Summary
Autophagy is a ubiquitous eukaryotic cytoplasmic quality and quantity control pathway. The role of autophagy in cytoplasmic homeostasis seamlessly extends to cell-autonomous defense against intracellular microbes. Recent studies also point to fully integrated, multitiered regulatory and effector connections between autophagy and nearly all facets of innate and adaptive immunity. Autophagy in the immune system as a whole confers measured immune responses; on the flip side, alterations in autophagy can lead to inflammation and tissue damage, as evidenced by Crohn's disease predisposition polymorphisms in autophagy basal apparatus (Atg16L) and regulatory (IRGM) genes. Polymorphisms in the IRGM gene in human populations have also been linked to predisposition to tuberculosis. There are several areas of most recent growth: (i) links between autophagy regulators and infectious disease predisposition in human populations; (iii) demonstration of autophagy role in infection control in vivo in animal models; (ii) the definition of specific anti-autophagic defenses in highly evolved pathogens; and (iii) recognition of connections between the ubiquitin system and autophagy of bacteria (and interestingly mitochondria, which are incidentally organelles of bacterial evolutionary origin) via a growing list of modifier and adapter proteins including p62/SQSTM1, NDP52, Atg32, Parkin and Nix/BNIP3L.
Introduction: autophagy in infection
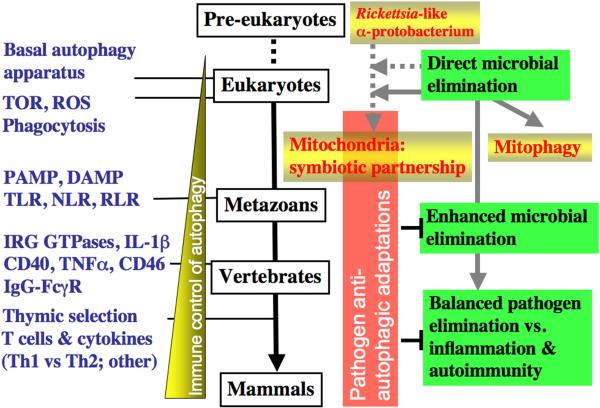
Macroautophagy, or sensu stricto autophagy, is a cytoplasmic quality and quantity control process whereby cytoplasmic components are sequestered into double membrane organelles termed autophagosomes and delivered for degradation and elimination to lysosomes (Fig. 1). The scope of autophagy (and its associated processes of chaperone-mediated-autophagy and microautophagy) includes not only the removal of defective or surplus organelles and large macromolecular aggregates but extends to the now well recognized roles in immunity [Deretic, 2009 #12838]. Evolutionarily the most ancient presentation of autophagy in immunity, its action as a cell cell-autonomous defense against intracellular microbes, is preserved to modern days (Fig. 2). This primal role was likely one of the initial or early acquired functions of autophagy that has subsequently co-evolved along with the various aspects of immune defenses listed in Fig. 2. The ancient nature of the processes of direct capture and elimination of microbes that manage to invade the cytoplasm is perhaps reflected in the present-day autophagic removal of defective or surplus mitochondria in a process that has earned a special term “mitophagy”. Mitochondria are organelles that have evolved from a Rickettsia-like α-protobacterium endosymbiont (incidentally, the initial 1984 observations of autophagy following bacterial infection were on Rickettsia in neutrophils) (reviewed in [Deretic, 2009 #12838]). A vestige of these relationships is perhaps retained in the regulatory interplay of cell survival and death tied to mitochondria as we will discuss at the end of this review.
Figure 1. Proposed evolutionary relationships between autophagy and infection and immunity.
Abreviations: ______
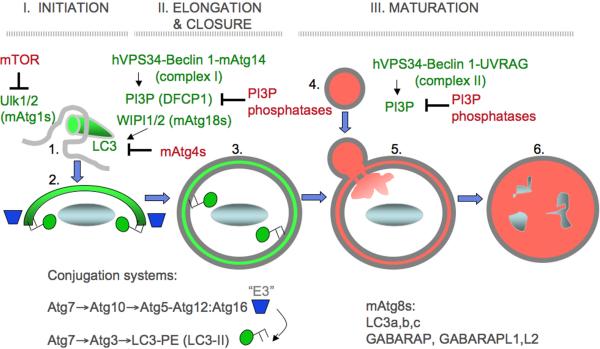
Fig. 2. Autophagy morphological stages.
