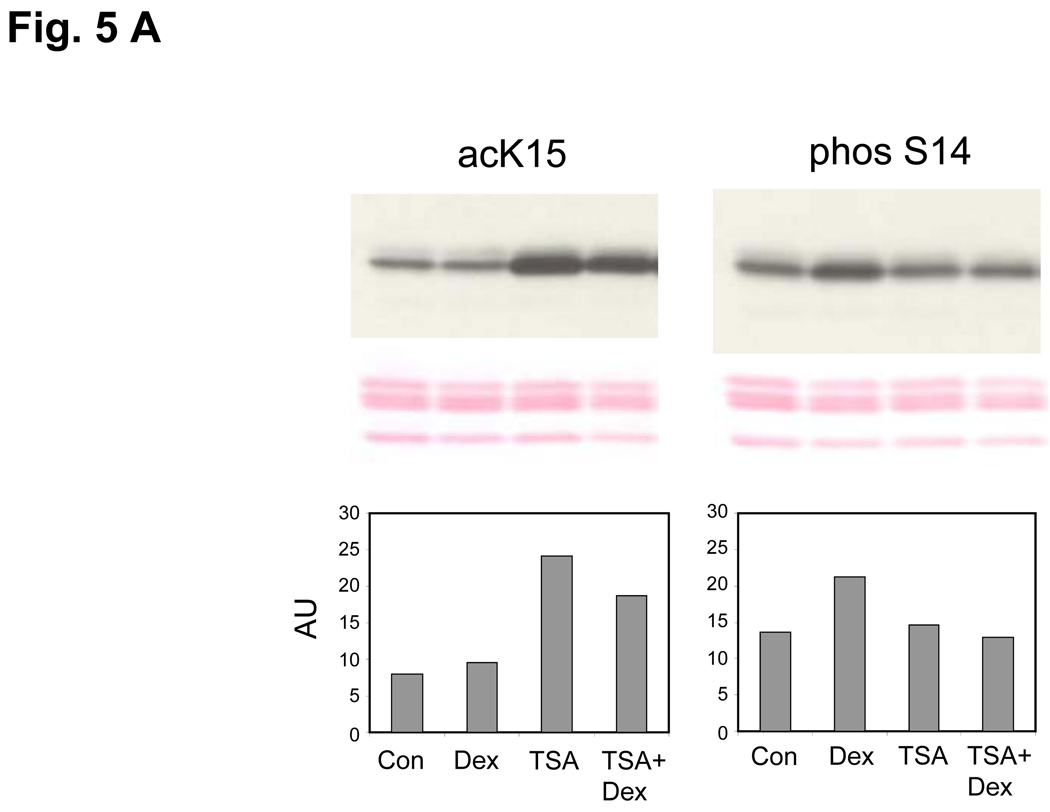
In an attempt to understand components of the mechanisms involved in glucocorticoid regulation of epigenetic chromosomal remodeling, we examined the effect of TSA, an inhibitor of histone deacetylases, on the acK15 and phosS14 in thymocytes (Fig. 5A). In thymocytes treated with TSA alone, acK15 was markedly increased (Fig. 5A, left). In addition, the amount of acK15 remained high in cells treated with TSA plus dexamethsone, whereas phosS14 increased in the dexamethasone treated thymocytes but remained at control levels in cells treated with dexamethasone plus TSA (Fig. 5A, right). Our data suggest that a histone deacetylase is either activated or induced in glucocorticoid treated thymocytes and consequently acK15 is deacetylated. Thus deacetylation of K15 allows the phosphorylation of S14, further suggesting that they have a reciprocal relationship.
