Abstract
Precisely directed cleavage and polyadenylation of mRNA is a fundamental part of eukaryotic gene expression. Yet, 3′ end heterogeneity has been documented for thousands of mammalian genes, and usage of one cleavage and polyadenylation signal over another has been shown to impact gene expression in many cases. Building upon the rich biochemical and genetic understanding of the 3′ end formation, recent genomic studies have begun to suggest that widespread changes in mRNA cleavage and polyadenylation may be a part of large, dynamic gene regulatory programs. In this review, we begin with a modest overview of the studies that defined the mechanisms of mammalian 3′ end formation, and then discuss how recent genomic studies intersect with these more traditional approaches, showing that both will be crucial for expanding our understanding of this facet of gene regulation.
Introduction
The 3′ end cleavage and polyadenylation event is critical for transcriptional termination, export of mRNA from the nucleus, and significantly impacts the stability and translation of a given mRNA in the cytoplasm [1]. Yet, this event carries more importance than merely being the way in which a mRNA transcript is “finished.” It has long been known that differential usage of cleavage and polyadenylation signals (PASs) associated with a gene can alter the coding sequence of the protein encoded by that gene [2–4] or dictate how much protein is made from a transcript [5]. The observation that in some systems, the 3′ untranslated region (UTR) is able to confer similar temporo-spatial expression on a reporter gene in roughly 80% of cases tested [6] particularly underscores the notion that differential PAS usage has the potential to significantly impact gene function even if a particular switch does not alter the protein coding sequence of the transcript produced. The contribution of mutations disrupting cleavage and polyadenylation to human disease has been known for some time [7–9] and that other mutations in these and other elements within 3′ UTR may impact human disease is beginning to be appreciated [10–12].
Over the last three decades, several approaches have been utilized to better understand constitutive and differential transcript 3′ end formation for multiple classes of genes in several organisms. This review will focus on the 3′ end formation of non-histone protein coding genes in mammalian cells. We will first provide a modest overview of the maturation of this field prior to the point at which genomic studies rooted in computational analysis began to appear in the literature. We will next survey the relevant genomic studies of the last decade, comparing and contrasting the lessons learned from this work with more traditional biochemical and genetic studies that appeared during the same period. Finally, we will make an attempt to summarize our current understanding of the role of alternative cleavage and polyadenylation in mammalian cells, and provide one vision of what the future may hold for our understanding of this facet of gene regulation.
The Foundation
The fundamental difference between mRNA 3′ end formation in bacteria and mammalian cells was first described in 1978. While the 3′ end of bacterial mRNA transcripts was known to correspond precisely to the site of transcriptional termination seminal work by Nevins and Darnell demonstrated that transcription proceeded well downstream of the 3′ end of the mature Ad2 transcript, and that this mature 3′ end was generated by an endonucleolytic cleavage event [13].
There was already a clue as to what the signal for this endonucleolytic cleavage event might be. Sequence analysis of six distinct cDNAs had identified a consensus hexamer (AAUAAA) located approximately 20 nucleotides 5′ of the site of poly(A) addition [14]. A second putative element had also been noticed. Researchers comparing the DNA sequences of several recently cloned genes noticed a tendency for TTTT and TTGT sequences to appear downstream of the mRNA cleavage and polyadenylation site [15]. The importance of these elements for proper 3′ end processing was established in deletion and point mutation studies utilizing viral and endogenous PASs [16–23]. Deletions of either element disrupted both cleavage and polyadenylation. As might have been expected individual point mutations within the more highly conserved AAUAAA hexamer impacted these processes much more strongly than point mutations engineered in the downstream U/GU-rich sequence.
These studies complemented the discovery that mutations in the core AAUAAA hexamer were associated with human alpha- and beta-thallasemias. Study of the affected locus in cells from patients with these diseases revealed decreased levels of mature mRNA from the mutated genes and the presence of abnormally long “readthrough” transcripts derived from these loci [7,8]. This and subsequent work demonstrated that cleavage and polyadenylation were required for proper Pol II transcriptional termination in mammalian cells [24], with the most dramatic demonstrations being provided in studies substituting or deleting PASs on circular viral genomes [25,26].
The identification of core sequence elements at the site of the cleavage and polyadenylation reactions and the observation that mutation of these sequences disrupted this reaction suggested that these elements functioned as binding sites for the enzymes controlling these reactions. Preliminary studies in nuclear extracts first established that the reactions were separable [27,28], and then demonstrated that they were dependent upon the sequence elements that had previously been defined [19,23,29–32]. These early mechanistic studies laid the foundation for the biochemical purification of the five factors that together formed the core polyadenylation machinery. A four-protein complex termed Cleavage and Polyadenylation Stimulation Factor (CPSF) was found to interact directly with the core AAUAAA hexamer through its 160 kDa subunit [33–35]. The same 160 kDa subunit of CPSF also bound directly to two other factors involved in the cleavage and polyadenylation reaction - the poly(A) polymerase (PAP) [35–37] and the 77 kDa subunit of the Cleavage Stimulation Factor (Cstf). A second 64 kDa component of Cstf bound the GU-rich region downstream from the cleavage and polyadenylation site [35,38,39]. The final two factors required for cleavage, Cleavage Factors I and II, (CF-I(m) and CF-II(m)) were definitively characterized later on [40–44]. That deficiencies in PAP or either of two individual components of the Cstf complex had been demonstrated to be cell lethal in yeast, Drosophila, and vertebrate cells [45–47] highlighted the fundamental and universal importance of the core polyadenylation complex in gene expression. However, differential usage of multiple PASs within a single transcription unit had long been observed in the context of several viral and mammalian genes. While use of one PAS over another could often be ascribed to the relative “strength” of the core elements of a given PAS, it was clear that auxiliary sequences and factors must play some role in influencing PAS choice in various contexts. Two laboratories studying this phenomenon in viral systems provided the first description of how this might occur.