Shown are the execution stages of autophagy, controlled by an upstream signaling cascade centered around the Ser/Thr kinase Tor, a metabolic regulator of autophagy, and a cascade of Atg factors, starting with Atg1. 1. Initiation. Upstream signaling brings about complex changes in autophagic initiation machinery leading to the formation of membranous precursors of autophagosome. (a) Signaling: nutrient or growth factor starvation signals via Ser/Thr kinase Tor; immunological stimuli (e.g. TLR activation with PAMPs) act through protein complexes containing MyD88 and Beclin 1 (Atg6), a key regulator of autophagy. (b) Beclin 1 is in an inhibitory complex with Bcl-2. Following stimulation, JNK-1 phosphorylates Bcl-2 (Bcl 2-Pi) releasing Beclin 1 from the inhibitory complex with Bcl-2. (c) Activated Beclin 1, in a tri-partite complex with type III phosphatidylinositol 3-kinase hVPS34 and Atg14, cooperates with other Atg factors to initiate autophagosome formation. (d) Atg16L1 (a genetic risk locus for inflammatory Crohn's disease) is in a noncovalent complex with the Atg5–Atg12 protein conjugate Atg16L1 marks the site of phosphatidylinositol 3-phosphate (PI3P) -dependent initiation of autophagosome formation and sets off phases e, f and g (shown only in elongation phase for clarity). Autophagic isolation membrane (phagophore) wraps around the cytoplasmic target (cytosol, protein aggregates, mitochondria, peroxisomes, microbes, etc.). 2. Elongation. Phagophore enlarges and closes to form a double membrane organelle termed the autophagosome. (e,f) Atg16L1/Atg5–Atg12 (e) acts as an E3 enzyme to stimulates conversion of LC3 I into the LC3 II form (f) via a protein-protein and protein-lipid conjugation cascade (not shown) LC3 I has a free C-terminal Gly residue, whereas LC3 II C-terminal Gly is lipidated with phosphatidylethanolamine (PE) leading to membrane association. This stage is pre-proteolytic and the lumen is not yet acidified. (g) Atg4 reverses LC3 II into LC3 I by removing PE. Atg4 is sensitive to reactive oxygen species (ROS) released from mitochondria and may be one of the signals promoting initiation+elongation stages. 3. Maturation. Autophagosome matures into autolysosome, where the captured cargo is degraded. (h) The maturation phase is controlled by another Beclin 1-interacting factor, UVRAG. A second tri-partite complex with Beclin 1 and hVPS34 has been postulated (indicated by parentheses), containing UVRAG (Vps38) as a subunit in place of the initiation-specific factor Atg14; however, published evidence suggests that UVRAG acts independent of Beclin 1 at this stage. UVRAG interacts with HOPS which acts as a tethering and Rab gunanine nucleotide exchange factor (GEF). (i) HOPS with Vps39 stimulates activation of Rab7 by loading Rab7 with GTP via the Rab7 Vps39 (GEF) associated with HOPS complexes. Rab7 is a small GTPase controlling trafficking and identity of late endosomal/lysosomal compartments. (j) Maturation occurs through fusion with late endosomal/lysosomal organelles or delivery of trafficking intermediates carrying H+ ATPase components and lysosomal hydrolytic enzymes. The maturing autophagosome becomes acidified and is converted into a degradative organelle (autolysosome) with single delimiting membrane (the inner of the two membranes is dissolved) containing degraded material including internal membranes originating form the captured cytoplasmic material.
Autophagy affects the development and function of many aspects of innate and adaptive by two principal means: (a) as a generic factor influencing fundamental cellular functions to which immune cells are subject just like any other cell type in the body, and (b) as an immunity-specialized regulator and effector. Here we briefly review the process of autophagy and provide an update on the already recognized key principles of autophagy in infection and immunity. Next, we cover the latest developments concerning autophagy in immunity and inflammation in human populations, summarize studies of autophagy in animal infection models, and conclude by presenting interesting molecular adapter parallels between bacteria [Yoshikawa, 2009 #12927;Zheng, 2009 #12928;Thurston, 2009 #12920] and mitochondria [Novak, 2009 #12926;Kanki, 2009 #12895;Okamoto, 2009 #12934] as they are targeted for autophagic removal.
Autophagy as a fundamental cell biological process affecting all cell types including immune cells
The key morphological feature of autophagy is the emergence within the cytosol of membranous organelles called autophagosomes that capture various cytoplasmic targets and deliver them for lysosomal degradation in autolysosomes. All cells carry out some basal, constitutive autophagy wheras elicited autophagy is a cell's adaptation response to various nutritional and stress agonists including immunological stimuli. An elicited autophagic response in principle involves three stages (Fig. 1): (i) initiation, (ii) elongation (i) and maturation. During the initiation, Atg1 (mammalian Ulk1 and Ulk2) in cooperation with the type III phosphatidylinositol 3 kinase hVPS34 in association with Beclin 1 and Atg14 (Complex I), lead to the formation of nascent autophagosomes from rough endoplasmic reticulum (rER) [Hayashi-Nishino, 2009 #12942] or possibly other membranes. The sites on the rER, dubbed “ER cradle” or omegasome when visualized using a PI3P-binding/ER-targeting marker DFCP-1 [Axe, 2008 #6103], are in the vicinity of phosphatidylinositol 3-phosphate (PI3P) generated by hVPS34. The PI3P-bidning effectors WIPI-1 and WIPI-2 (mammalian Atg18) promote or allow the formation of autophagic isolation membranes (phagophores) marked by LC3 (mammalian Atg8). Both the elongation of the phagosphore and its closure to form a double membrane autophagosome [Fujita, 2008 #6113] are supported by multiple subsets of Atg8 paralogs in mammalian cells, known as LC3A, LC3B, LC3C, GABARAP, GABARAPL1, and GATE16/GABARAPL2. These Atg8 equivalents are C-terminally lapidated by phosphatidylethanolamine (PE) as a result of a ubiquitination-like protein-protein and protein-lipid conjugation cascades, resulting in the Atg5-Atg12 conjugate noncovalently associated with Atg16, and LC3-PE (Atg5-Arg12/Atg16 serves as an “E3-like enzyme” guiding localized LC3-PE production). The generation of LC3-II is opposed by different Atg4s, which act as reactive oxygen species-sensitive delipidating enzymes, [Scherz-Shouval, 2007 #4670]. Maturation is the final, degradative stage of the pathway when closed autophagosomes fuse with late endosomal/lysosomal organelles or carrier inetremediates, generating autolysosomes, delimited by a single membrane. Maturation depends on hVPS34-Belcin 1 in association with UVRAG (Vps38) (Complex II). The captured material is degraded in autolysosomes. Sometimes autophagosomal organeles eliminate the captured material by exocytosis, such as when mitochondria are removed en masse during cellular differentiation [Zhang, 2009 #12959].