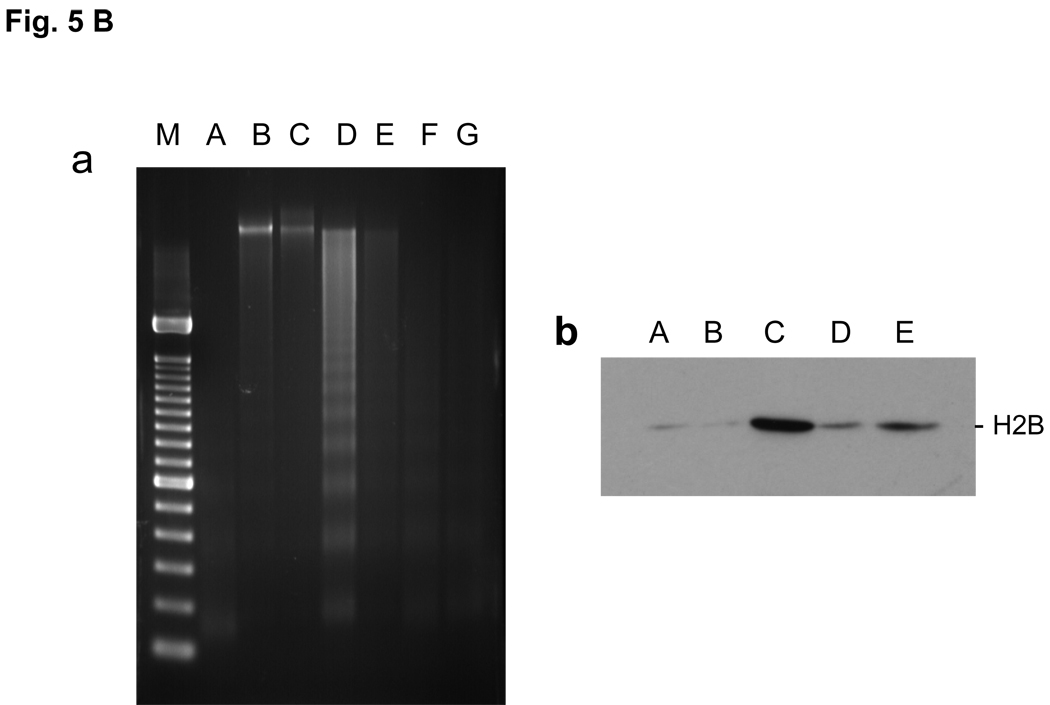
FIG. 5. Effect of TSA on histone K15 acetylation, S14 phosphorylation and DNA degradation.
(A) Enhancement of K15 and suppression of S14 phosphorylation by a HDAC inhibitor (TSA). Dex, Thymocytes were treated with 1 µM dexamethasone; TSA, 1 µM TSA for 3 h, TSA+Dex, pretreated with 1 µM TSA for 1 h and treated with 1 µM Dexamethasone for 3 h. Histones were extracted and western blotted against acK15 (left) and phosS14 (right). Histones obtained from whole chromatin show a smaller decrease of acK15 in the dexamethasone treated thymocytes as compared to Fig. 4B, which was obtained from soluble chromatin from apoptotic cells. The data shown represents three experiments.
(B) TSA prevents Dex induced DNA degradation and histone release in soluble chromatin
a. Prevention of DNA degradation. Thymocytes were incubated at 37°C for 4 hrs. Nuclei were isolated. The nuclei were treated with lysis buffer and the soluble chromatin was treated with RNase and proteinase K and run on 1.8% agarose gel electrophoresis. M, DNA size marker same as shown in Fig. 1C, A, Fresh thymocytes. B, Control thymocytes incubated without chemicals. C, thymocytes with 10 nM Dex. D, 100 nM Dex. E, 1 µM TSA. F, TSA with 10 nM Dex. G, TSA with 100 nM Dex.
b. Prevention of histone release. The soluble chromatin was obtained from thymocytes as described above (a). The soluble chromatin was run on SDS-PAGE gel then analyzed by western blot against anti-H2B. A Fresh thymocytes, B, Control thymocytes incubated without chemicals. C, 100 nM Dex, D, 1 µM TSA, E, TSA with 100 nM Dex.
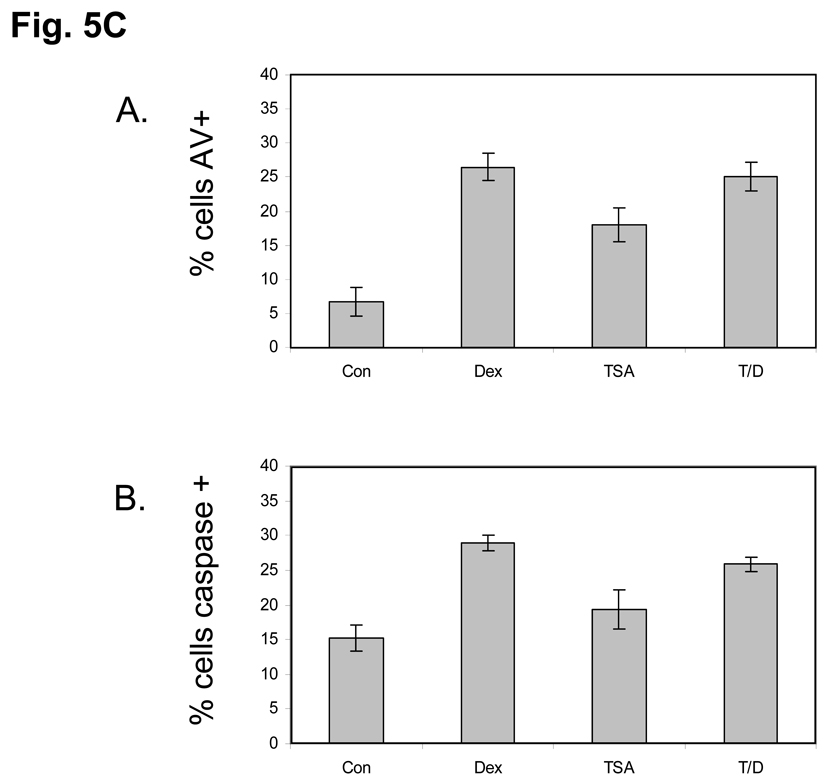
(C) TSA does not prevent apoptosis in Dex treated Thymocytes. Isolated rat thymocytes were treated with Dex 1µM, TSA 1 µM, TSA+Dex or left untreated for 6 hours. Annexin-V binding and caspase 3-like activity was analyzed by flow cytometry. Dead cells were not used in the analysis. A. Dex, TSA alone and TSA plus Dex treatment all caused an increase in the percentage of Annexin-positive cells. B. Dex, TSA alone and TSA plus Dex treatment all caused an increase in the percentage of cells with caspase 3-like activity.