In adenovirus, mutation of sequences upstream of the core elements of the promoter-proximal L1 PAS within the major late transcription unit reduced cleavage and polyadenylation at this site [48]. A similar observation was reported in the SV40 system, where the efficiency of the SV40 late PAS was dramatically reduced by deletion of a sequence upstream of the core hexameric element [49]. Such elements were termed upstream elements (USEs). Further work demonstrated that the USE of the SV40 late PAS was bound by the U1A protein of the U1 snRP [50], and that a direct interaction between U1A and the 160 kDa subunit of CPSF increased the efficiency of cleavage and polyadenylation at this site [51]. Interestingly, U1A had also been shown to inhibit polyadenylation of its own mRNA by binding to a conserved site within the 3′ UTR [52,53]. Thus, this protein was documented to have both stimulatory and repressive effects on 3′ end formation of distinct PASs.
The implication of the U1 snRP as a modulator of cleavage and polyadenylation was reminiscent of earlier mechanistic studies [27] and dovetailed with additional studies that were demonstrating a linkage between splicing and 3′ end formation. The presence of an upstream intron was known to facilitate cleavage and polyadenylation both in vitro and in vivo [54,55], and insertion of a 5′ splice acceptor between the 3′ splice acceptor of the terminal intron and the PAS markedly decreased these reactions [56]. Further studies reinforced the involvement of U1 in 3′ end formation, and found that additional known splicing factors such as SFRS1, SRp20, PTB, and other hnRNPs could also modulate PAS selection through either by binding to additional non-core sequence elements or competing for known core elements [57–59]. This evidence for the coupling between the splicing and 3′ end formation machineries was complemented by demonstration of core cleavage and polyadenylation factors in transcription initiation complexes, and that both the cap-binding complex and the C-terminal domain of RNA Polymerase II modulated cleavage and polyadenylation both in vitro and in vivo [29,60–63].
Thus, the second decade of molecular research into 3′ end formation ended with the core cis-elements and trans-factors involved in mammalian cleavage and polyadenylation having been unambiguously identified, and the intimate inter-connection of these processes with transcription and other RNA processing events firmly established [64,65]. Over a hundred examples of alternative PAS usage in mammals were known to occur in a tissue-specific manner or in response to extracellular stimuli [66]. The impact of the 3′ UTR on post-transcriptional control of gene expression was already a major theme in developmental biology [67,68], so the demonstrated and potential impacts of alternative 3′ end formation on gene expression, even in the absence of changes in protein coding sequence, was broadly recognized. It was at this time that larger-scale computational analyses, taking advantage of the accumulation of information in mRNA and EST databases, dramatically expanded the documentation of alternative cleavage and polyadenylation in the mammalian transcriptome.
Large-scale identification of 3′ ends
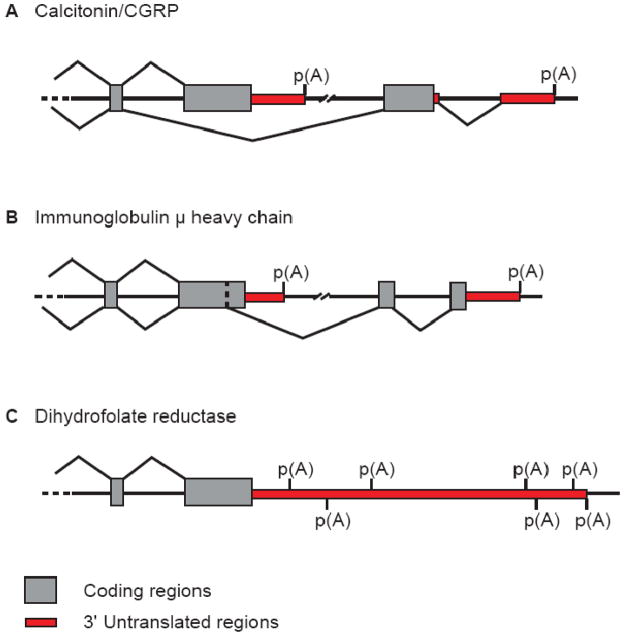
A richer appreciation of the heterogeneity of mRNA 3′ end formation came from large-scale analyses of the sequence data that had been accumulating in public databases. These analyses, no doubt motivated by published surveys of the experimental literature [66], clustered EST sequences and mapped the 3′ end of these sequences to first establish the prevalence of alternative cleavage and polyadenylation. A first attempt, clustering 164,000 ESTs, identified several novel 3′ ends, and documented alternative cleavage and polyadenylation for 19% of the genes studied, often in a tissue-specific manner [69]. As time progressed, increases in the amount of publicly available sequence facilitated the discovery of two or more mappable cleavage and polyadenylation sites in thousands of genes. More recent studies indicate that over half of human genes have multiple mRNA 3′ ends [70,71]. These genes can be assigned into one of three classes (Figure 1). Skipped-exon associated 3′ UTRs utilize one of two mutually exclusive terminal exons, often altering the C-terminus of the encoded protein. Genes with composite exons are defined by competing splicing and polyadenylation events within a single exon, such that either event results in mRNA containing a part of that exon’s sequence. Finally, genes with tandem UTRs utilize multiple PASs in the terminal exon, with no direct influence on protein coding sequence. Tandem UTRs are the most abundant type of alternative cleavage and polyadenylation, accounting for more than half of all events [71].
Figure 1. Types of alternative polyadenylation.
There are three common types of alternative polyadenylation: (A) skipped-exon associated 3′ UTRs, as exemplified by the Calcitonin/CGRP gene locus, (B) composite exons associated 3′ UTRs, similar to the Immunoglobulin μ heavy chain, and (C) single terminal exons with two or more polyA sites (often called tandem UTRs), as illustrated by Dihydrofolate reductase and its seven mapped polyA sites.
Large-scale computational analyses of 3′ ends underscored the importance of the known consensus AAUAAA in mammalian PASs [72,73]. But at the same time, these studies revealed ten variant hexamers associated with 30% of mapped 3′ ends, fully three times more than had been previously thought [20]. Consistent with the alternative PASs that had been described in the literature, variations in the canonical AAUAAA were limited to a single base in each variant [72,73,1]. With this established, the focus turned to identifying why one PAS might be preferentially used over another.
The results of the analyses were remarkably consistent with the paradigms that had been set mechanistically. PASs containing the consensus core AAUAAA were used more frequently than the other variants [73]. Examination of sequences surrounding mapped cleavage and polyadenylation sites revealed an enrichment of U- and G-rich elements both upstream and downstream of these sites [74]. The most frequently used cleavage and polyadenylation sites showed the strongest enrichment of both downstream U-rich sequences as well as specific GU-rich hexamers [75], reinforcing the importance of recognition of the element by the Cstf complex in 3′ end formation. Given a clear correlation between the conformity of a PAS to established models of “strength” and the apparent usage of that PAS, it seemed that the existence of multiple cognate CPSF-like complexes was unlikely [73].