Downregulation of elicited autophagy is the least studied aspect the autophagic pathway. The best studied negative regulator of autophagy is the Ser/Thr kinase Tor, which in response to nutritional signals controls Atg1 (Ulk1/2). It is possible that provision of energy and nutrients(generated by autophagy) leads to a gradual return to the baseline level following mTOR reactivation leading to re-binding of Ulk1 and Ulk2 and repression of autophagy. The numerous PI3P phosphatases, termed myotubularins may contribute to this process or adjust the strength of autophagic induction. A recent report on one of the myotubularins [Vergne, 2009 #12887], Jumpy, as a PI3P phosphatase required for downregulation of autophagy and maintenance of proper muscle function (mutations in Jumpy can cause centronucelar myopathy) suggests that lipid phosphatases countering PI3P production may modulate or even downregulate autophagy (Fig. 2). Myotubularins may also adjust the level of constitutive autophagy. Another mechanism for limiting autophagy has been recently reported, whereby cellular FLICE-like inhibitor protein (cFLIP) binds Atg3 to limit the Atg3-mediated step of LC3 conjugation to downregulate autophagosome biogenesis [Lee, 2009 #12931]. Other mechanisms (e.g. self-limiting nature of LC3 consumption during autophagy, intersections with other pathways that may terminate autophagy while initiating other processes) are likely to play a role or be more important.
The Drosophila salivary gland example (where autophagy leads to cell death) notwithstanding, autophagy is now believed to act primarily in support of cellular survival and ensuring proper function of the cell [Kroemer, 2008 #6101]. For instance, cells employ autophagy to remove leaky or irreversibly depolarized mitochondria or to autodigest their own cytosol for energy and amino acids to preserve essential anabolic functions during times of nutrient or growth factor deprivation. In its life-preserving function, autophagy can prevent energetic catastrophe and necrotic cell death, a feature that becomes apparent when apoptosis is disabled [Kroemer, 2008 #6101]. Cell survival and death is one area where autophagy significantly affects immune cells, as in the case of mature T cells in the periphery [Feng, 2008 #6098;Pua, 2009 #9197] and certain subsets of B cells [Miller, 2008 #5240], most likely in a generic way as any other cell type would be affected. This includes developmentally programmed reduction in the number of mitochondria in T cells [Stephenson, 2009 #12951;Pua, 2009 #9197] following T cell exit from the thymus.
Specialized regulatory and effector roles of autophagy in immunity
The autophagy roles in innate and adaptive immunity represent a complex set of connections between canonical autophagy or related at nearly all aspects of the immune system [Deretic, 2009 #12838]. The canonical autophagy engagement includes the following: (i) Direct autophagic capture of intracellular microbes and their elimination via autolysosomes in the process sometimes referred to as xenophagy. (ii) Positive and negative regulation of innate immunity responses by acting upstream and downstream of pattern recognition receptors (PRR) (reviewed in [Deretic, 2009 #12838]). (iii) Innate immunity effector functions by acting (with notable variances [Saitoh, 2008 #6193]) downstream of pathogen associated molecular patterns (PAMP) sensed by PRRs such as Toll-like receptors (TLR) [Delgado, 2008 #4830], cytoplasmic Nod-like receptors represented by Drosophila PE-PGRS [Yano, 2008 #5212], Nod1 and Nod2 [Travassos, 2009 #12964], and the RIG-I-like receptor MDA-5 [Tormo, 2009 #12925]. (iv) Autophagy plays effector or regulatory functions downstream of systems sensing danger signals/alarmins, also known as damage associated molecular patterns (DAMP), such as ATP [Biswas, 2008 #5317] and self-DNA-containing complexes [Chaturvedi, 2008 #5147]. (v) Autophagy is an effector downstream of cell surface receptors such as CD46-Cyt-1, which engages an adapter protein (GOPC) interacting with the key autophagy regulator Beclin 1 [Joubert, 2009 #12932] reminiscent of the engagement of Beclin 1 complexes by MyD88 and TRIF downstream of TLRs [Shi, 2008 #9332]. (vi) Autophagy is a downstream effector or a parallel/upstream co-effector of cell-autonomous defenses dependent on immunity related GTPases (IRG), such as human IRGM (reviewed in [Deretic, 2009 #12838]) and two murine IRGs, Irgm1 (LRG-47) [Feng, 2008 #5215] and Irga6 (IIGP1) [Zhao, 2008 #6094], although other significant functions for IRGs have been proposed [Tiwari, 2009 #12933]. (vii) Autophagy is an effector of Th1/Th2 cytokines produced following T cell polarization, with Th1 cytokines, such as IFN-γ, inducing autophagy whereas Th2 cytokines, such as IL-4 and IL-13 inhibiting autophagy thus explaining in part why Th1 cytokines afford protection against intracellular pathogens, while Th2 cytokines (albeit protective against the metazoan parasites, helminthes) are permissive to intracellular protozoan parasites and bacterial pathogens (reviewed in [Deretic, 2009 #12838]). (viii) Autophagy enhances endogenous cytoplasmic antigen presentation via MHC II [Munz, 2009 #9694] and possibly MHC I [English, 2009 #9210]. (ix) In cells of the reticulo-endothelial system, autophagy elicits a collection of immune processes collectively termed APMA (autophagic macrophage activation) (reviewed in [Deretic, 2009 #12838]). (x) Autophagy also plays a key role in negative and positive selection of CD4 T cells in the thymus [Nedjic, 2008 #6302] most likely by enabling autophagic endogenous antigen presentation by stromal thymic epithelial cells that play a key role in thymic selection of naïve T cell repertoires.
