Abstract
Hsp90 is an essential eukaryotic molecular chaperone that stabilizes a large set of client proteins, many of which are involved in various cellular signaling pathways. The current list of Hsp90 interactors comprises about 200 proteins and this number is growing steadily. In this paper, we report on the application of three complementary proteomic approaches directed towards identification of novel proteins that interact with Hsp90. These methods are coimmunoprecipitation, pull down with biotinylated geldanamycin, and immobilization of Hsp90β on sepharose. In all, this study led to the identification of 42 proteins, including 18 proteins that had not been previously characterized as Hsp90 interactors. These novel Hsp90 partners not only represent abundant protein species, but several proteins were identified at low levels, among which signaling kinase Cdk3 and putative transcription factor tripartite motif-containing protein 29. Identification of tetratricopeptide-repeat-containing mitochondrial import receptor protein Tom34 suggests the involvement of Hsp90 in the early steps of translocation of mitochondrial preproteins. Taken together, our data expand the knowledge of the Hsp90 interactome and provide a further step in our understanding of the Hsp90 chaperone system.
Keywords: Heat shock protein 90, Identification, Immobilization, Interactome, Kinase, Partners
Introduction
Hsp90 is a ubiquitous eukaryotic chaperone protein. Comprising about 1% of all cellular proteins, Hsp90 is involved in diverse processes ranging from processing and maintenance of RNA to protein sorting and assembly of the tubulin-based cytoskeleton network (Te et al. 2007; Lotz et al. 2008; Zhao et al. 2008). However, one of the most exciting roles of Hsp90 is the stabilization of a set of client proteins. The two biggest coherent protein classes among Hsp90 clients are kinases and transcription factors (Pratt and Toft 2003). Together with various cochaperones, such as Hsp70 and Cdc37, Hsp90 stabilizes and activates its clients, facilitating the execution of many essential signaling pathways. Inhibition of Hsp90 leads to proteasomal degradation of its clientele and subsequent disruption of prosurvival signaling, which ultimately results in growth arrest and apoptotic cell death. Many Hsp90 clients such as Akt/PKB, Raf, and Bcr-Abl are oncoproteins that are either mutated or overexpressed in cancer cells, which in turn depend on these proteins for growth and proliferation (Weinstein and Joe 2006). This makes Hsp90 an attractive target for cancer therapy and its inhibitors were promising in clinical trials (Modi et al. 2007; Ramalingam et al. 2008).
To date, about 200 proteins have been shown to interact with Hsp90 (Picard 2008). Thus, in spite of being a very abundant protein, Hsp90 seems to interact with other proteins in a selective manner. Even though several crystal structures of Hsp90 are now available, the broad variety of its clientele makes it challenging to resolve the molecular mechanisms of Hsp90’s selectivity (Ali et al. 2006; Pearl et al. 2008; Shiau et al. 2006). In addition, the number of proteins interacting with Hsp90 is continuously growing, challenging the existing concepts and suggesting new hypotheses for the selectivity of Hsp90 (Citri et al. 2006; Prince and Matts 2004). Thus, identification of Hsp90-interacting proteins and cellular processes in which Hsp90 is involved remains a key question in the Hsp90 field. Comprehensive knowledge about the roles of Hsp90 in various aspects of cell life would not only improve our understanding of cell biology but might also lead to the design of alternative strategies for treatment of certain diseases, for example, cancer.
So far, proteins have been found to interact with Hsp90 occasionally, mainly by coimmunoadsorption. However, several recent studies have focused on identification of Hsp90 partners using large-scale proteomic approaches, such as immunoprecipitation, immobilization of the C-domain of Hsp90α, genome-wide two-hybrid screens, and proteome analysis of tumor cells subjected to treatment with Hsp90 inhibitor (Falsone et al. 2005; Millson et al. 2005; Zhao et al. 2005; McClellan et al. 2007; Schumacher et al. 2007; Te et al. 2007). These studies have led to the identification of a number of novel Hsp90-interacting partners, including cochaperones and client proteins, and suggested several previously unknown functions of Hsp90, for example, cellular transport, cytokinesis, and epigenetic gene regulation. Remarkably, the current progress in revealing the Hsp90 interactome could only be achieved by integration of the results from various strategies directed towards identification of Hsp90-interacting proteins. This suggests that further investigations in different experimental settings will possibly shed more light on the Hsp90 interactome and extend our knowledge of the Hsp90 chaperone machinery.
The aim of our research was to isolate and identify novel Hsp90-binding partners by the combined use of three substantially different proteomic approaches, such as coimmunoprecipitation, purification of Hsp90 protein complexes with biotinylated geldanamycin, and, for the first time, immobilization of a full-length Hsp90β. We identified 18 novel putative Hsp90-binding partners, including Tom34, Cdk3, and tripartite motif-containing protein 29 (TRIM29), and 24 proteins that have been shown to interact with Hsp90 before.
Materials and methods
Materials
The antibodies used for coimmunoprecipitation and Western blotting were: anti-Hsp90 (F-8) mouse monoclonal antibody, anti-Cdc37 (H-271) rabbit polyclonal antibody, and anti-Hsp70 mouse monoclonal antibody (all from Santa Cruz Biotechnology). A protease inhibitor cocktail was from Sigma; biotinylated geldanamycin was from InvivoGen; dithiobis[succinimidyl propionate] (DSP) was from Pierce; Protein G-Sepharose and cyanogen bromide (CNBr)-activated Sepharose were from GE Healthcare. Hsp90β protein was purified as described (Rüdiger et al. 2002).
Cell culture
The human epidermoid carcinoma cells A431 were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, penicillin (100 units per milliliter), streptomycin (100 μg/ml), and amphotericin B (0.25 μg/ml) in 5% CO2, 95% air at 37°C in a humidified incubator. In all experiments, 75−85% confluent cells were used.