Nonetheless, it was obvious that alternative or tissue-specific cleavage and polyadenylation events were far from rare. Interestingly, these alternative events were often associated with non-canonical PASs, and these PASs were located proximal to the promoter as compared to more consitutively used PASs that most closely resembled the established consensus [73]. This suggested that usage of alternative PASs is likely dependent upon additional factors for efficient utilization, and prompted further investigation aimed at better understanding how these variant sites were used.
In-depth analyses of EST libraries revealed over a thousand human mRNAs characterized by tissue- or disease-biased alternative PAS usage [76], and stage-specific differences in the differentiation of individual cell lineages [77]. Broad generalizations about global patterns of PAS usage could be made – for example, ESTs from the brain were more likely to be derived from distal PASs (and therefore had longer 3′ UTRs) as compared to other tissues [78]. Thus, even though analyses of EST libraries were handicapped by non-standardized annotations, shallow depth of coverage, and uneven coverage across samples these studies were able to reinforce the idea PAS usage was actively regulated, and differed according to cell type, developmental stage, or disease state.
However, one important aspect of alternative PAS usage remained to be addressed: were multiple PAS signals in a transcription unit conserved among species? Comparing ESTs derived from orthologous human and mouse genes revealed a significant similarity in the number of 3′ ends mapped for each gene [70,79], suggesting that alternative cleavage and polyadenylation events are actively selected for through mammalian evolution. Interestingly however, the majority of tissue-specific and non-canonical sites were not themselves conserved [79]. This might be explained by the notion that the exact position of regulatory elements need not be as constrained as sequences coding for a protein. Thus, similar regulation of the same gene via alternative cleavage and polyadenylation in different species may be achieved by utilizing closely located but species-specific non-canonical polyA sites.
Genomic studies of active 3′ end variation
Analyses of static EST collections against the genome laid the framework for a broad understanding of the general principles of constitutive and alternative polyadenylation, but the next leap in our understanding of these processes was made possible with the development of methods for genome-wide querying of UTR expression using microarrays and RNA-sequencing. While individual cell-type and cell-state dependent usage of PAS had been established, the global scope of these changes within a well-characterized gene expression had not been specifically addressed.
The first study taking advantage of newer tools to address this question found surprising answers in the context of primary T lymphocyte activation [80]. The study found that tandem UTRs (Figure 1C), the largest class of alternative polyadenylation events, showed coordinated regulation at 48 hours after stimulation, with a pronounced shift toward expression of mRNA isoforms with shorter 3′ UTRs. This observation was in some ways supportive of previous work that documented an increased efficiency of the core polyadenylation machinery in activated and proliferating cells, in part due to increases in expression of the 64 kDa subunit of the Cstf complex [81,82]. However, it was clear that this model could not completely explain the observed changes for two reasons. First, although a global shift towards upstream PASs was observed in genes with tandem UTRs, certain events were remarkably more robust than the overall trend. Secondly, in cases of splicing-coupled alternative cleavage and polyadenylation events, switches between PASs were equivalently bidirectional.
With carefully controlled analysis on previous generation genomic platforms, the global tendency toward expression of shorter UTR isoforms after activation could be generalized to other murine and human immune cells. The finding that both mitogenic cellular activation signals and transformed cancer cell lines expressed markedly shorter 3′ UTRs supported the notion that a global use of proximal PAS could be more generally associated with cellular proliferation. Meta-analysis of publicly available gene expression data with a novel gene expression-based proliferation index revealed that 3′ UTR length is inversely correlated with cellular proliferation [80]. Reporter assays experimentally determined that shorter 3′ UTR isoforms conferred increases in protein expression, and that the negative effects conferred by longer isoforms could be alleviated by specific deletion of microRNA sites in the distal portion of the 3′ UTR [80].
The general principles suggested by this work were confirmed and extended in several subsequent studies. Work focusing primarily on breast cancer cell lines documented that usage of upstream PASs could be correlated to increases in expression of known oncogenes at the protein level [83]. Another study utilized relative mRNA 3′ UTR expressions to prospectively classify distinct tumors derived from several mouse models of lymphoma [84]. This study identified differential mRNA isoform expressions for several hundreds of genes for each cancer subtype, and these signatures could classify cancer subtypes with 74% accuracy [84]. However, it is unclear whether this analysis unequivocally ruled out the contribution of transcriptional increases or decreases in their metrics of 3′ ends for individual transcripts, which would have been expected to contribute to these classifications. Regardless, consistent with the previous studies [80,83], the lymphoma samples tended to express mRNAs with shorter 3′ UTRs than mature B cells.
That finding that 3′ UTRs were found to shorten in highly proliferative and transformed cells was complemented by meta-analysis of microarray data derived from several models of mammalian differentiation. During the first ~8 days of mouse embryonic development, a progressive lengthening of 3′ UTRs was observed, perhaps reflecting progressively increased differentiation [85]. Differentiation of proliferative C2C12 muscle cell lines to myotubes resulted in the expression of mRNAs with longer 3′ UTRs than the myoblast cells [85]. In contrast, induction of pluripotent stem cells from diverse differentiated cellular origins was found to correlate with a shortening of 3′ UTRs [86]. Together, these studies suggested that differentiation is associated with a bias towards more frequent use of longer mRNA isoforms, presumably through the use of more distal PASs. Since both early embryonic cells and pluripotent cells are rapidly dividing, it is very likely that proliferation rates are also changing during these in vivo and in vitro processes. Therefore, it will be important to demonstrate whether differentiation per se affects alternative polyadenylation in a model system where proliferation and differentiation are uncoupled.
Activity-induced changes in PAS usage have also been documented in the neuronal system. Stimulation of hippocampal neuron explants with potassium chloride revealed ~70 genes undergoing alternative polyadenylation events that would be predicted to lead to changes in peptide sequence [87]. For most of those genes, neuronal activation favored mRNA isoforms with truncated coding sequences, predicted to have major consequences in regards to the function of the encoded proteins [87]. For example, one event produced a shorter mRNA isoform of the gene homer1, producing a dominant negative form of the protein that restricts excitatory synapse number [88].