Autophagy role in infection and inflammatory disease in human populations
The role of autophagy in human immunity has been validated through unbiased genetic analyses (genome wide association screens) of predisposition loci associated with Crohn's disease (CD), a form of inflammatory bowel disease. These studies have linked the genes for the core autophagy factor ATG16L1 and an autophagy-related factor IRGM [McCarroll, 2008 #9171]. The polymorphism ATG16L1*300A, conferring risk for Crohn's, appears to reduce the ability to autophagically clear CD-associated microbes such as adherent-invasive E. coli (AIEC) [Lapaquette, 2009 #12937], although the reported differences in stability or activity between the *300A (risk) vs *300T (protective) alleles of ATG16L1 have been recently questioned [Fujita, 2009 #12938]. In murine models of chemically-induced colitis, which has been used to model IBD albeit with a caveat that it does not in full reflect CD, transgenic mice deficient or hypomorphic in Atg16L1 have shown either increased pro-inflammatory (IL-1β) signaling [Saitoh, 2008 #6193] or Paneth cell deficiency [Cadwell, 2008 #6192], previously implciated in human CD. Most recently, a connection has been established between autophagy and Nod2, a risk locus identified early on by tracking familial predisposition to Crohn's disease [Travassos, 2009 #12964]. In that study, Nod2 appeared to recruit ATG16L1 to the bacterial entry site at the plasma membrane, while Crohn's disease-risk Nod2 variant failed to do so [Travassos, 2009 #12964].
The other autophagy-related CD risk locus IRGM [McCarroll, 2008 #9171], encodes the sole human member of a family of immunity related GTPases (IRG). IRGs have been known for a long time to play a role in cell-autonomous antimicrobial defenses but have been orphaned for the exact innate imunity function [Bekpen, 2009 #9351], although parasitophorous phagosome maturation models have been proposed [Tiwari, 2009 #12933]. The association of IRGM polymorphisms with CD has been replicated in a number of diverse populations including some specificity for the ileal form of CD. The initial CD-associated SNPs in IRGM (not altering IRGM amino acid sequence) have been later on shown to be a part of a haplotype carrying a large, 20.1 kb deletion in the promoter region of IRGM [McCarroll, 2008 #9171]. Like ATG16L1, IRGM has been reported to play a role in clearance of AIEC [Lapaquette, 2009 #12937].
IRGM has recently been identified as a risk locus for tuberculosis in African populations [Intemann, 2009 #12939]. Just like with the CD risk alleles, the IRGM polymorphisms that confer risk of developing pulmonary tuberculosis are located in the promoter region of IRGM and are linked to the 20.1 kb deletion haplotype, but the specific SNPs (e.g. −261T/protection vs −261C/risk) differ from those associated with CD. IRGM has been initially shown to induce autophagy in human macrophages to control Mycobacterium tuberculosis (reviewed in [Deretic, 2009 #12838]). One of the stumbling blocks in studying the role of IRGs in vivo is that the mouse is not suitable as a model system to study human IRGM, since mice have over 20 IRG genes (Irgm1–3, Irga1–8, Irgb1–10, and Irgd) while humans and chimpanzees have only IRGM [Bekpen, 2009 #9351]. Based on syntenic relationships it was initially assumed that the human IRGM gene corresponds to one of the murine IRGs, and a more recent analysis has shown that the human IRGM evolved through a series of steps from the pro-simian IRGM9, which may have had roots corresponding to the mouse Irgm1 [Bekpen, 2009 #9351].
Additional factors previously implicated in the control of intracellular M. tuberculosis but with less understood effector function, such as extracellular ATP (DAMP agonist of P2X7 receptor), have now been shown to induce autophagy to control intracellular mycobacteria [Biswas, 2008 #5317]. The role of autophagy in control of M. tuberculosis has been further affirmed by findings that the previously well appreciated anti-tuberculosis factor 1,25-dihydroxyvitamin D3 (calcitriol) confers its anti-tuberculosis action by inducing autophagy [Yuk, 2009 #12940]. Calcitriol serum levels correlate with active disease outcome and vitamin D receptor polymorphisms have been associated with risk for tuberculosis in human populations. Calcitriol has been previously shown to induce autophagy in cancer studies, and now this has been extended to antimicrobial action via autophagy in tuberculosis [Yuk, 2009 #12940].