Coimmunoprecipitation
All manipulations with cell lysates were carried out at 4°C. A431 cells were lysed in immunoprecipitation (IP) buffer containing 20 mM Tris, pH 7.4, 100 mM NaCl, 0.5% TX-100, and protease inhibitors (1 ml of lysis buffer per 100 cm2 of 80% confluent cells). Lysates were centrifuged at 20,000×g for 15 min and supernatants were collected. After preclearing with Protein G-Sepharose beads, 500 µl of lysates was incubated overnight with 12 µl of anti-Hsp90 antibody (F-8). Subsequently, solutions were supplemented with 20 µl of Protein G-Sepharose beads and incubated while shaking for 1 h. The beads were washed three times with IP buffer and boiled in 100 µl of sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer to liberate bound proteins. Protein samples were separated by SDS-PAGE, followed by either Coomassie staining or Western blotting.
Purification of Hsp90 protein complexes using biotinylated geldanamycin
A431 cells were lysed in phosphate-buffered saline (PBS) buffer containing 0.25% NP-40 and protease inhibitors (1 ml of lysis buffer per 100 cm2 of 80% confluent cells). Lysates were centrifuged at 20,000×g for 15 min and supernatants were collected. Twenty-millimolar DSP in dimethyl sulfoxide was added at indicated final concentrations and the mixtures were incubated for 15 min at room temperature. DSP-mediated cross-linking was blocked by the addition of 1 M Tris, pH 7.5, buffer to the samples at 20-mM final concentration and 15-min incubation at room temperature. Biotinylated geldanamycin (biotin-GA) was added at 1 mM to all samples except for the control followed by overnight incubation at 4ºC. To remove unbound biotin-GA, samples were dialyzed against PBS buffer. Cross-linked Hsp90 protein complexes containing biotin-GA were purified by incubation with 10% (v/v) of NeutrAvidin agarose beads overnight at 4ºC. After washing of the beads four times with 50 mM Tris, 8 M urea, and 200 mM NaCl, pH 8, and twice with PBS, proteins were eluted with twofold PAGE loading buffer and separated on 10% SDS-PAGE, followed by Coomassie or silver staining.
Immobilization of Hsp90β
One hundred milligrams of CNBr-activated Sepharose beads were washed five times with 2.5 ml cold 1 mM HCl and twice with 2.5-ml coupling buffer containing 100 mM NaHCO3, 0.5 M NaCl, pH 8.3. Subsequently, 1 ml of 2 mg/ml Hsp90β in coupling buffer was added to the beads, followed by overnight incubation at 4ºC. The beads were incubated for 4 h with 1 ml of 100 mM NaHCO3, 1 M ethanolamine, pH 9, to block unreacted functional groups. Finally, the beads were washed three times with 50 mM Tris, 1 M NaCl, pH 8, and twice with PBS and stored at 4ºC. The control samples were prepared using the same procedure, except addition of Hsp90β was omitted. All the buffers used for immobilization of Hsp90β were supplemented with protease inhibitors to prevent proteolytic degradation of Hsp90β.
Purification of Hsp90-binding proteins with immobilized Hsp90β
A431 cells were lysed in lysis buffer containing 20 mM Tris, 100 mM NaCl, 0.5% TX-100, pH 7.4, and protease inhibitors. Lysates were centrifuged at 20,000×g for 15 min, and collected supernatants were incubated for 1 h with 10% (v/v) CNBr-activated Sepharose beads blocked with ethanolamine. Precleared lysates were then incubated overnight with 10% (v/v) immobilized Hsp90β at 4ºC. The beads were washed four times with lysis buffer supplemented with 20% glycerol and 0.5% Tween-20. Bound proteins were eluted by boiling for 10 min with twofold PAGE loading buffer and separated on 10% SDS-PAGE, followed by Coomassie staining.
In-gel digestion
Each lane from Coomassie-stained gel was cut into 20 slices of equal size. Each slice was cut into small cubes prior to digestion. The gel pieces were placed in a 0.5-ml tube and washed with 250 µl of water, followed by 15-min dehydration with 100% acetonitrile (ACN). Proteins were reduced with 100 µl 10 mM dithiothreitol (DTT) in 50 mM NH4HCO3 for 1 h at 60°C, followed by dehydration with 100% ACN and incubation in 100 µl 55 mM iodoacetamide in 50 mM NH4HCO3 in the dark. Then, the gel pieces were dehydrated twice with 100 µl 100% ACN, and 20 µl of 12 mg/ml trypsin in 50 mM NH4HCO3 was added to each sample, followed by 30-min incubation at 4°C. After removal of excess trypsin, samples were incubated in 20 µl of 50 mM NH4HCO3 overnight at 37°C, and the supernatants were transferred to new tubes. Peptides were extracted from the gel pieces with 5% formic acid for 2 min at 65 °C, followed by shaking for 20 min. Supernatants were collected and combined with the ones from the previous step.
LC-LTQ mass spectrometry analysis
Nanoscale liquid chromatography tandem mass spectrometry (MS/MS) was performed by coupling an Agilent 1100 Series LC system to an LTQ quadrupole ion trap mass spectrometer (Finnigan, San Jose, CA, USA). Peptide mixtures were concentrated and desalted using an online C18 trap column (OD 375 μm, ID 100 μm packed with 20 mm of 5 μm AQUA C18, RP particles (Phenomenex)) and further separation was achieved by gradient elution of peptides onto a C18 reverse-phase column (OD 375 μm, ID 50 μm packed with 15 cm of 3 μm C18, RP particles (Reprosil)). MS detection in the LTQ was achieved by directly spraying the column eluent into the electrospray ionization source of the mass spectrometer via a butt-connected nanoelectrospray ionization emitter (New Objective). A linear 60-min gradient (10–45% B) was applied for peptide elution into the MS at a final flow rate of 100 nl/min. The total analysis time was 1 h. Mobile-phase buffers were 0.1 M acetic acid; 80% ACN, 0.1 M acetic acid.