While activation of both neuronal and immune cells results in some shared physiological outcomes e.g. marked changes in intracellular calcium levels, the abovementioned studies support the notion that dynamic utilization of alternative PAS may be common in response to many external cues given to normal, primary cells. It is an intriguing possibility that modulation of the visibility of gene products to post-transcriptional regulators reflects a second strategy by which gene expression is largely controlled in mammalian cells, perhaps as important and widespread as changes in transcript copy number. Several isoform-sensitive microarray studies have shown that the genes regulated by alternative splicing in a particular gene expression program are distinct from the genes that are simultaneously undergoing changes in transcript copy number (e.g. [89]), but similar systematic analyses of alternative polyandenylation programs remain to be performed.
Inter-relationships among 3′ end variation, transcription and pre-mRNA splicing
The preliminary evidence that transcript variation via alternative splicing is uncoupled from transcript copy number stands in stark contrast to the well-known coupling of 3′ end formation to transcriptional initiation, pre-mRNA splicing, and other post-transcriptional modes of regulation [90]. The last decade has seen an explosion in the number of examples of defined cis-auxiliary PAS determinants, the trans-factors that interact with these elements, and the mechanisms by which these interactions promote or inhibit cleavage and polyadenylation at a given site. There are too many of these studies to discuss here, but they have been recently well-reviewed elsewhere [91]. The aggregate of these studies can be broadly summarized to describe competition with or recruitment of core factors of the cleavage and polyadenylation complex via established sequence motifs, direct inhibition of this complex via protein-protein interactions, or altering reaction kinetics to favor competing processes. Although some alternative cleavage and polyadenylation events can be ascribed to specific expression of non-canonical subunits of one of the core complexes involved in these processes [92,93] the vast majority of trans-factors mechanistically demonstrated to be involved in regulated polyadenylation are known players in constitutive and alternative splicing.
Recent genomic work has complemented these studies, provided additional evidence for the interrelationship of alternative splicing and regulated PAS usage. A study defining transcriptome-wide association of the Nova protein, a known modulator of alternative splicing, implicated this protein in the regulation of alternative cleavage and polyadenylation in the brain [94]. A broader analysis of high-throughput sequencing data across several tissue types revealed that in genes with tandem UTRs, conserved binding motifs for several tissue-specific splicing factors could be found in the more distal regions of the last exon [95]. The same study also found correlations in patterns of alternative splicing and alternative cleavage and polyadenylation across tissues, suggesting these two modes of regulation are coordinated.
Genomic analyses of transcriptome dynamics are not the only approaches that are revealing the level of interconnectedness of 3′ end formation with other processes. A recent study in which the human 3′ processing complex was isolated on a viral cleavage and polyadenylation signal and characterized by multi-dimensional protein identification technology (MudPIT) revealed a staggering 85 proteins, less than half of which were known components of the core cleavage and polyadenylation complex or the splicing machinery [96]. A second study linked cleavage and polyadenylation with nucleosome positioning, demonstrating a decrease in nucleosome density at known cleavage and polyadenylation sites, irrespective of site usage and transcription [97]. Interestingly, increased nucleosome density was found downstream of actively utilized cleavage and polyadenylation sites, but this was correlated only with site usage, and not with the “strength” of the site as judged by conformity to known sequence motifs [97]. These results support a kinetic model of the regulation of cleavage and polyadenylation, where nucleosome positioning and/or specific histone-associated proteins may slow the rate of polymerase elongation or recruit modulators of the 3′ end machinery. Studies such as these, taken together with recent work identifying and characterizing non-canonical mechanisms of 3′ end formation [98], suggest that there is still much to be learned in regards to the mechanisms of constitutive and regulated 3′ end formation, and how these mechanisms interface with upstream and downstream cellular processes.
Future Perspectives
In the coming years, it is likely that high-throughput sequencing based methods will play an even larger role in developing our understanding of several aspects of 3′ end formation. Crosslinked and immunoprecipitated RNA approaches (CLIP-seq or HITS-CLIP) will likely lead to a better understanding of the binding specificities of core and accessory factors involved in constitutive and regulated cleavage and polyadenylation. Newer sequencing-based approaches also allow direct identification and quantitation of mRNA 3′ ends. Thus, the repertoire of canonical and non-canonical PASs in human and mouse can be catalogued, even to the point of assessing heterogeneity in individual cleavage sites. As these methods are extended to estimate 3′ end frequencies across panels of in vivo cells and tissues, or in response to external cues, cell and cell-state specific alternative polyadenylation events will be better defined. Precise collections of PASs associated with distinct genetic programs may pave the way for improved a priori identification of cis-elements directing alternative cleavage and polyadenylation programs.
Yet, while future studies employing newer genomic methods have the potential to revolutionize our understanding of the complexity of alternative cleavage and polyadenylation, it is likely that these studies will echo and extend the observations that have been previously made in the biochemical, molecular, and genetic studies that built the foundation of this field. The importance of these established approaches will remain critical in understanding the mechanisms by which this aspect of gene regulation is effected. Perhaps the major hurdle in pressing forward is the same one that been there from the beginning: the perceived and demonstrated universality of the core factors controlling the cleavage and polyadenylation reactions has largely stymied genetic approaches in mammals. This obstacle will need to be overcome, since the amount of biological material required for classical biochemical approaches precludes analysis of these processes in many developmental systems and primary cell types. While routine usage of viral PASs transient transfection, and minigene construts have yielded remarkable insights that have stood the test of time, the knowledge that promoter identity is known to control downstream RNA processing events [99], suggests that direct experimental manipulations of genes known to undergo differential cleavage and polyadenylation may best performed with cis- elements of the gene in the context of that gene’s endogenous locus. While this is an arduous task in any mammalian system, necessity remains the mother of invention. If future endeavors in this field can recapitulate the heroic and innovate efforts that have been made over the last three decades, there is no doubt that the future yet holds many surprises in the regulation and impact of this facet of gene regulation.