Animal models of autophagy role in infection
In addition to human population genetic studies that have now established the significance of autophagy in infectious and inflammatory diseases, the role of autophagy in infection has been tested in a number of animal models. Models ranging from nematodes to mammals have shown that autophagy plays a role in direct elimination of pathogens [Jia, 2009 #12941;Yano, 2008 #5212;Shelly, 2009 #12907;Zhao, 2008 #6094]. Murine models of autophagy function within central immunological organs (thymus) [Nedjic, 2008 #6302] and in the immunological effector periphery [Cadwell, 2008 #6192;Saitoh, 2008 #6193]. have uncovered the primarily anti-inflammatory role of autophagy.
In the study by Jia et al., [Jia, 2009 #12941] the Caenorhabditis elegans model was used along with the social amoeba Dictyostelium discoideum to test whether autophagy genes (C. elegans bec-1 and lgg, corresponding to Beclin 1/Atg6 and Atg8; D. discoideum ATG1, ATG6 and ATG7) conferred protection against the enteric pathogen Salmonella typhimurim. In C. elegans, S. typhymurium can cause persistent infection in the lumen of the intestinal tract but never replicates intracellularly. In D. discoideum, Salmonella is taken up by the amoebae (that have long been popular for study of phagocytosis) but also fails to replicate intracellularly. However, mutations in autophagy genes rendered these two hosts highly susceptible to invasive bacterial infection and permitted intracellular bacterial replication [Jia, 2009 #12941]. In these studies, the role of autophagy was also tested in daf-2e1370 background. The C. elegans daf-2 mutants have extended life-span due to reduced insulin/IGF-1-like signaling in the nematode, and also show heightened resistance against bacterial infection for unknown reasons. This increased resistance to bacteria in daf-2e1370 worms turned out to be due to autophagy [Jia, 2009 #12941]. These elegant studies in C. elegans link pathways relevant for aging with innate immunity to infections. Whether and how insulin signaling, caloric restriction, and longevity intersect with autophagy in its anti-infection and anti-inflammatory roles in mammalian animal models and humans is an interesting but unexplored area.
A number of studies in Drosophila show that autophagy downstream of PRR stimulation plays an in vivo role in innate immunity defense against viruses and bacteria [Yano, 2008 #5212;Shelly, 2009 #12907]. The theme of an antagonism between insulin signaling and cell-autonomous defense role of autophagy seen in C. elegans [Jia, 2009 #12941] extends to the fly, but with an interesting twist: infection with the virus prevented insulin-dependent activation of inhibitors (e.g. Akt) of autophagy [Shelly, 2009 #12907]. This indicates that processes induced during active infection may trump insulin signaling to allow autophagy to proceed unabated and defend cells against pathogens. Another Drosophila study [Yano, 2008 #5212] showed that autophagy is induced in vivo downstream of PRRs, in this case the Drosophila NLR cytosolic PRR equivalent PG-PGRS responding to diaminopimelic acid-type peptidoglycan released from bacteria. Finally, murine animal models have shown that autophagy can protect in vivo against bacterial and protozoan parasites [Zhao, 2008 #6094]. Autophagy helps contain inflammation in immune effector tissues such as the murine intestinal tract [Saitoh, 2008 #6193;Cadwell, 2008 #6192] and through central tolerance by dictating positive and negative selection of naïve T cells in the thymus [Nedjic, 2008 #6302]. The latter process appears to involve a carefully orchestrated matchmaking between the autophagic self-antigen presentation by thymus stromal cells during thymic selection of naïve T cell repertoires and the autophagic activity in peripheral tissues.
Collectively, these in vivo studies establish the role of autophagy in defense against infection and prevention of excessive inflammation. Finally, possibly the most promising use of animal models of autophagy in infection and immunity is the application to vaccine development. A recent study [Jagannath, 2009 #9195] has indicated that autophagy may break through some well known vaccine barriers, such as a frustrating inability to come up with a vaccine against tuberculosis that is better than BCG. It turns out that rapamycin-treated dendritic cells infected ex vivo with the tuberculosis vaccine strain BCG reintroduced into mice, show enhanced (relative to BCG alone) Th1-mediated protection when challenged with virulent M. tuberculosis [Jagannath, 2009 #9195].