The LTQ was operated in positive ion mode, and peptides were fragmented in data-dependent mode. One mass spectrometry survey zoom scan was followed by three data-dependent MS/MS scans. Target ions already selected for MS/MS were dynamically excluded for 30 s.
Database searches
Tandem mass spectra were extracted and charge state deconvoluted by BioWorks version 3.3. All MS/MS samples were analyzed using Mascot (Matrix Science, London, UK; version 2.1.02). Mascot was set up to search the fragment mass spectra against IPI_HUMAN_v3.36 database. The database was searched with a parent ion tolerance of 0.5 Da and a fragment ion mass tolerance of 0.9 Da. Fixed and variable modifications were the iodoacetamide derivative of cysteine and oxidation of methionine, respectively. Probability assessment of peptide assignments and protein identifications was made using Scaffold (version Scaffold-1.6, Proteome Software Inc., Portland, OR, USA). Only peptides with ≥90% probability were considered. Criteria for protein identification included detection of at least two uniquely identified peptides and a probability score of ≥99%.
Results
Identification of Hsp90 partners by coimmunoprecipitation
To isolate and identify new Hsp90-interacting proteins, we used three different proteomic approaches, i.e., co-IP with the antibody against Hsp90α/β, purification of Hsp90 protein complexes using biotinylated Hsp90 inhibitor GA, and immobilization of Hsp90β on the Sepharose support, followed by purification of binding partners from cell lysates.
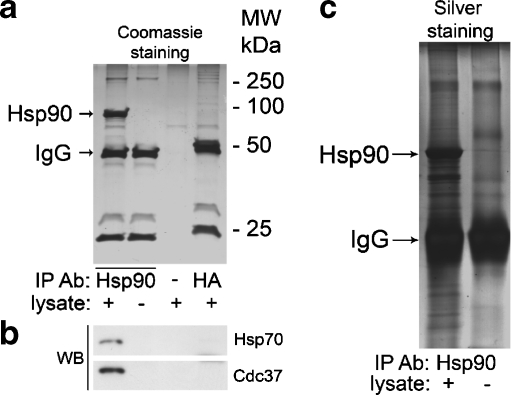
In the first approach, Hsp90 protein complexes were immunoprecipitated with the monoclonal antibody against Hsp90α/β. Samples without antibody or with an “irrelevant” antibody against HA-tag were used as controls (Fig. 1). Notably, Hsp90 was immunoprecipitated only in the sample containing both A431 cell lysate and anti-Hsp90 antibody (lane 1), whereas no nonspecific binding of Hsp90 to the protein G-beads (lane 3) or to anti-HA antibody (lane 4) was observed, as shown in Fig. 1a. This demonstrates the relevance of the selected controls, suggesting that proteins, which are present only in lane 1 but not in lanes 3 and 4, are coprecipitated specifically with Hsp90 and, therefore, represent putative Hsp90-binding partners.
Fig. 1.
a Coomassie staining of the proteins coimmunoprecipitated with Hsp90 (lane 1), and appropriate controls, including only antibody (lane 2), only A431 cell lysate (lane 3), and A431 cell lysate with an “irrelevant” antibody against HA-tag (lane 4). b Cochaperones of Hsp90, Hsp70, and Cdc37 were coprecipitated only in the sample containing both A431 cell lysate and antibody against Hsp90 (lane 1) but not in the control samples (lanes 2–4), as detected by Western blotting. c Silver staining of IP samples, including control without lysate
Coomassie staining shows that Hsp90 is the major protein that immunoprecipitates with anti-Hsp90 antibody (Fig. 1a, lane 1). Apart from several proteins in the molecular weight region between 50 and 100 kDa (Fig. 1a, lane 1), other intensively stained protein bands either belong to the antibody (Fig. 1a, lane 2) or represent the proteins nonspecifically bound to the Protein G-beads (Fig. 1a, lane 3). To demonstrate that other proteins, which could not be detected by Coomassie staining, coimmunoprecipitate with Hsp90, we performed Western blot detection of two well-known major Hsp90 cochaperones, Hsp70 and Cdc37. Both Hsp70 and Cdc37 were specifically coimmunoprecipitated with Hsp90, as shown in Fig. 1b.
The amounts of coimmunoprecipitated proteins, however, were lower than the detection limit of Coomassie staining, explaining our inability to visualize these proteins in Fig. 1a. Silver staining of the same samples resulted in the detection of several proteins present only in the Hsp90 co-IP sample, as shown in Fig. 1c for the sample and the control without lysate. Since silver staining is more destructive for proteins, Coomassie-stained gels were used for further analysis.
To identify coimmunoprecipitated proteins, the sample of interest (Fig. 1a, lane 1) and the appropriate controls (Fig. 1a, lanes 3 and 4) were subjected to mass spectrometric analysis. This led to the identification of 31 proteins exclusively present in the sample of interest. Identified proteins are listed in Table 1. The monoclonal antibody used for IP recognizes both α and β isoforms of Hsp90, which resulted in precipitation of both Hsp90α and Hsp90β (Table 1). Consequently, this approach does not distinguish between Hsp90α and Hsp90β partners among the identified proteins.
Table 1.