Acknowledgments
We apologize to those whose work was omitted due to space constraints, in particular for the omission of work in non-mammalian systems has contributed so much to the understanding of this field. Research in our laboratories is supported by grants from the National Cancer Institute (JRN), the Cancer Prevention and Research Institute of Texas (JRN), Swedish Cancer Foundation (RS) and Swedish Research Council (RS).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zhao J, Hyman L, Moore C. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev. 1999;63:405–45. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alt FW, Bothwell AL, Knapp M, Siden E, Mather E, Koshland M, Baltimore D. Synthesis of secreted and membrane-bound immunoglobulin mu heavy chains is directed by mRNAs that differ at their 3′ ends. Cell. 1980;20:293–301. doi: 10.1016/0092-8674(80)90615-7. [DOI] [PubMed] [Google Scholar]
- 3.Early P, Rogers J, Davis M, Calame K, Bond M, Wall R, Hood L. Two mRNAs can be produced from a single immunoglobulin mu gene by alternative RNA processing pathways. Cell. 1980;20:313–9. doi: 10.1016/0092-8674(80)90617-0. [DOI] [PubMed] [Google Scholar]
- 4.Rosenfeld MG, Lin CR, Amara SG, Stolarsky L, Roos BA, Ong ES, Evans RM. Calcitonin mRNA polymorphism: peptide switching associated with alternative RNA splicing events. Proceedings of the National Academy of Sciences of the United States of America. 1982;79:1717–1721. doi: 10.1073/pnas.79.6.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaufman RJ, Sharp PA. Growth-dependent expression of dihydrofolate reductase mRNA from modular cDNA genes. Mol Cell Biol. 1983;3:1598–608. doi: 10.1128/mcb.3.9.1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Merritt C, Rasoloson D, Ko D, Seydoux G. 3′ UTRs are the primary regulators of gene expression in the C. elegans germline. Curr Biol. 2008;18:1476–82. doi: 10.1016/j.cub.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Higgs DR, Goodbourn SE, Lamb J, Clegg JB, Weatherall DJ, Proudfoot NJ. Alpha-thalassaemia caused by a polyadenylation signal mutation. Nature. 1983;306:398–400. doi: 10.1038/306398a0. [DOI] [PubMed] [Google Scholar]
- 8.Orkin SH, Cheng TC, Antonarakis SE, Kazazian HH. Thalassemia due to a mutation in the cleavage-polyadenylation signal of the human beta-globin gene. EMBO J. 1985;4:453–6. doi: 10.1002/j.1460-2075.1985.tb03650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gehring NH, Frede U, Neu-Yilik G, Hundsdoerfer P, Vetter B, Hentze MW, Kulozik AE. Increased efficiency of mRNA 3′ end formation: a new genetic mechanism contributing to hereditary thrombophilia. Nat Genet. 2001;28:389–92. doi: 10.1038/ng578. [DOI] [PubMed] [Google Scholar]
- 10.Danckwardt S, Hentze MW, Kulozik AE. 3′ end mRNA processing: molecular mechanisms and implications for health and disease. EMBO J. 2008;27:482–98. doi: 10.1038/sj.emboj.7601932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen J, Férec C, Cooper DN. A systematic analysis of disease-associated variants in the 3′ regulatory regions of human protein-coding genes I: general principles and overview. Hum Genet. 2006;120:1–21. doi: 10.1007/s00439-006-0180-7. [DOI] [PubMed] [Google Scholar]
- 12.Chen J, Férec C, Cooper DN. A systematic analysis of disease-associated variants in the 3′ regulatory regions of human protein-coding genes II: the importance of mRNA secondary structure in assessing the functionality of 3′ UTR variants. Hum Genet. 2006;120:301–33. doi: 10.1007/s00439-006-0218-x. [DOI] [PubMed] [Google Scholar]
- 13.Nevins JR, Darnell JE. Steps in the processing of Ad2 mRNA: poly(A)+ nuclear sequences are conserved and poly(A) addition precedes splicing. Cell. 1978;15:1477–93. doi: 10.1016/0092-8674(78)90071-5. [DOI] [PubMed] [Google Scholar]
- 14.Proudfoot NJ, Brownlee GG. 3′ non-coding region sequences in eukaryotic messenger RNA. Nature. 1976;263:211–4. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- 15.Lai EC, Stein JP, Catterall JF, Woo SL, Mace ML, Means AR, O’Malley BW. Molecular structure and flanking nucleotide sequences of the natural chicken ovomucoid gene. Cell. 1979;18:829–42. doi: 10.1016/0092-8674(79)90135-1. [DOI] [PubMed] [Google Scholar]
- 16.Fitzgerald M, Shenk T. The sequence 5′-AAUAAA-3′forms parts of the recognition site for polyadenylation of late SV40 mRNAs. Cell. 1981;24:251–60. doi: 10.1016/0092-8674(81)90521-3. [DOI] [PubMed] [Google Scholar]
- 17.Montell C, Fisher EF, Caruthers MH, Berk AJ. Inhibition of RNA cleavage but not polyadenylation by a point mutation in mRNA 3′ consensus sequence AAUAAA. Nature. 1983;305:600–5. doi: 10.1038/305600a0. [DOI] [PubMed] [Google Scholar]
- 18.McDevitt MA, Imperiale MJ, Ali H, Nevins JR. Requirement of a downstream sequence for generation of a poly(A) addition site. Cell. 1984;37:993–9. doi: 10.1016/0092-8674(84)90433-1. [DOI] [PubMed] [Google Scholar]
- 19.McDevitt MA, Hart RP, Wong WW, Nevins JR. Sequences capable of restoring poly(A) site function define two distinct downstream elements. EMBO J. 1986;5:2907–13. doi: 10.1002/j.1460-2075.1986.tb04586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wickens M, Stephenson P. Role of the conserved AAUAAA sequence: four AAUAAA point mutants prevent messenger RNA 3′ end formation. Science. 1984;226:1045–51. doi: 10.1126/science.6208611. [DOI] [PubMed] [Google Scholar]
- 21.Gil A, Proudfoot NJ. A sequence downstream of AAUAAA is required for rabbit beta-globin mRNA 3′-end formation. Nature. 1984;312:473–4. doi: 10.1038/312473a0. [DOI] [PubMed] [Google Scholar]
- 22.Gil A, Proudfoot NJ. Position-dependent sequence elements downstream of AAUAAA are required for efficient rabbit beta-globin mRNA 3′ end formation. Cell. 1987;49:399–406. doi: 10.1016/0092-8674(87)90292-3. [DOI] [PubMed] [Google Scholar]