Specific viral and bacterial factors inhibiting autophagic degradation
One conclusion from model infection studies in lower metazoans is that autophagy is particularly good in controlling microbes when a host-pathogen pair tested has not been perfected through co-evolution [Deretic, 2009 #9352]. In other words, autophagy, in its cell-autonomous defense role, effectively trumps the ability of a microbe to invade and survive in a new host. The flip side of this is that when a microbe is highly adapted it often has specific adaptations to deal with or counter autophagy. Detailed molecular analyses of specific pathogen effectors targeting autophagic machinery targets is well in viruses, have been recently reviewed [Deretic, 2009 #9352]. There very clear common themes that are developiong form the detailed mechanistic studies: (i) Many of the viral factors characterized so far target Beclin 1 or other parts of the core autophagy machinery to inhibit autophagy maturation as in the case of HIV Nef [Kyei, 2009 #12888] and influenza A virus M2 proteins [Gannage, 2009 #12943]. Herpes simplex virus 1 protein ICP34.5 also binds to Beclin 1 but it inhibits autophagy altogether (reviewed in [Deretic, 2009 #9352]). How can Beclin 1-interacting factors have different effects - ICP34.5 inhibits autophagy including initiation while Nef and M2 inhibit maturation but do not suppress initiation? This is possible since Beclin 1 is engaged in two different hVPS34 complexes (Fig. 1) acting at separate steps: complex 1 (Atg14-Beclin 1-hVPS34) functioning at initiation, and complex 2 (UVRAG-Beclin 1- hVPS34) playing a role during maturation. This complex 2-focused action makes easy sense in the context of HIV. Paradoxically, induction of early stages of autophagy in HIV-infected macrophages promotes Gag processing and enhances infectious HIV virion yields [Kyei, 2009 #12888]. Thus, inhibition of maturation only ensures the benefits of early autophagy stages but prevents autophagic degradation [Kyei, 2009 #12888]. In the report on influenza A virus effects on autophagy, no benefit in virus yields have been observed; nevertheless, inhibition of autophagy by M2 via Beclin 1 enhanced apoptotic effects of infection, which could contribute to pathogenesis [Gannage, 2009 #12943]. In the case of HIV, T cells (which undergo cell death, unlike macrophages that are normally spared) cell death has also been reported as related to autophagic events [Espert, 2009 #12944].
Beclin 1 is a target for another mechanism of inhibition of autophagy by γ herpesvirus encoded Bcl-2 like proteins (vBcl-2) via binding to the BH3–like domain of Beclin 1 [Ku, 2008 #12947;Sinha, 2008 #6202;Xiaofei, 2009 #12946]. This is akin to the normal regulation of autophagy via cellular Bcl-2, which binds (albeit weakly) to Beclin 1 and dissociates from it upon agonist signals transduced via JNK-1 [Wei, 2008 #5204] or DAPK [Zalckvar, 2009 #9155]. In the case of murine γ herpesvirus 68 vBcl-2 protein referred to as M11, anti-apoptotic and anti-autophagic functions could be mutationally separated, and it was shown that autophagy plays a role in maintenance of latent infection in vivo [Xiaofei, 2009 #12946]. However, Beclin 1 is not the only autophagic target for manipulation by viruses. Herpesviruses such as Kaposi's sarcoma-associated herpesvirus, encode viral FLIP (vFLIP) with similar action to cFLIP. Like cFLIP, vFLIPs inhibits autophagy by preventing Atg3 from interacting with and processing LC3 [Lee, 2009 #12931]. Atg3 acts as an E2-like enzyme in the cascade of LC3 activation en route to its C-terminal conjugation to PE. Thus, vFLIP may affect elongation and closure stage of autophagosomal formation. IN conclusion, viruses show a great deal of specific adaptations affecting initiation, elongation/closure, and maturation stages of the execution phase of autophagy.
The currently welldelineated examples of specific bacterial factors suppressing autophagy are limited to the members of an elite group of intracellular bacterial pathogens (Listeria, Shigella, Rittsketsia) that escape into the cytosol and come in contact directly with the host cell cytosol [Yoshikawa, 2009 #12927]. An emerging common theme here is that proteins IcsB (Shigella) and ActA (Listeria), otherwise well known for their role in actin-based motility (enabling these bacteria to ride atop actin tails throughout the host cell cytoplasm) also inhibit autophagy. Shigella IcsA caps IcsB (VirG) and protects it from recognition by the autophagic factor Atg5 (reviewed in [Deretic, 2009 #12838]). Listerial ActA interacts with cytosolic actin polymerization machinery (Arp2/3, VASP and actin), which not only provides motility but also generates a surface coating composed of cell's own proteins thus camouflaging against autophagic capture. This function of ActA, independently of the AktA intracellular motility role, is necessary to ensure optimal listerial intracellular survival at 2–4 h postinfection [Yoshikawa, 2009 #12927]. When artificially forced, ActA can confer protection from autophagic capture to non-cognate protein aggregates known to be typical autophagic targets [Yoshikawa, 2009 #12927]. It would be of interest to know whether Rickettsie possess similar methods of protection from autophagy to complete the cytosolic bacterial pathogens “trifecta”.
Many bacterial, protozoan, and fungal intracellular pathogens remaining enclosed within the phagosome and may utilize different methods of protection. This is exemplified by Listeria remaining in the phagosome (potentially associated with persistent infection) when it interferes differently with autophagy – it blocks maturation by low level listeriolysin O action poking a limited number of holes in phagosomal membranes (without causing complete escape into the cytosol) thus preventing acidification. In principle, this might potentially apply to other intra-phagosomal intracellular pathogens such as M. tuberculosis and Histoplasma capsulatum, given that they prevent acidification of their respective parasitophorous vacuoles.