Hsp90 partners identified by immunoprecipitation (IP), purification with biotinylated geldanamycin (Bio-GA), and purification with immobilized Hsp90β (Imm. Hsp90β)
| Protein name | Acc. no. | MW, Da | IP | Bio-GA | Imm. Hsp90β | Function |
|---|---|---|---|---|---|---|
| Heat shock protein Hsp90β | IPI00414676 | 83118.1 | × | × | × | |
| Heat shock protein Hsp90α | IPI00382470 | 98147.1 | × | × | ||
| Heat shock cognate 71-kDa protein | IPI00003865 | 71,082 | × | × | Chaperone | |
| Heat shock 70-kDa protein 1 | IPI00304925 | 70,022 | × | Chaperone | ||
| Stress-70 protein, mitochondrial precursor | IPI00007765 | 73,920 | × | Chaperone | ||
| Hsp90 cochaperone Cdc37 | IPI00013122 | 44,450.2 | × | Chaperone | ||
| Cell division cycle 37 homolog-like 1 | IPI00302028 | 38,816.6 | × | Chaperone | ||
| 60-kDa heat shock protein precursor | IPI00784154 | 61,037.7 | × | × | Chaperone | |
| DnaJ homolog subfamily A member 1 | IPI00012535 | 44,850.6 | × | Chaperone | ||
| Hsc70/Hsp90-organizing protein (Hop) | IPI00013894 | 62,624.1 | × | × | Chaperone | |
| Mitochondrial import receptor Tom70 | IPI00015602 | 68,096 | × | × | Chaperone | |
| Mitochondrial import receptor Tom34 | IPI00009946 | 34,542.3 | × | × | Chaperone | |
| TTC4 tetratricopeptide repeat protein 4 | IPI00000606 | 44,662.1 | × | Chaperone | ||
| CHIP Isoform 1 of STIP1 homology and U box-containing protein 1 | IPI00025156 | 34,839 | × | Chaperone | ||
| FKBP52 FK506-binding protein 4 | IPI00219005 | 51,787.9 | × | Immunophilin | ||
| Actin, cytoplasmic 1 | IPI00021439 | 41,719.8 | × | × | Structural | |
| Filamin A | IPI00302592 | 279,990.7 | × | × | Structural | |
| Tubulin beta-2C chain | IPI00007752 | 49,812.7 | × | × | Structural | |
| Isoform 1 of vinculin | IPI00291175 | 116,706.3 | × | Structural | ||
| Myosin-9 | IPI00019502 | 226,519.5 | × | Structural | ||
| Kinesin heavy chain | IPI00012837 | 109,668.3 | × | Transport | ||
| Isoform 1 of epidermal growth factor receptor precursor | IPI00018274 | 134,261.2 | × | × | Signaling | |
| Serine/threonine protein phosphatase 5 | IPI00019812 | 56,862.3 | × | Signaling | ||
| Cell division protein kinase 3 (Cdk3) | IPI00023503 | 37,597.1 | × | Signaling | ||
| Fatty acid synthase | IPI00026781 | 273,382 | × | × | Metabolism | |
| D-3-phosphoglycerate dehydrogenase | IPI00011200 | 56,501.4 | × | Metabolism | ||
| Glyceraldehyde-3-phosphate dehydrogenase | IPI00219018 | 36,035.3 | × | Metabolism | ||
| l-lactate dehydrogenase B chain | IPI00219217 | 36,620.6 | × | Metabolism | ||
| Isoform 1 of heterogeneous nuclear ribonucleoprotein M | IPI00171903 | 77,499.3 | × | × | RNA processing | |
| Isoform B1 of heterogeneous nuclear ribonucleoproteins A2/B1 | IPI00396378 | 37,412.3 | × | RNA processing | ||
| Isoform 1 of heterogeneous nuclear ribonucleoprotein K | IPI00216049 | 50,960.5 | × | RNA processing | ||
| Ewing sarcoma breakpoint region 1 | IPI00009841 | 68,947.8 | × | RNA processing | ||
| Isoform 1 of plasminogen activator inhibitor 1 RNA-binding protein | IPI00410693 | 44,947.8 | × | RNA processing | ||
| KH-type splicing regulatory protein | IPI00479786 | 73,097.6 | × | RNA processing | ||
| Isoform 1 of polyadenylate-binding protein 1 | IPI00008524 | 70,653.3 | × | RNA processing | ||
| Eukaryotic translation initiation factor 2 subunit 1 | IPI00219678 | 35,963.5 | × | Translation | ||
| Elongation factor 2 | IPI00186290 | 95,322.1 | × | Translation | ||
| 60S ribosomal protein L12 | IPI00024933 | 17,801.1 | × | Translation | ||
| 40S ribosomal protein S3a | IPI00419880 | 29,796.1 | × | Translation | ||
| 40S ribosomal protein S2 | IPI00013485 | 31,307.2 | × | Translation | ||
| 40S ribosomal protein S3 | IPI00011253 | 26,670.6 | × | Translation | ||
| Protein RCC2 | IPI00465044 | 56,066.8 | × | Chromosome condensation | ||
| TRIM29 Isoform alpha of tripartite motif-containing protein 29 | IPI00073096 | 65,817.7 | × | Transcription | ||
| Hypothetical short protein | IPI00795193 | 5,159.9 | × | Unknown |
Novel Hsp90 partners are shown in italic.
Identification of Hsp90 partners using biotinylated geldanamycin
Geldanamycin (GA) is an inhibitor of Hsp90, which has been shown to bind with high affinity to the N-domain of Hsp90 (Stebbins et al. 1997). On this basis, we used a biotinylated derivative of GA (biotin-GA) for selective affinity purification of Hsp90 in complex with its interacting proteins. Our preliminary experiments demonstrated that incubation with biotin-GA followed by affinity purification with NeutrAvidin agarose efficiently isolated Hsp90 from A431 cell lysates. However, the proteins bound to Hsp90 were washed away during the preparative steps (data not shown). To prevent the loss of Hsp90-interacting proteins, lysates were subjected to cross-linking with DSP, a homobifunctional amino-reactive DTT-reversible cross-linker, followed by incubation with biotin-GA.