- 23.Conway L, Wickens M. Analysis of mRNA 3′ end formation by modification interference: the only modifications which prevent processing lie in AAUAAA and the poly(A) site. EMBO J. 1987;6:4177–84. doi: 10.1002/j.1460-2075.1987.tb02764.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whitelaw E, Proudfoot N. Alpha-thalassaemia caused by a poly(A) site mutation reveals that transcriptional termination is linked to 3′ end processing in the human alpha 2 globin gene. The EMBO Journal. 1986;5:2915–2922. doi: 10.1002/j.1460-2075.1986.tb04587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Connelly S, Manley JL. A functional mRNA polyadenylation signal is required for transcription termination by RNA polymerase II. Genes Dev. 1988;2:440–52. doi: 10.1101/gad.2.4.440. [DOI] [PubMed] [Google Scholar]
- 26.Lanoix J, Acheson NH. A rabbit beta-globin polyadenylation signal directs efficient termination of transcription of polyomavirus DNA. EMBO J. 1988;7:2515–22. doi: 10.1002/j.1460-2075.1988.tb03099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moore CL, Sharp PA. Site-specific polyadenylation in a cell-free reaction. Cell. 1984;36:581–91. doi: 10.1016/0092-8674(84)90337-4. [DOI] [PubMed] [Google Scholar]
- 28.Moore CL, Sharp PA. Accurate cleavage and polyadenylation of exogenous RNA substrate. Cell. 1985;41:845–55. doi: 10.1016/s0092-8674(85)80065-9. [DOI] [PubMed] [Google Scholar]
- 29.Hart RP, McDevitt MA, Nevins JR. Poly(A) site cleavage in a HeLa nuclear extract is dependent on downstream sequences. Cell. 1985;43:677–83. doi: 10.1016/0092-8674(85)90240-5. [DOI] [PubMed] [Google Scholar]
- 30.Sperry AO, Berget SM. In vitro cleavage of the simian virus 40 early polyadenylation site adjacent to a required downstream TG sequence. Mol Cell Biol. 1986;6:4734–41. doi: 10.1128/mcb.6.12.4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zarkower D, Wickens M. Formation of mRNA 3′ termini: stability and dissociation of a complex involving the AAUAAA sequence. The EMBO Journal. 1987;6:177–186. doi: 10.1002/j.1460-2075.1987.tb04736.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Humphrey T, Christofori G, Lucijanic V, Keller W. Cleavage and polyadenylation of messenger RNA precursors in vitro occurs within large and specific 3′ processing complexes. EMBO J. 1987;6:4159–68. doi: 10.1002/j.1460-2075.1987.tb02762.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bienroth S, Wahle E, Suter-Crazzolara C, Keller W. Purification of the cleavage and polyadenylation factor involved in the 3′-processing of messenger RNA precursors. J Biol Chem. 1991;266:19768–76. [PubMed] [Google Scholar]
- 34.Murthy KG, Manley JL. Characterization of the multisubunit cleavage-polyadenylation specificity factor from calf thymus. J Biol Chem. 1992;267:14804–11. [PubMed] [Google Scholar]
- 35.Murthy KG, Manley JL. The 160-kD subunit of human cleavage-polyadenylation specificity factor coordinates pre-mRNA 3′-end formation. Genes Dev. 1995;9:2672–83. doi: 10.1101/gad.9.21.2672. [DOI] [PubMed] [Google Scholar]
- 36.Ryner LC, Takagaki Y, Manley JL. Multiple forms of poly(A) polymerases purified from HeLa cells function in specific mRNA 3′-end formation. Mol Cell Biol. 1989;9:4229–38. doi: 10.1128/mcb.9.10.4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bardwell VJ, Zarkower D, Edmonds M, Wickens M. The enzyme that adds poly(A) to mRNAs is a classical poly(A) polymerase. Mol Cell Biol. 1990;10:846–9. doi: 10.1128/mcb.10.2.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takagaki Y, Manley JL, MacDonald CC, Wilusz J, Shenk T. A multisubunit factor, CstF, is required for polyadenylation of mammalian pre-mRNAs. Genes Dev. 1990;4:2112–20. doi: 10.1101/gad.4.12a.2112. [DOI] [PubMed] [Google Scholar]
- 39.MacDonald CC, Wilusz J, Shenk T. The 64-kilodalton subunit of the CstF polyadenylation factor binds to pre-mRNAs downstream of the cleavage site and influences cleavage site location. Mol Cell Biol. 1994;14:6647–54. doi: 10.1128/mcb.14.10.6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Christofori G, Keller W. 3′ cleavage and polyadenylation of mRNA precursors in vitro requires a poly(A) polymerase, a cleavage factor, and a snRNP. Cell. 1988;54:875–89. doi: 10.1016/s0092-8674(88)91263-9. [DOI] [PubMed] [Google Scholar]
- 41.Takagaki Y, Ryner LC, Manley JL. Four factors are required for 3′-end cleavage of pre-mRNAs. Genes Dev. 1989;3:1711–24. doi: 10.1101/gad.3.11.1711. [DOI] [PubMed] [Google Scholar]
- 42.Gilmartin GM, Nevins JR. An ordered pathway of assembly of components required for polyadenylation site recognition and processing. Genes Dev. 1989;3:2180–90. doi: 10.1101/gad.3.12b.2180. [DOI] [PubMed] [Google Scholar]
- 43.Rüegsegger U, Blank D, Keller W. Human pre-mRNA cleavage factor Im is related to spliceosomal SR proteins and can be reconstituted in vitro from recombinant subunits. Mol Cell. 1998;1:243–53. doi: 10.1016/s1097-2765(00)80025-8. [DOI] [PubMed] [Google Scholar]
- 44.Vries HD, Rüegsegger U, Hübner W, Friedlein A, Langen H, Keller W. Human pre-mRNA cleavage factor II(m) contains homologs of yeast proteins and bridges two other cleavage factors. EMBO J. 2000;19:5895–904. doi: 10.1093/emboj/19.21.5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lingner J, Kellermann J, Keller W. Cloning and expression of the essential gene for poly(A) polymerase from S. cerevisiae. Nature. 1991;354:496–498. doi: 10.1038/354496a0. [DOI] [PubMed] [Google Scholar]
- 46.Schalet A, Lefevre G. The Genetics and Biology of Drosophila. New York: Academic Press; The proximal region of the X-chromosome; pp. 848–902. [Google Scholar]