We also examined the DNA degradation and histone release in the soluble chromatin fraction obtained from isolated nuclei (Figure 5B). DNA from cells treated with dexamethasone displays a ladder pattern indicating internucleosomal degradation (lane C in the Fig. 5B left), which is not seen in the two controls (lanes A and B). However, the cells treated with either TSA alone (lane D) or dexamethasone plus TSA showed almost no internucleosomal degradation (lanes E and F). The histone release assay also showed that TSA treatment alone had very little effect on the release of histones (lanes D and E in the Fig. 5B, right). This lack of internucleosomal degradation under these conditions was not the result of arresting the apoptotic program since cells treated with TSA alone, Dex alone or TSA plus Dex for 6 hours still underwent apoptosis as judged by annexin-V binding and caspase activation (Fig. 5C). Together these data indicate that deacetylation of histones is crucial to internucleosomal cleavage of DNA during apoptosis.
Phosphorylation assay of synthetic H2B peptides
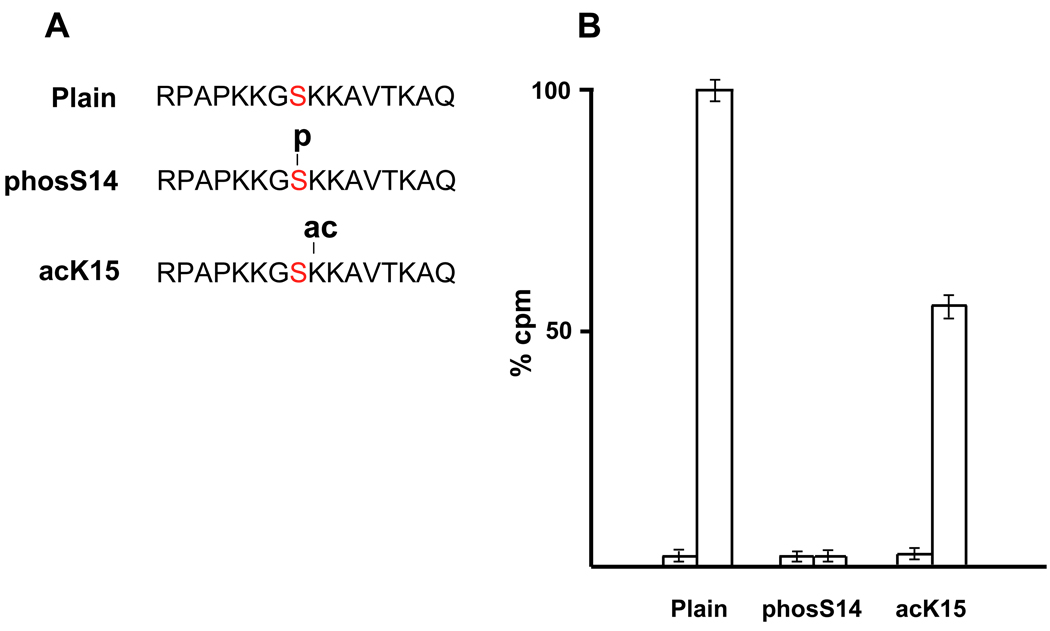
To gain further insight into the kinases and mechanisms involved in S14 phosphorylation we examined the effect of acetylation of K15 on the ability of S14 to be phosphorylated using synthetic H2B peptides in vitro. Figure 6A shows the 16 amino acid peptide substrates of H2B from the arginine 7 (R7) to glutamine 22 (Q22). One peptide, acK15, is acetylated at K15, and one, phosS14, is phosphorylated at S14. The control peptide has no modification. These three peptides were tested for their ability to be phosphorylated by PKC, which is a protein kinase known to be responsible for the phosphorylation of the N-terminal H2B6,9. Figure 6B indicates the incorporation of 32P into the 3 different synthetic H2B peptides by the PKC phosphorylation assay. The incorporation of 32P into phosS14 peptide was negligible (1.7 %), since the S14 site was already phosphorylated. The phosphorylation was suppressed nearly 35% in the acK15 peptide. These data suggest that acetylation at K15 can inhibit the phosphorylation of S14 in vitro.