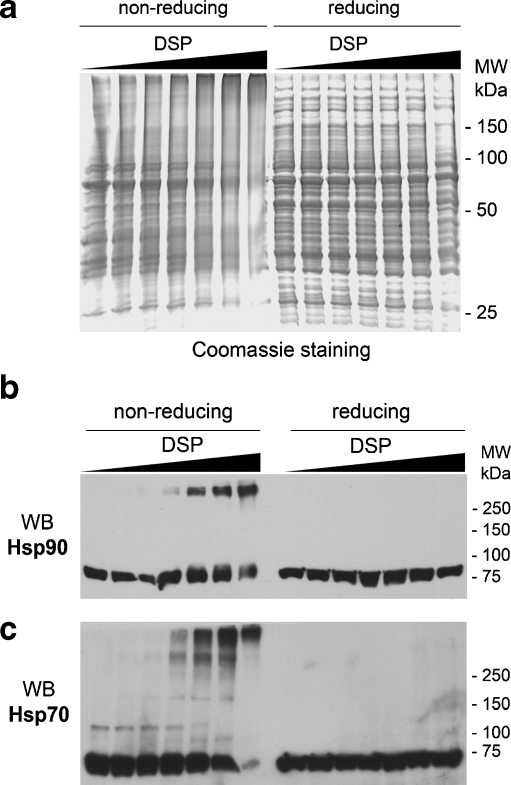
Despite its wide application for stabilizing low-affinity protein–protein interactions, excessive cross-linking may introduce false positives. To determine the optimal cross-linking conditions, A431 cell lysates were incubated with various concentrations of DSP. Separation on nonreducing SDS-PAGE (Fig. 2a, left panel) shows that DSP induces concentration-dependent cross-linking of cellular proteins, which is completely reversed by DTT present in reducing loading buffer (Fig. 2a, right panel), demonstrating the DSP-mediated nature of the cross-linking. To monitor the extent of the cross-linking of Hsp90 and its cochaperone Hsp70, these proteins were detected by Western blotting (Fig. 2b, c). Both proteins undergo DSP-mediated cross-linking. While 1 mM DSP cross-links virtually all cellular Hsp70 and most of Hsp90, likely giving rise to nonspecific coupling, DSP concentrations below 0.5 mM, which cross-link only small fractions of Hsp90 and Hsp70, were chosen for purification experiments.
Fig. 2.
a A431 cell lysates subjected to various concentrations of DSP were separated on nonreducing (without DTT, left panel) and reducing (with DTT, right panel) SDS-PAGE and detected by Coomassie staining. DSP concentrations are: 0, 0.03, 0.06, 0.125, 0.25, 0.5, and 1 mM. The same samples as in a were transferred onto the nitrocellulose membrane, followed by Western blotting detection of Hsp90 (b) and its cochaperone Hsp70 (c)
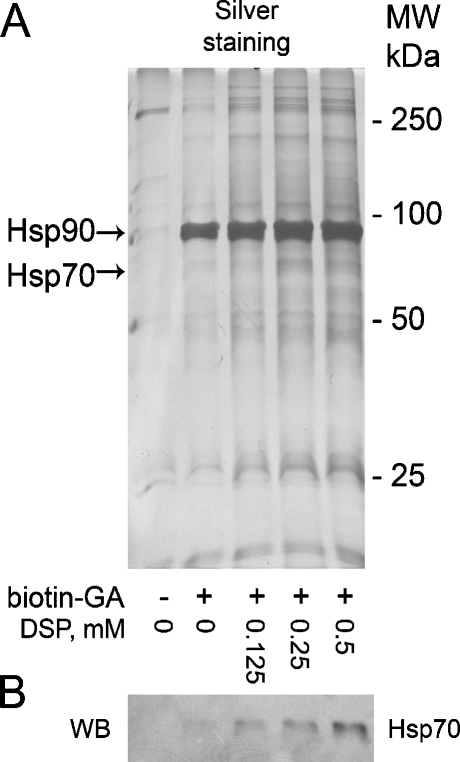
To purify Hsp90 complexes, A431 cell lysates cross-linked with the selected concentrations of DSP were incubated with biotin-GA, followed by purification with NeutrAvidin agarose. Obtained samples were separated on SDS-PAGE and proteins were detected by silver staining. Figure 3a shows that the applied approach results in purification of Hsp90 and some other proteins, which amount is significantly higher in the cross-linked samples. Notably, no Hsp90 was detected in the control sample (no biotin-GA).
Fig. 3.
a A431 cell lysates were cross-linked with DSP and incubated with biotin-GA, followed by purification of biotinylated protein complexes with NeutrAvidin agarose, separation on 10% SDS-PAGE and silver staining. b Western blotting detection of Hsp70 demonstrates the need to perform cross-linking to purify Hsp90 partners
To check the efficiency of the method under different cross-linking conditions, copurification of Hsp70, the Hsp90 cochaperone, was detected by Western blotting. While no Hsp70 was detected in the control sample, addition of biotin-GA results in a DSP-dependent increase in the amount of copurified Hsp70, as shown in Fig. 3b. Thus, the highest Hsp70 signal is detected in the sample prepared using biotin-GA and 0.5 mM DSP, suggesting these conditions to be optimal for copurification of Hsp90 partners. On this basis, the sample containing biotin-GA and 0.5 mM DSP was chosen for mass spectrometric analysis, while the sample containing no biotin-GA was used as a control. Mass spectrometric analysis led to identification of 16 proteins present exclusively in the sample of interest but not in the control sample. As with coimmunoprecipitation, both α and β isoforms of Hsp90 were purified using biotin-GA. All identified proteins are listed in Table 1.