Adapter molecules targeting bacteria for autophagy
Recent reports [Yoshikawa, 2009 #12927;Zheng, 2009 #12928;Thurston, 2009 #12920] addressing how intracellular bacteria are earmarked and captured by autophagosomes show some unexpected fundamental similarities with (just as recently discovered) processes of mitochondrial tagging for autophagy and removal [Novak, 2009 #12926;Kanki, 2009 #12895;Okamoto, 2009 #12934]. The literal linchpin to autophagic uptake of either cytosolic bacetria or mitochondria is the interaction between adapter proteins (which recognize molecular tags - such as polyubiquitin - on bacteria or mitochondria) and LC3, most often through the LC3 interaction region (LIR) initially recognized as the WXXL motif (Fig. 3). Whether a bacterium or a mitochondrion, a LIR-containing adapter bridges these targets with nascent LC3-positive autophagosomes (Fig. 3).
When Listeria lacks ActA, it becomes coated with pulyubiquitin upon its escape into the cytsool. The polyubiquitin tags are then recognized by a key adapter protein p62 (also known as sequestosome 1 or SQSTM1). This adapter was initially identified as a bridge [Kirkin, 2009 #9203] between LC3 and polyubiquitinated protein aggregates destined for autophagic removal. Once listeria-associated polyubiquitin is recognized by p62, the bacteria become decorated with polyubiquitin-p62-LC3, since the p62 LIR sequence WXXL binds to LC3, thus bridging ubiquitin-tagged bacteria with forming autophagosomes [Yoshikawa, 2009 #12927].
A similar bridging mechanism functions in the context of Shigella infection, but relates to the phagosomal shrouds left after disruption and escape of Shigella into the cytosol [Dupont, 2009 #12949]. These membrane remnants, if not promptly removed by autophagy [Dupont, 2009 #12949], gather and accumulate proinflammatory signaling molecules, such as TRAF6. Interestingly, p62 may play a role in both the removal of Shigella phagosomal remnants via polyubiquitin-p62-LC3 bridge and in inflammatory signals. The phagosomal remnants left after Shigella escape are polyubiquitinated and their removal depends on ubiqutin K48 and p62 [Dupont, 2009 #12949]. Thus, damaged phagosomal membranes can be targeted and removed by autophagic degradation using the ubiqutin-p62-LC3 system, ridding the cytoplasm of defective membranous organelles [Dupont, 2009 #12949]. It is not known whether this process manages to capture any Shigellae before they fully escape from the phagosome. Nevertheless, TRAF6 accumulates on the phagosomal remnants, activates IKK-γ (NEMO), and promotes inflammatory outputs (e.g. IL-8), possibly as a second tier defense that can recruit immune cells to clear the infection. TRAF6 is known to directly interact with p62, and TRAF6 pro-inflammatory signaling was exaggerated when autophagy was blocked [Dupont, 2009 #12949]. This indicates a second role for p62 in the inflammatory signaling, possibly when not promptly removed by autophagy, as seen in other unrelated instances [Komatsu, 2007 #5273;Alirezaei, 2008 #9392;Mathew, 2009 #12918]. Nevertheless, some TRAF6 signaling was taking place even when mutant K48 ubiquitin was overexpressed in cells, leading the authors to propose a p62-independent component for TRAF6 recruitment [Dupont, 2009 #12949]. Finally, when autophagy was artificially abrogated in Shigella-infected cells, cells eventually underwent cell death either by caspase 1-dependent pyroptosis or mitochondrial membrane depolarization-associated necrosis in the wake of the inability to clear the danger signal sent by the phagosomal remnants. This may be a third tier response, limiting infection spread by cell death, when pathogen clearance by autophagy misses its mark.
Although Salmonella normally does not escape into the cytosol like Shigella does, 25% of Salmonella containing vacuoles can be damaged and such bacteria colocalize with polyubiquitinated complexes. Thes, most likely cytosolic, Salmonellae have been reported as being delivered to autophagosomes via p62 and LC3 [Zheng, 2009 #12928] as in the case of Listeria and Shigella phagosomal remnants. However, p62 is not the only participant in these processes. NDP52, a newly characterized polyubiquitin- and LC3-interacting factor, turned out to be key for control of cytosolic Salmonella prolyferation by delivery to autophagosomes [Thurston, 2009 #12920]. NDP52 was identified in an unbiased study following the reports on the ubiquitinated Salmonella fraction in the cytosol, and is an interacting partner (indirectly, via Nap1 and Sintbad) of TBK1, a non-cannonical member of the IKK family that affects the integriity of Salmonella vacuoles [Thurston, 2009 #12920]. Moreover, NDP52 was found to play a role in autophagy of Streptococcus pyogenes, thus indicating its key role in autophagic control of bacteria that reach cell's cytosol but are not properly equipped with anti-autophagy factors.
Adapter molecules targeting bacteria and mitochondria for autophagy
Interestingly, the principles of an adapter-based delivery to autophagosomes described above for bacteria also apply to mitochondria, albeit with inevitable differences regarding the specific factors involved. In yeast, mitochondria are subjected to autophagic degradation upon recruitment of Atg8 (the yeast equivalent of LC3) by the mitochondrial outer membrane-anchored protein Atg32 [Kanki, 2009 #12895;Okamoto, 2009 #12934] that contains the Atg8/LC3 motif WXXI. Atg32 is induced in yeast during respiratory growth and may lead to mitophagy in response to oxidative stress [Okamoto, 2009 #12934] and starvation [Kanki, 2009 #12895].