FIG. 6. Phosphorylation assay using synthetic H2B peptides.
(A) Three kinds of H2B peptides were produced (Plain, phosS14 and acK15).
(B) The PKC induced incorporation of 32P (cpm) into the modified peptides is shown as percent of unmodified peptides (100%). The left columns for each peptide indicate the control value without PKC. Right columns: incorporation of 32P with active PKC used in the assay system.
DISCUSSION
Our results indicate that the phosphorylation of S14 in histone H2B occurs during apoptosis of thymocytes and is abundant in the soluble or degraded chromatin fraction of apoptotic cells. In contrast, very little S14 H2B phosphorylation is found in the insoluble chromatin fraction of apoptotic or normal control cells. We also show that K15 is acetylated in normal cells but this modification is greatly diminished during the activation of apoptosis. In addition we present evidence that K15 deacetylation is necessary to allow S14 H2B phosphorylation and inhibiting this deacetylation step inhibits internucleosomal DNA degradation.
There are two major steps of DNA degradation during apoptosis. One is the production of high molecular weight (HMW) DNA fragments and the other is internucleosomal DNA degradation. In the early stage of apoptosis, HMW DNA fragments are produced and this is necessary for the subsequent internucleosomal DNA degradation. Our findings indicate that deacetylation of K15 and S14 phosphorylation associates with internucleosomal DNA degradation. Other histone modifications are known to occur during apoptosis such as H4 acetylation20, H4 trimethylation21, H3 methylation and phosphorylation22,23 and H1 dephosphorylation24. Together with H2B phosphorylation, these histone modifications may occur in the same chromatin domain and may work cooperatively.
Our data also reveal that K15 is acetylated in normal rat thymocytes but during the activation of apoptosis this acetylation is decreased as the phosphorylation of S14 is increased. It has been reported that the N-terminal tail of H2B itself is crucial for chromatin condensation25. Moreover, the H2B acetylation at the N-terminal end is important to maintain a stable structure of H2A–H2B dimers in the nucleosome26,27. Since the N-terminal domain of H2B including the S14 site exists outside of the nucleosome structure28, it could be susceptible to modification by phosphatases or kinases if left unprotected. Deacetylation of K15 seems to be necessary for the phosphorylation of S14, whereas acetylation of K15 may prevent the S14 phosphorylation. These observations suggest that acetylation of K15 in normal cells could be functionally important in protecting cells from irreversible apoptotic DNA degradation. In this regard we previously demonstrated in HL-60 cells that one of the kinases responsible for the S14 phosphorylation is Mst-1, although PKC-delta has also been reported to phosphorylate this site in immune cells8. Mst-1 is activated by caspase-38 and can then translocate from cytoplasm to nucleus29,30.
The current study demonstrates an interesting epigenetic switch between two amino-acid modifications in the N-terminus of H2B that occurs during apoptosis in mammalian cells. A similar relationship has been reported for the phosphorylation at Ser10 interacting with the adjacent K9 methylation sites in H331. Intriguingly, we have shown that the H2B phosphorylation could be a prerequisite for apoptosis execution in various mammalian cells resulting from several apoptotic agents6,9. Recently it has been shown that H2B phosphorylation occurs during oxidative stress in yeast at Ser10 which is homologous to S14 in vertebrates13. These authors suggest a specific deacetylation at K11 and phosphorylation at S10 at the histone H2B tail required for cell death in yeast14.
There are numerous reports that general histone deacetylase inhibition causes apoptosis32. Indeed in our studies using the non-specific histone deacetylase inhibitor TSA we show that it alone can cause cell death. However we also demonstrate that TSA prevented internucleosomal DNA degradation induced by glucocorticoids as well as hyperacetylating K15H2B and inhibiting S14H2B phosphorylation. Thus during apoptosis K15 is likely deacetylated by endogenous HDAC’s in a more specific manner.
Previously, we have shown that CAD (−/−) DT40 chicken cells treated with VP16 had a pSer14 positive reaction which could be detected by immunocytochemistry9. Within cell nuclei, the pSer14 reaction was co-localized with DAPI dense chromatin suggesting that pSer14 is associated with chromatin condensation rather than caspase dependent internucleosomal DNA fragmentation. In the current studies using primary thymocytes from adrenalectomized rats we observed that phosS14 H2B during thymocyte apoptosis seemed mainly localized at the periphery of nuclei which does not yet show clear condensation (Fig. 1B). This difference in the structure of chromatin and the localization of phosS14 may be a reflection of cell type or the apoptotic agent used for the induction of cell death.
The epigenetic switch between the two histone modifications in H2B could induce a change of nucleosome structure in the H2B tail at specific regions of chromatin, and the remodeling of that chromatin domain would facilitate the accessibility for endonucleases. These findings are important for the structural analysis of nucleosomes during apoptosis, especially the interaction between covalent histone modification and internucleosomal DNA degradation.