Purification of Hsp90-binding proteins with immobilized Hsp90β
The third approach towards the identification of Hsp90 partners includes incubation of cellular proteins with immobilized Hsp90, followed by identification of purified proteins by mass spectrometry. Notably, the β isoform of Hsp90 was chosen for immobilization experiments. Thus, in contrast with coimmunoprecipitation and purification with biotin-GA that do not allow us to distinguish between Hsp90α- and Hsp90β-interacting proteins, this approach allows selective purification of only Hsp90β partners.
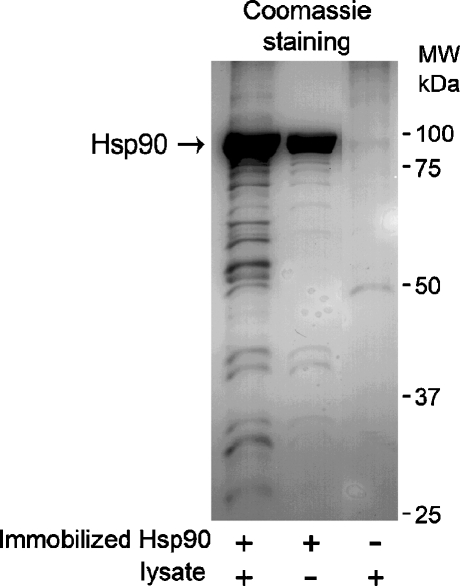
A431 cells were lysed and incubated with immobilized Hsp90β, followed by extensive washing and elution of the bound proteins from the beads with SDS-PAGE loading buffer. The eluates were separated on SDS-PAGE and proteins were detected by Coomassie staining. Figure 4 shows that in the presence of SDS-PAGE loading buffer immobilized Hsp90β elutes from the beads, causing possibly disadvantageous effect for further experimental steps, such as mass spectrometry. However, elution under conditions in which Hsp90β remains bound to the support, for example, using low pH or high-salt-containing buffers, was inefficient in our hands (not shown). Despite the unfavorable elution of Hsp90β, at least eight additional distinct protein bands are detected in Fig. 4, lane 1, which represent A431 cellular proteins bound to immobilized Hsp90β. These bands and the corresponding regions of the control sample (Fig. 4, lane 3) were subjected to mass spectrometry analysis, which resulted in identification of 11 proteins exclusively present in the sample prepared using immobilized Hsp90β and A431 cell lysates but not in the control sample. Identified proteins are listed in Table 1.
Fig. 4.
Immobilized Hsp90β allows purification of Hsp90β interacting proteins from A431 cell lysates, as detected by Coomassie staining of 10% SDS-PAGE. At least eight new distinct protein bands can be detected on the first lane of the gel, as compared to the control samples (without lysates and without immobilized Hsp90β)
Interestingly, the amount of Hsp90β eluted from the beads after incubation with cell lysates is significantly higher than that in the control sample, where incubation with lysates was omitted (Fig. 4, lanes 1 and 2). Densitometric analysis of the protein bands revealed a twofold difference in the amount of eluted Hsp90β between these two samples. This suggests that immobilized Hsp90β exists in the monomeric state and retains its ability to form dimers with Hsp90 from cell lysates. Notably, since virtually all immobilized Hsp90β forms homodimers upon incubation with the lysates, proteins purified using this method interact with dimeric Hsp90. In addition, mass spectrometric analysis showed that Hsp90α did not bind to immobilized Hsp90β (Table 1), supporting the evidence that Hsp90 mainly forms homodimers (ββ) but not heterodimers (αβ; Sreedhar et al. 2004). This also confirms that the approach selectively purifies Hsp90β-binding proteins.
Discussion
The general aim of this research was to obtain new insights into the Hsp90 interactome. The interactome comprises a diverse set of proteins ranging from highly abundant cochaperones, structural proteins, ribosomal subunits, and proteasomal proteins to signaling proteins, including over 70 kinases and transcription factors, such as p53 only present in low abundance. Notably, many substrates bind to a certain conformation of Hsp90 and dissociate upon the adenosine triphosphatase (ATPase)-driven conformational change of Hsp90 (Richter et al. 2003). Similarly, Hsp90’s kinase-targeting cochaperone Cdc37 binds to an Hsp90 dimer, temporally arrests the ATPase cycle, and is subsequently released upon the conformational rearrangements of the complex (Vaughan et al. 2006). Therefore, attempts to perform proteomic investigations of the Hsp90 interactome are facing several distinct difficulties such as the low abundance of client proteins and the transient mode of their interaction with Hsp90. Another general obstacle is the possibility of steric conflicts between an antibody used to isolate Hsp90 complexes and the partners of Hsp90. In an attempt to overcome these problems and to increase the coverage of the targeted interactome, we combined the use of three proteomic approaches, which should complement each other. In all, these methods led to the identification of 42 proteins, including 18 proteins that have not been previously characterized as Hsp90 interactors (Table 1). The presence of 24 well-established Hsp90 partners among the identified proteins suggests the high relevance of the obtained results.