- 47.Takagaki Y, Manley JL. Levels of Polyadenylation Factor CstF-64 Control IgM Heavy Chain mRNA Accumulation and Other Events Associated with B Cell Differentiation. Molecular Cell. 1998;2:761–771. doi: 10.1016/s1097-2765(00)80291-9. [DOI] [PubMed] [Google Scholar]
- 48.DeZazzo JD, Imperiale MJ. Sequences upstream of AAUAAA influence poly(A) site selection in a complex transcription unit. Mol Cell Biol. 1989;9:4951–61. doi: 10.1128/mcb.9.11.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carswell S, Alwine JC. Efficiency of utilization of the simian virus 40 late polyadenylation site: effects of upstream sequences. Mol Cell Biol. 1989;9:4248–58. doi: 10.1128/mcb.9.10.4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lutz CS, Alwine JC. Direct interaction of the U1 snRNP-A protein with the upstream efficiency element of the SV40 late polyadenylation signal. Genes Dev. 1994;8:576–86. doi: 10.1101/gad.8.5.576. [DOI] [PubMed] [Google Scholar]
- 51.Lutz CS, Murthy KG, Schek N, O’Connor JP, Manley JL, Alwine JC. Interaction between the U1 snRNP-A protein and the 160-kD subunit of cleavage-polyadenylation specificity factor increases polyadenylation efficiency in vitro. Genes Dev. 1996;10:325–37. doi: 10.1101/gad.10.3.325. [DOI] [PubMed] [Google Scholar]
- 52.Boelens WC, Jansen EJ, Venrooij WJV, Stripecke R, Mattaj IW, Gunderson SI. The human U1 snRNP-specific U1A protein inhibits polyadenylation of its own pre-mRNA. Cell. 1993;72:881–92. doi: 10.1016/0092-8674(93)90577-d. [DOI] [PubMed] [Google Scholar]
- 53.Gunderson SI, Beyer K, Martin G, Keller W, Boelens WC, Mattaj LW. The human U1A snRNP protein regulates polyadenylation via a direct interaction with poly(A) polymerase. Cell. 1994;76:531–41. doi: 10.1016/0092-8674(94)90116-3. [DOI] [PubMed] [Google Scholar]
- 54.Niwa M, Rose SD, Berget SM. In vitro polyadenylation is stimulated by the presence of an upstream intron. Genes Dev. 1990;4:1552–9. doi: 10.1101/gad.4.9.1552. [DOI] [PubMed] [Google Scholar]
- 55.Nesic D, Cheng J, Maquat LE. Sequences within the last intron function in RNA 3′-end formation in cultured cells. Mol Cell Biol. 1993;13:3359–69. doi: 10.1128/mcb.13.6.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Niwa M, MacDonald CC, Berget SM. Are vertebrate exons scanned during splice-site selection? Nature. 1992;360:277–80. doi: 10.1038/360277a0. [DOI] [PubMed] [Google Scholar]
- 57.Lou H, Yang Y, Cote GJ, Berget SM, Gagel RF. An intron enhancer containing a 5′ splice site sequence in the human calcitonin/calcitonin gene-related peptide gene. Mol Cell Biol. 1995;15:7135–42. doi: 10.1128/mcb.15.12.7135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moreira A, Takagaki Y, Brackenridge S, Wollerton M, Manley JL, Proudfoot NJ. The upstream sequence element of the C2 complement poly(A) signal activates mRNA 3′ end formation by two distinct mechanisms. Genes Dev. 1998;12:2522–34. doi: 10.1101/gad.12.16.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Veraldi KL, Arhin GK, Martincic K, Chung-Ganster LH, Wilusz J, Milcarek C. hnRNP F influences binding of a 64-kilodalton subunit of cleavage stimulation factor to mRNA precursors in mouse B cells. Mol Cell Biol. 2001;21:1228–38. doi: 10.1128/MCB.21.4.1228-1238.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson SD, Wickens M, Bentley DL. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385:357–61. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- 61.Dantonel JC, Murthy KG, Manley JL, Tora L. Transcription factor TFIID recruits factor CPSF for formation of 3′ end of mRNA. Nature. 1997;389:399–402. doi: 10.1038/38763. [DOI] [PubMed] [Google Scholar]
- 62.Flaherty SM, Fortes P, Izaurralde E, Mattaj IW, Gilmartin GM. Participation of the nuclear cap binding complex in pre-mRNA 3′ processing. Proc Natl Acad Sci USA. 1997;94:11893–8. doi: 10.1073/pnas.94.22.11893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hirose Y, Manley JL. RNA polymerase II is an essential mRNA polyadenylation factor. Nature. 1998;395:93–6. doi: 10.1038/25786. [DOI] [PubMed] [Google Scholar]
- 64.Proudfoot N. Connecting transcription to messenger RNA processing. Trends Biochem Sci. 2000;25:290–3. doi: 10.1016/s0968-0004(00)01591-7. [DOI] [PubMed] [Google Scholar]
- 65.Hirose Y, Manley JL. RNA polymerase II and the integration of nuclear events. Genes Dev. 2000;14:1415–29. [PubMed] [Google Scholar]
- 66.Edwalds-Gilbert G, Veraldi KL, Milcarek C. Alternative poly(A) site selection in complex transcription units: means to an end? Nucleic Acids Res. 1997;25:2547–61. doi: 10.1093/nar/25.13.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jansen RP. mRNA localization: message on the move. Nat Rev Mol Cell Biol. 2001;2:247–56. doi: 10.1038/35067016. [DOI] [PubMed] [Google Scholar]
- 68.Kuersten S, Goodwin EB. The power of the 3′ UTR: translational control and development. Nature Reviews Genetics. 2003;4:626–637. doi: 10.1038/nrg1125. [DOI] [PubMed] [Google Scholar]
- 69.Gautheret D, Poirot O, Lopez F, Audic S, Claverie JM. Alternate polyadenylation in human mRNAs: a large-scale analysis by EST clustering. Genome Res. 1998;8:524–30. doi: 10.1101/gr.8.5.524. [DOI] [PubMed] [Google Scholar]