In mammalian cells, mitochondria are removed by mitophagy either (i) developmentally, e.g. during reticulocyte maturation following exit form the bone marrow [Zhang, 2009 #12959], or T cell maturation following exit from the thymus [Pua, 2009 #9197;Stephenson, 2009 #12951]) or (ii) as a continuous housekeeping process of removing irreversibly depolarized or damaged mitochondria that cannot be repaired by fusion with the mitochondrial network. It turned out that the developmental removal of mitochondria during reticulocyte maturation, a process that at least in part may include exocytosis of mitochondria captured by autophagosomes [Zhang, 2009 #12959], is carried out by an adapter protein Nix, which delivers mitochondria to autophagosomes [Schweers, 2007 #5050;Novak, 2009 #12926], in a mechanistically similar fashion to Atg32, NDP52, and p62. Nix has a number of functions assigned in earlier studies. The basis for this is that Nix has a LIR motif (WVEL), and interacts with the LC3 family members [Novak, 2009 #12926;Schwarten, 2009 #12957] (Fig. 3___). Nix has been identified as a BH3-only protein with a transmembrane domain localizing to mitochondria [Zhang, 2009 #12959], which can affect mitochondrial transmembrane potential [Sandoval, 2008 #5294] and promote cell death via a mechanism surprisingly less reliant on its weak BH3 domain or cytochrome c release. Nix's pivotal role in developmental removal of mitochondria from reticulocytes [Zhang, 2009 #12959], acting as an adapter between nascent autophagosomes and mitochondira destined to be eliminated, might be linked to and perhaps be an extension of the other known mitochondrial functions of Nix [Sandoval, 2008 #5294]. It has also been reported that Nix-dependent elimination of mitochondria may be only partially affected by the loss of Atg7 [Zhang, 2009 #12961]. This appears paradoxical, although recently a non-canonical, Atg5/Atg7-independent autophagy pathway has been proposed [Nishida, 2009 #12962]. Non-developmental autophagy of mitochondria depends on the cytosolic E3 ubiquitin ligase Parkin, recruited to the surface of dysfunctional mitochondria [Narendra, 2008 #12902] possibly in cooperation with the mitochondria associated Ser/Thr kinase Pink1 [Dagda, 2009 #12963]. The mitochondrial targets ubiquitinated by Parkin are not known. It also remains to be determined whether the mitochondrial or mitochondria-associated targets are recognized by adapter proteins such as p62, NBR1, NDP52, or other LIR-containing proteins that remain to be characterized.
Conclusions and opinions
The role of autophagy in infection and immunity has now been firmly established although additional sub-themes remain to be uncovered and many details remain to be delineated. In this review we have emphasized some of the latest advances topped by the most recent evidence for autophagy in infection, immunity, and inflammation in human populations. Animal studies, ex vivo, in vitro, and molecular work, all attest to the fact that autophagy and immune defense against infection are deeply intertwined. The recent studies on autophagic targeting of mitochondria and bacteria in the cytosol of eukaryotic cells represent the latest eidence in support of the potential common roots for the process. As an opinion, we here offer a proposal that autophagy may have evolved as one of the first cell-autonomous defenses against invading microbes. We furthermore propose that mitochondira of today may be viewed as “soft fossil” evedence of the primordial relationships, reflecting both autophagic removal of invading microbes and capture and removal of mitochondria by autophagy. We submit that this can be of use in understanding not only the roots of immune functions of autophagy but also shedding some light on mitochondria-centered regulatory events. In this model, autophagy and cell death may have been two stages of the early evolution of defense against infection, in the following binary sequence of events: (i) successful autophagic clearance of bacteria and cell survuival; (ii) autophagic clearance being overwhelmed by infection triggering cell death to preserve the species or the whole organism from infection spread. If this model is applied to non-immune functions, it might help us glean a better understanding of how cell death is triggered or regulated in cancer, inflammation, and other health and disease states.
Supplementary Material
Molecular adapters targeting bacteria and mitochondria for autophagic degradation Abbreviations: ______.
Acknowledgments
This work was supported by grants RC1AI086845, AI069345, AI42999 from National Institutes of Health, 107160-44-RGRL from amfAR, a Bill and Melinda Gates Grand Challenge Explorations grant and a grant from Crohn's & Colitis Foundation of America
References
- Nix is a selective autophagy receptor for mitochondrial clearance Ivana Novak1, Vladimir Kirkin2,*, David G. McEwan2, Ji Zhang3, Philipp Wild2, Vladimir Rogov4,5, Alexis Rozenknop2,4, Frank Löhr4, Doris Popovic1,2, Angelo Occhipinti6, Andreas S. Reichert6, Janos Terzic1,7, Volker Dötsch4, Paul A. Ney3, Ivan Dikic1,2,7. [DOI] [PMC free article] [PubMed]
- A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation Mitsuko Hayashi-Nishino, Naonobu Fujita, Takeshi Noda, Akihito Yamaguchi, Tamotsu Yoshimori and Akitsugu Yamamoto NCB in press. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Molecular adapters targeting bacteria and mitochondria for autophagic degradation Abbreviations: ______.