Among the proteins listed in Table 1 are highly abundant proteins, including major Hsp90 cochaperones, structural proteins, ribosomal subunits, and metabolic and RNA-processing proteins. Also, novel Hsp90 substrates at relatively low abundance were identified, such as the signaling proteins cell division protein kinase 3 (Cdk3) and TRIM29 (Table 1). An essential element of the evaluation of our findings is their comparison with the results of related studies. A recent study reports on the application of IP with endogenous Hsp90, which yielded 39 interaction partners of Hsp90 (Falsone et al. 2005). From these proteins, only nine were previously established Hsp90 partners. Characteristically, virtually all of the identified proteins were highly abundant cytosolic proteins, raising the possibility of their nonspecific copurification with Hsp90. The relevance of these results must be further confirmed either by independent proteomic approaches or by conventional biochemical methods. Another related study utilized Hsp90 immunoadsorption and immobilization of a recombinant C-terminal domain of Hsp90α (Hsp90CT; Te et al. 2007). These two assays resulted in the identification of largely overlapping sets of proteins, which were composed mainly of known Hsp90 cochaperones. Besides the known Hsp90 partners, two tetratricopeptide repeat (TPR)-domain-containing proteins, Ttc1 and FLJ21908, were isolated with immobilized Hsp90CT, which was consistent with the design of the assay. Although the use of Hsp90CT excludes the specific isolation of Cdc37, which binds to the N-domain of Hsp90 (Roe et al. 2004), immunoprecipitation also failed to isolate Cdc37 (Te et al. 2007). Notably, Falsone et al. could not coimmunoprecipitate Cdc37, which may have resulted from overlapping binding sites of the antibody and Cdc37 on Hsp90 (Falsone et al. 2005). In contrast, IP of Hsp90 with the antibody used in this study led to copurification of several functionally different Hsp90 cochaperones, including Cdc37, Hsp70, Hsp40 (DnaJA1), and Hop. As these chaperones in various combinations are generally required for the stabilization of most of the Hsp90 substrates, their presence in the immunoprecipitated fraction facilitates the purification of Hsp90-interacting proteins and strengthens the relevance of the identified proteins.
Analysis of the putative Hsp90 partners identified in this work revealed that most proteins (29 of them) were obtained by IP, while the use of biotin-GA and immobilized Hsp90β led to a further identification of 14 and ten proteins, respectively. Each method resulted in identification of a set of proteins containing a large fraction of well-established Hsp90 partners (55%, 64%, and 70% for IP, biotin-GA, and immobilized Hsp90β, respectively). At the same time, a modest overlap between the three sets suggests the usefulness of combining several complementary assays. While most types of Hsp90 cochaperones were purified by IP, immobilized Hsp90β mainly captured TPR-domain-containing chaperones but not Cdc37, being consistent with the use of immobilized Hsp90CT (Te et al. 2007). The use of biotin-GA excludes the possibility to purify Cdc37 as we recently showed that GA disrupts the Hsp90/Cdc37 complexes in A431 cells (Tsaytler et al., unpublished).
One of the TPR-domain-containing proteins identified using immobilized Hsp90β was TTC4. Crevel et al. have recently demonstrated the interaction between the TPR domain of TTC4 and Hsp90 in HeLa cells and suggested that TTC4 links Hsp90 activity and DNA replication in human cells (Crevel et al. 2008). Furthermore, it was proposed that TTC4 might contribute to the development of a variety of different tumors. Our findings provide further evidence in line with the hypothesis that TTC4 is a genuine Hsp90 interactor in human cancer cells.
Using IP and immobilized Hsp90β, we identified two mitochondrial import receptor proteins Tom70 and Tom34. Tom70 is localized in the outer mitochondrial membrane and is required for recognition and translocation of mitochondrial preproteins from the cytoplasm into the mitochondria. It has been shown that Tom70 interacts with Hsp90 via its TPR domains located in the cytosolic portion of the protein (Young et al. 2003) and that Hsp90 is directly involved in preprotein targeting and transport (Fan et al. 2006). In contrast, not much is known about Tom34. While the role of Tom34 in preprotein import remains elusive, it was suggested to be a cytosolic protein that might function as a chaperone-like protein during protein translocation (Yang and Weiner 2002). Using a yeast two-hybrid screen, Young et al. demonstrated the interaction between the 12-kDa C-terminal domain of Hsp90α and Tom34 (Young et al. 1998). Moreover, the analysis of the phylogenetic tree of TPR domains of different proteins suggested the recognition of Hsp90 by Tom34 (Schlegel et al. 2007). Here, we isolated and identified Tom34 using immobilized Hsp90β and immunoprecipitation with Hsp90. While the immunoprecipitation with Hsp90α/β antibody does not distinguish between the isoforms, the assay with immobilized Hsp90β specifically revealed the interaction of Tom34 with the full-length Hsp90β isoform. Although the GCUNC45 protein has been recently shown to interact specifically with the beta isoform of Hsp90 (Chadli et al. 2008), the functional difference between two cytosolic Hsp90 isoforms is generally unknown. Together with the observation of the interaction between C-domain of Hsp90α and Tom34, our data provide strong evidence that Tom34 forms stable complexes with both cytosolic isoforms of Hsp90 in mammalian cells. Furthermore, our results suggest that Tom34 is a novel TPR-containing cytosolic cochaperone of Hsp90α/β, which forms complexes with Hsp90 that are likely involved in the early steps of translocation of mitochondrial preproteins.
The ability of the antibody used for immunoprecipitation to purify Hsp90/Cdc37 complexes led to the identification of cyclin-dependent kinase 3 (Cdk3). To date, only five members of the Cdk protein family (Cdk2, Cdk4, Cdk6, Cdk9, and Cdk11) have been shown to interact with Hsp90 (Citri et al. 2006). It was, therefore, surprising that we identified Cdk3, perhaps the least well-studied protein among all Cdks (Ye et al. 2001), whereas none of the well-established Hsp90 client kinases were found. Similar to most of the Hsp90 client kinases, Cdks are recruited to Hsp90 by Cdc37 (Caplan et al. 2007). Interestingly, early studies devoted to the interactions between Hsp90/Cdc37 and the members of Cdk family identified Cdk4 and Cdk6, whereas other Cdks failed to interact with Hsp90/Cdc37 (Stepanova et al. 1996; Lamphere et al. 1997). Only recently, Cdk2 has been shown to be a genuine client kinase of Hsp90/Cdc37 (Prince et al. 2006). Interestingly, Cdk3 and Cdk2 are the closest homologs among the proteins of Cdk family (76% identical). While some features of kinases, such as the presence of conserved glycine-rich loops (Terasawa et al. 2006), are recognized by Hsp90/Cdc37, these features are present in many kinases that are not Hsp90 clients (Vaughan et al. 2006). Therefore, the nature of the specific interaction between Hsp90 and client kinases remains obscure. Thus, it is not possible to predict the Cdk3/Cdc37/Hsp90 interaction based on the amino acid sequence of Cdk3. As many Hsp90 client kinases, including Cdk2 and Cdk4, undergo Hsp90 inhibition-mediated downregulation, treatment of cells with Hsp90 inhibitor GA would represent an ideal assay to test whether Cdk3 is dependent on Hsp90. However, several tested commercial anti-Cdk3 antibodies failed to detect Cdk3. Therefore, we are currently establishing the tagged-Cdk3 protein expression system to perform biochemical binding assays aiming to validate our data that suggest that Cdk3 is a novel Hsp90-interacting kinase.