MATERIALS AND METHODS
Isolation of thymocytes and induction of apoptosis
Male Sprague Dawley rats (2–3 month of age) were used and the animals were bilaterally adrenalectomized by the provider at least 5 days before use. They were maintained under controlled conditions of temperature (25°C) and lighting, and allowed free access to food and 0.85% saline. All experimental protocols were approved by the animal review committee at NIEHS/NIH and were performed in accordance with the guidelines set forth in the NIH Guide for the Care and Use of Laboratory Animals published by the USPHS. Animals were sacrificed by decapitation, and the thymus was surgically removed. The thymus was minced with scissors in RPMI 1460 medium containing 10% heat-inactivated fetal calf serum, 2 mM glutamine, 75 U/ml streptomycin, and 100 U/ml penicillin at room temperature. The minced tissues were placed in a tube and shaken by hand until free thymocytes were released.
For the isolation of nuclei, the thymocytes were washed once in isolation buffer (20 mM Hepes pH 7.5, 10 mM KCl, 3 mM MgCl2, 0.25 M sucrose, 1 mM DTT and 1 mM PMSF). The cells were re-suspended in isolation buffer, and treated with 0.25 % NP-40 in buffer for 10 min at 4°C. The buffer containing 1.2 M sucrose was underlayed as a cushion and centrifuged. The nuclear pellet was washed once in isolation buffer.
Thymocytes, 2 × 106 cells/ml, were incubated in RPMI 1640 medium at 37°C, 5% CO2 atmosphere with 1 µM dexamethasone for various time points depending on experiments. The cells were harvested at room temperature and were washed twice with PBS solution. For the studies using a histone deacetylase inhibitor, cells were pre-treated for 1 hour with 1 µM of TSA33. HTC cells, a rat hepatoma cell line, were cultured in DMEM-H medium containing 5% heat-inactivated fetal calf serum, 2 mM glutamine, 75 U/ml streptomycin, and 100 U/ml penicillin at 37°C, 5% CO2 atmosphere19. For the induction of apoptosis in this cell line, the HTC cells were irradiated with 90 mJ/cm2 of UV-C and assayed 24 hrs later or treated with 50 ng/ml of Fas ligand (Kamiya Biomedical) for 18 hrs. HL-60, a human promyelocytic leukemia cell line, were cultured in RPMI 1640 medium at 37°C, 5% CO2 atmosphere. For the induction of apoptosis of HL-60 cells, the cells were treated with 20µg/ml of VP-16 (etoposide) for 16 hrs6.
Flow cytometric analysis of apoptotic thymocytes
The thymocytes (1 × 106) treated with dexamethasone as described above were analyzed by flow cytometry using a Becton Dickinson FACSort. A change in cell size was determined by measuring the cells ability to scatter light in the forward direction. An increase or decrease in forward light scatter indicates an increase or decrease in cell size, respectively. Cell density or granularity is determined by side scattered light. Data shown are contour plots of forward-scattered light versus side-scattered light. Propidium iodide (10 mg/ml) was added as a vital dye to gate out cells which had lost membrane integrity.
Annexin-V and Caspatag assay: Thymocytes were isolated as described and treated with either dexamethasone 1 µM, TSA 1 µM, or TSA plus Dex (TSA pretreatment), or left untreated for 6 hours. The cells were then stained with Annexin-V and propidium iodide using manufacturers protocol (Trevigen, Gaithersburg, MD). Briefly, 1 × 106 cells are incubated with fluorescently tagged annexin-V and then analyzed by flow cytometry using a FACSort (Becton-Dickinson, CA). For determining caspase activity cells were incubated with a fluorescent caspase 3 substrate following the manufacturer’s protocol (Caspatag, Millipore, Billerica, MA) and then analyzed by flow cytometry. Propidium iodide (10 mg/ml) was added as a vital dye to gate out cells which had lost membrane integrity.
Electron microscopy
For electron microscopy, thymocytes treated with 1 µM dexamethasone in vitro for 4 hrs were harvested and gently centrifuged and washed twice with PBS. The pellets were fixed in a modified Karnovsky buffer (2.0% paraformaldehyde/ 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer, pH 7.4). The materials were post-fixed with 2% OsO4 for 1h. The cells were passed through a series of alcohol washes for dehydration and then embedded in Epon. Ultra thin sections were mounted and stained with 5% uranyl acetate followed by Reynold’s lead citrate and observed on a JEOL 100S transmission electron microscope.