- 70.Tian B, Hu J, Zhang H, Lutz CS. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–12. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yan J, Marr TG. Computational analysis of 3′-ends of ESTs shows four classes of alternative polyadenylation in human, mouse, and rat. Genome Res. 2005;15:369–75. doi: 10.1101/gr.3109605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Graber JH, Cantor CR, Mohr SC, Smith TF. In silico detection of control signals: mRNA 3′-end-processing sequences in diverse species. Proc Natl Acad Sci USA. 1999;96:14055–60. doi: 10.1073/pnas.96.24.14055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Beaudoing E, Freier S, Wyatt JR, Claverie JM, Gautheret D. Patterns of variant polyadenylation signal usage in human genes. Genome Res. 2000;10:1001–10. doi: 10.1101/gr.10.7.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Legendre M, Gautheret D. Sequence determinants in human polyadenylation site selection. BMC Genomics. 2003;4:7. doi: 10.1186/1471-2164-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hu J, Lutz CS, Wilusz J, Tian B. Bioinformatic identification of candidate cis-regulatory elements involved in human mRNA polyadenylation. RNA. 2005;11:1485–93. doi: 10.1261/rna.2107305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Beaudoing E, Gautheret D. Identification of alternate polyadenylation sites and analysis of their tissue distribution using EST data. Genome Res. 2001;11:1520–6. doi: 10.1101/gr.190501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu D, Brockman JM, Dass B, Hutchins LN, Singh P, McCarrey JR, MacDonald CC, Graber JH. Systematic variation in mRNA 3′-processing signals during mouse spermatogenesis. Nucleic Acids Res. 2007;35:234–46. doi: 10.1093/nar/gkl919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang H, Lee JY, Tian B. Biased alternative polyadenylation in human tissues. Genome Biol. 2005;6:R100. doi: 10.1186/gb-2005-6-12-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ara T, Lopez F, Ritchie W, Benech P, Gautheret D. Conservation of alternative polyadenylation patterns in mammalian genes. BMC Genomics. 2006;7:189. doi: 10.1186/1471-2164-7-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–7. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Edwalds-Gilbert G, Milcarek C. Regulation of poly(A) site use during mouse B-cell development involves a change in the binding of a general polyadenylation factor in a B-cell stage-specific manner. Molecular and Cellular Biology. 1995;15:6420–6429. doi: 10.1128/mcb.15.11.6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Takagaki Y, Seipelt RL, Peterson ML, Manley JL. The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell. 1996;87:941–52. doi: 10.1016/s0092-8674(00)82000-0. [DOI] [PubMed] [Google Scholar]
- 83.Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–84. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Singh P, Alley T, Wright S, Kamdar S, Schott W, Wilpan R, Mills K, Graber J. Global Changes in Processing of mRNA 3′ Untranslated Regions Characterize Clinically Distinct Cancer Subtypes. Cancer Res. 2009 doi: 10.1158/0008-5472.CAN-09-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ji Z, Lee JY, Pan Z, Jiang B, Tian B. Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc Natl Acad Sci USA. 2009;106:7028–33. doi: 10.1073/pnas.0900028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ji Z, Tian B. Reprogramming of 3′ untranslated regions of mRNAs by alternative polyadenylation in generation of pluripotent stem cells from different cell types. PLoS ONE. 2009;4:e8419. doi: 10.1371/journal.pone.0008419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Flavell SW, Kim T, Gray JM, Harmin DA, Hemberg M, Hong EJ, Markenscoff-Papadimitriou E, Bear DM, Greenberg ME. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron. 2008;60:1022–38. doi: 10.1016/j.neuron.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sala C, Futai K, Yamamoto K, Worley PF, Hayashi Y, Sheng M. Inhibition of dendritic spine morphogenesis and synaptic transmission by activity-inducible protein Homer1a. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience. 2003;23:6327–6337. doi: 10.1523/JNEUROSCI.23-15-06327.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pan Q, Shai O, Misquitta C, Zhang W, Saltzman AL, Mohammad N, Babak T, Siu H, Hughes TR, Morris QD, Frey BJ, Blencowe BJ. Revealing global regulatory features of mammalian alternative splicing using a quantitative microarray platform. Mol Cell. 2004;16:929–41. doi: 10.1016/j.molcel.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 90.Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 91.Millevoi S, Vagner S. Molecular mechanisms of eukaryotic pre-mRNA 3′ end processing regulation. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kubo T, Wada T, Yamaguchi Y, Shimizu A, Handa H. Knock-down of 25 kDa subunit of cleavage factor Im in Hela cells alters alternative polyadenylation within 3′-UTRs. Nucleic Acids Res. 2006;34:6264–71. doi: 10.1093/nar/gkl794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dass B, Tardif S, Park JY, Tian B, Weitlauf HM, Hess RA, Carnes K, Griswold MD, Small CL, Macdonald CC. Loss of polyadenylation protein tauCstF-64 causes spermatogenic defects and male infertility. Proc Natl Acad Sci USA. 2007;104:20374–9. doi: 10.1073/pnas.0707589104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, Darnell JC, Darnell RB. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–9. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shi Y, Giammartino DCD, Taylor D, Sarkeshik A, Rice WJ, Yates JR, Frank J, Manley JL. Molecular architecture of the human pre-mRNA 3′ processing complex. Mol Cell. 2009;33:365–76. doi: 10.1016/j.molcel.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Spies N, Nielsen CB, Padgett RA, Burge CB. Biased chromatin signatures around polyadenylation sites and exons. Mol Cell. 2009;36:245–54. doi: 10.1016/j.molcel.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wilusz JE, Spector DL. An unexpected ending: noncanonical 3′ end processing mechanisms. RNA. 2010;16:259–66. doi: 10.1261/rna.1907510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kornblihtt AR. Coupling transcription and alternative splicing. Adv Exp Med Biol. 2007;623:175–89. doi: 10.1007/978-0-387-77374-2_11. [DOI] [PubMed] [Google Scholar]