Isolation of soluble chromatin and DNA analysis
For the preparation of soluble chromatin from apoptotic cells, nuclei isolated from apoptotic cells were incubated with a lysis buffer (10 mM Tris-HCl, pH.7.4, 10 mM EDTA and 0.5% Triton X-100) for 10 min at 4°C. The nuclei were centrifuged and the supernatant collected as soluble chromatin (approximately 5% of total chromatin from apoptotic cells) and the pellet as insoluble chromatin. For the analysis of DNA degradation, cells were isolated and harvested after the incubation at 37°C with 1 µM dexamethasone and were treated with 1 mg/ml of DNase-free RNase for 1 hour at 37°C and then 400 µg/ml proteinase K was added with overnight incubation at 37°C. DNA (1 or 2 µg) was run on 1.8% agarose gels at 100 volts with a size marker of DNA (100 base pairs of DNA ladder, Ready-Load™, Invitrogen, Carlsbad, CA). The DNA was stained with 0.25 µg/ml of ethidium bromide and photographed with UV-transilluminator.
Western blotting, Immuno-fluorescent analysis and Tunel assay
Histones were prepared as described previously34 and the proteins (5 µg) were analyzed. For western blot analysis, antibodies of site specific histone H2B modifications were purchased from Upstate (now Millipore). Methods for immuno-fluorescent analysis were described earlier9. Briefly, dexamethasone treated thymocytes (1 × 104) were fixed with 4% paraformaldehyde for 10 min. After washing with PBS, the cells were plated on cover slips. The cells were dried at 4°C and permeabilized with 0.2% Triton X-100. The cells were equilibrated with 2% goat serum in PBS and were incubated with anti-phosS14 antibody at 37°C for 1 h in a moisture chamber. After washing, the cells were further treated with Cy3. The nuclei were counterstained with DAPI. For the detection of cleaved DNA in nuclei, a fluorescent terminal deoxynucleotidyl transferase dUTP end labeling (TUNEL) assay was conducted (Promega). Images were taken by phase contrast microscopy (Olympus, IX70).
Immunohistochemical analysis of histone H2B phosphorylation and acetylation in thymus tissues
Adrenalectomized rats were injected with dexamethasone (1 mg in 0.9% saline per animal) intra-peritoneally. The control animals were injected with a same amount of saline only. After 18 h, the animals were decapitated and the thymus was surgically removed and fixed in 4% paraformaldehyde. Thin sections of the thymus tissue were obtained and placed on a glass and further processed for immuno-histochemistry. The slides were treated with anti-phosS14 or anti-acetyl K15 antibodies and conjugated with an anti-rabbit IgG horseradish peroxidase from donkey as a secondary antibody. For each experiment, sections stained without primary antibody were examined simultaneously as a control.
Phosphorylation of synthetic H2B peptides
Three H2B peptides were synthesized, containing 16 amino acid residues from arginine 7 (R7) to glutamine 22 (Q22) (AnaSpec, Inc.). Two peptides were modified with acetylation at K15 or phosphorylation at S14. One was a non-modified peptide as shown in the Fig. 4A. They were assayed with a PKC phosphorylation assay (Upstate-Millipore) in a reaction mixture (30 µl) containing Ca2+ and Mg2+ ions, lipid activator, PKA/CaM inhibitors, 1 µM TSA, 100µM synthetic peptides, [32P-gamma]ATP and active PKC enzyme and buffer. The mixtures were incubated at 30°C for 10 min. The 32P-labeled peptides were trapped with a p81 filter (Millipore Co), and washed once with acetone. Radioactivity was assessed with a scintillation counter.
Antibody recognition of epitopes when adjacent residues are modified
A double modified peptide with phosphorylation and acetylation at serine 14 and lysine 15 was developed, phosS14/acK15: RPAPKKGS(p)K(ac)KAVTKAQ, and synthetic peptides described above were loaded on a nitrocellulose membrane using Dot Dolt loading apparatus (Biorad). The amount of peptides ranged from 0.08 to 20 µg per 200 µl TBS solution per well. The peptides on the membrane were washed twice with 200 µl of Tween-TBS (TTBS) and were processed similarly to a western blot method. After blocking with 5% milk for 1h, the membrane was treated with two antibodies (phosS14 and acK15) at 1/800 and 1/1,000 respectively. After washing the membrane with TTBS, the membrane was treated with secondary antibody (1/2,000 of HRP) for 40 min. Pictures of a peroxidase reaction in dots were taken by Las 3000 MINI (FUJIFILM). For ninhydrin staining, 0.2 µg of each H2B peptide was spotted on a Whatman 3MM paper and sprayed with 2% Ninhydrin Spray Reagent (SIGMA). The paper was heated in an oven at 120°C for 10 min.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Tiwanda Marsh for the assistance in histochemical analysis. We thank for members of our laboratory for useful discussion. This research was supported by grant number 1Z01ES090057 of the Intramural Research Program of the NIEHS, NIH.
Abbreviations
- HTC
hepatoma cell line
- phosS14
phosphorylation at Serine 14
- acK15
adjacent Lysine 15
- HMW
high molecular weight
- At
attached to a glass substratum
- Dt
detached floating in the medium
REFERENCES
- 1.Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 2.Isenberg I. Histones. Ann. Rev. Biochem. 1979;48:159–191. doi: 10.1146/annurev.bi.48.070179.001111. [DOI] [PubMed] [Google Scholar]
- 3.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 4.Wyllie AH, Morris RG, Smith AL, Dunlop D. Chromatin cleavage in apoptosis: association with condensed chromatin morphology and dependence on macromolecular synthesis. J. Pathol. 1984;142:67–77. doi: 10.1002/path.1711420112. [DOI] [PubMed] [Google Scholar]
- 5.Compton MM. A biochemical hallmark of apoptosis: internucleosomal degradation of the genome. Cancer Metastasis Rev. 1992;11:105–119. doi: 10.1007/BF00048058. [DOI] [PubMed] [Google Scholar]
- 6.Ajiro K. Histone H2B phosphorylation in mammalian apoptotic cells: An association with DNA fragmentation. J. Biol. Chem. 2000;275:439–443. doi: 10.1074/jbc.275.1.439. [DOI] [PubMed] [Google Scholar]
- 7.Ajiro K, Th'ng J, Yau J, Nishi Y. Isolation and characterization of mammalian cells that are undergoing apoptosis by a bovine serum albumin density gradient. Anal. Biochem. 2004;332:226–233. doi: 10.1016/j.ab.2004.05.035. [DOI] [PubMed] [Google Scholar]
- 8.Hu Y, Liu Z, Yang SJ, Ye K. Acinus-provoked protein kinase C delta isoform activation is essential for apoptotic chromatin condensation. Cell Death Differ. 2007;14:2035–2046. doi: 10.1038/sj.cdd.4402214. [DOI] [PubMed] [Google Scholar]
- 9.Cheung WL, Ajiro K, Samejima K, Kloc M, Cheung P, Mizzen CA, et al. Apoptotic phosphorylation of histone H2B is mediated by mammalian sterile twenty kinase. Cell. 2003;113:507–517. doi: 10.1016/s0092-8674(03)00355-6. [DOI] [PubMed] [Google Scholar]
- 10.Mecklenbrauker I, Kalled SL, Leitges M, Mackay F, Tarakhovsky A. Regulation of B-cell survival by BAFF-dependent PKCdelta-mediated nuclear signalling. Nature. 2004;431:456–461. doi: 10.1038/nature02955. [DOI] [PubMed] [Google Scholar]
- 11.Fernandez-Capetillo O, Allis CD, Nussenzweig A. Phosphorylation of histone H2B at DNA double-strand breaks. J. Exp. Med. 2004;199:1671–1677. doi: 10.1084/jem.20032247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thorne AW, Kmiciek D, Mitchelson K, Sautiere P, Crane-Robinson C. Patterns of histone acetylation. Eur. J. Biochem. 1990;193:701–713. doi: 10.1111/j.1432-1033.1990.tb19390.x. [DOI] [PubMed] [Google Scholar]
- 13.Ahn SH, Cheung WL, Hsu JY, Diaz RL, Smith MM, Allis CD. Sterile 20 kinase phosphorylates histone H2B at serine 10 during hydrogen peroxide-induced apoptosis in S. cerevisiae. Cell. 2005;120:25–36. doi: 10.1016/j.cell.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 14.Ahn SH, Diaz RL, Grunstein M, Allis CD. Histone H2B Deacetylation at Lysine 11 Is Required for Yeast Apoptosis Induced by Phosphorylation of H2B at Serine 10. Mol. Cell. 2006;24:211–220. doi: 10.1016/j.molcel.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 15.Wyllie AH. Glucocorticoid-induced thymocyte apoptosis is associated with endogenous endonuclease activation. Nature. 1980;284:555–556. doi: 10.1038/284555a0. [DOI] [PubMed] [Google Scholar]
- 16.Mann CL, Hughes FM, Jr, Cidlowski JA. Delineation of the signaling pathways involved in glucocorticoid-induced and spontaneous apoptosis of rat thymocytes. Endocrinology. 2000;141:528–538. doi: 10.1210/endo.141.2.7314. [DOI] [PubMed] [Google Scholar]
- 17.Ajiro K, Bortner CA, Westmoreland J, Cidlowski JA. An endogenous calcium-dependent, caspase-independent intranuclear degradation pathway in thymocyte nuclei: Antagonism by physiological concentrations of K+ ions. Exp. Cell Res. 2008;314:1237–1249. doi: 10.1016/j.yexcr.2007.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tadakuma T, Kizaki H, Odaka C, Kubota R, Ishimura Y, Yagita, et al. CD4+ CD8+ thymocytes are susceptible to DNA fragmentation induced by phorbol ester, calcium ionophore and anti-CD3 antibody. Eur. J. Immunol. 1990;20:779–784. doi: 10.1002/eji.1830200411. [DOI] [PubMed] [Google Scholar]
- 19.Scoltock AB, Heimlich G, Cidlowski JA. Glucocorticoids inhibit the apoptotic actions of UV-C but not Fas ligand in hepatoma cells: direct evidence for a critical role of Bcl-x(L) Cell Death Differ. 2007;14:840–850. doi: 10.1038/sj.cdd.4402071. [DOI] [PubMed] [Google Scholar]
- 20.Allera C, Lazzarini G, Patrone E, Alberti I, Barboro P, Sanna P, et al. The condensation of chromatin in apoptotic thymocytes shows a specific structural change. J. Biol. Chem. 1997;272:10817–10822. doi: 10.1074/jbc.272.16.10817. [DOI] [PubMed] [Google Scholar]
- 21.Boix-Chornet M, Fraga MF, Villar-Garea A, Caballero R, Espada J, Nunez, et al. Release of hypoacetylated and trimethylated histone H4 is an epigenetic marker of early apoptosis. J. Biol. Chem. 2006;281:13540–13547. doi: 10.1074/jbc.M601136200. [DOI] [PubMed] [Google Scholar]
- 22.Hurd PJ, Bannister AJ, Halls K, Dawson MA, Vermeulen M, Olsen JV, Ismail H, Somers J, Mann M, Owen-Hughes T, Gout I, Kouzarides T. Phosphorylation of histone H3 Thr-45 is linked to apoptosis. J Biol Chem. 2009;284:16575–16583. doi: 10.1074/jbc.M109.005421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng MF, Lee CH, Hsia KT, Huang GS, Lee HS. Methylation of histone H3 lysine 27 associated with apoptosis in osteosarcoma cells induced by staurosporine. Histol Histopathol. 2009;24:1105–1111. doi: 10.14670/HH-24.1105. [DOI] [PubMed] [Google Scholar]
- 24.Kratzmeier M, Albig W, Hanecke K, Doenecke D. Rapid dephosphorylation of H1 histones after apoptosis induction. J. Biol. Chem. 2000;275:30478–30486. doi: 10.1074/jbc.M003956200. [DOI] [PubMed] [Google Scholar]
- 25.de la Barre AE, Angelov D, Molla A, Dimitrov S. The N-terminus of histone H2B, but not that of histone H3 or its phosphorylation, is essential for chromosome condensation. EMBO J. 2001;20:6383–6393. doi: 10.1093/emboj/20.22.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brower-Toland B, Wacker DA, Fulbright RM, Lis JT, Kraus WL, Wang MD. Specific contributions of histone tails and their acetylation to the mechanical stability of nucleosomes. J. Mol. Biol. 2005;346:135–146. doi: 10.1016/j.jmb.2004.11.056. [DOI] [PubMed] [Google Scholar]
- 27.Wunsch A, Jackson V. Histone release during transcription: acetylation stabilizes the interaction of the H2A-H2B dimer with the H3-H4 tetramer in nucleosomes that are on highly positively coiled DNA. Biochemistry. 2005;44:16351–16364. doi: 10.1021/bi050876o. [DOI] [PubMed] [Google Scholar]
- 28.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 29.Ura S, Masuyama N, Graves JD, Gotoh Y. Caspase cleavage of MST1 promotes nuclear translocation and chromatin condensation. Proc. Natl. Acad. Sci. USA. 2001;98:10148–10153. doi: 10.1073/pnas.181161698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Graves JD, Draves KE, Gotoh Y, Krebs EG, Clark EA. Both phosphorylation and caspase-mediated cleavage contribute to regulation of the Ste20-like protein kinase Mst1 during CD95/Fas-induced apoptosis. J. Biol. Chem. 2001;276:14909–14915. doi: 10.1074/jbc.M010905200. [DOI] [PubMed] [Google Scholar]
- 31.Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 32.Carew JS, Giles FJ, Nawrocki ST. Histone deacetylase inhibitors: Mechanisms of cell death and promise in combination cancer therapy. Cancer Lett. 2008;269:7–17. doi: 10.1016/j.canlet.2008.03.037. [DOI] [PubMed] [Google Scholar]
- 33.Yoshida M, Horinouchi S. Trichostatin and leptomycin. Inhibition of histone deacetylation and signal-dependent nuclear export. Ann. N. Y. Acad. Sci. 1999;886:23–36. doi: 10.1111/j.1749-6632.1999.tb09397.x. [DOI] [PubMed] [Google Scholar]
- 34.Ajiro K, Borun TW, Shulman SD, McFadden GM, Cohen LH. Comparison of the structures of human histone 1A and 1B and their intramolecular phosphorylation sites during the HeLa S-3 cell cycle. Biochemistry. 1981;17:1454–1464. doi: 10.1021/bi00509a008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.