TRIM29 belongs to the TRIM protein family, which members are involved in cell proliferation, development, antiviral defense, oncogenesis, and apoptosis (Nisole et al. 2005). TRIM29 has multiple zinc finger motifs and a leucine zipper motif and therefore binds to nucleic acids. As TRIM29 is overexpressed in certain types of cancer and is associated with increased tumor size and poor survival, it may act as a transcriptional regulatory factor involved in carcinogenesis and proliferation (Kosaka et al. 2007). The interaction of Hsp90 with TRIM29 implies that the role of Hsp90 in transcription regulation is not restricted to association with known Hsp90-binding transcription factors, such as p53 and HSF-1, but is perhaps more general.
Identification of cytoskeletal proteins, ribosomal subunits, and metabolic and RNA-processing proteins strengthen the hypothesis that, besides the regulation of a specific set of proteins, Hsp90 has a central function in several fundamental cellular processes (McClellan et al. 2007; Lotz et al. 2008). Thus, the IP- and biotin-GA-mediated purification of structural proteins, including tubulin and kinesin, provides further evidence for the involvement of Hsp90 in the assembly of the tubulin-based cytoskeleton network, cytokinesis, and cellular transport (McClellan et al. 2007; Te et al. 2007). Isolation of RNA-binding proteins and ribosomal subunits points to the suggested role of Hsp90 in ribosomal subunit nuclear export and RNA processing and maintenance (Schlatter et al. 2002; Zhao et al. 2008). Similar to Falsone et al., we identified several metabolic enzymes (Falsone et al. 2005). The relevance of their interactions with Hsp90 remains obscure, as these proteins can interact with Hsp90 in a nonspecific manner due to their high abundance in the cytosol. At the same time, Hsp90, via its interaction with glycolytic enzymes, might control ATP production levels (Falsone et al. 2005).
To summarize, our complementary proteomic approaches identified 18 novel probable Hsp90 partner proteins, among which are Cdk3 kinase, TRIM29, and Tom34, providing an essential step towards our understanding of the role of Hsp90 system in the regulation of various cellular processes, such as folding of signal transduction proteins, transcription, and mitochondrial protein translocation.
Acknowledgments
S.R. was supported by a Marie-Curie-Excellent Grant of the EU, a VIDI grant of the Netherlands Organization for Science (NWO), and a High Potential Grant of Utrecht University. The authors thank the Netherlands Proteomics Center for financial support. The authors thank Dimitrios Argyropoulos for his assistance in experimental work.
Abbreviations
- ACN
acetonitrile
- Cdk3
cyclin-dependent kinase 3
- CNBr
cyanogen bromide
- DSP
Dithiobis[succinimidyl propionate]
- GA
geldanamycin
Footnotes
Concise summary
This paper reports on identification of 18 novel Hsp90 partners, including signaling kinase Cdk3, putative transcription factor TRIM29, and Tom34. Presented data expand the knowledge of the Hsp90 interactome and provide a further step in our understanding of the Hsp90 chaperone system.
References
- Ali MM, Roe SM, Vaughan CK, Meyer P, Panaretou B, Piper PW, Prodromou C, Pearl LH. Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex. Nature. 2006;440:1013–1017. doi: 10.1038/nature04716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan AJ, Mandal AK, Theodoraki MA. Molecular chaperones and protein kinase quality control. Trends Cell Biol. 2007;17:87–92. doi: 10.1016/j.tcb.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Chadli A, Felts SJ, Toft DO. GCUNC45 is the first Hsp90 co-chaperone to show alpha/beta isoform specificity. J Biol Chem. 2008;283:9509–9512. doi: 10.1074/jbc.C800017200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citri A, Harari D, Shohat G, Ramakrishnan P, Gan J, Lavi S, Eisenstein M, Kimchi A, Wallach D, Pietrokovski S, Yarden Y. Hsp90 recognizes a common surface on client kinases. J Biol Chem. 2006;281:14361–14369. doi: 10.1074/jbc.M512613200. [DOI] [PubMed] [Google Scholar]
- Crevel G, Bennet D, Cotteril S. The human TPR protein TTC4 is a putative Hsp90 co-chaperone which interacts with Cdc6 and shows alterations in transformed cells. PLoS One. 2008;3:1–10. doi: 10.1371/journal.pone.0001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falsone SF, Gesslbauer B, Tirk F, Piccinini A, Kungl AJ. A proteomic snapshot of the human heat shock protein 90 interactome. FEBS Lett. 2005;579:6350–6354. doi: 10.1016/j.febslet.2005.10.020. [DOI] [PubMed] [Google Scholar]
- Fan ACY, Bhangoo MK, Young JC. Hsp90 functions in the targeting and outer membrane translocation steps of Tom70-mediated mitochondrial import. J Biol Chem. 2006;281:33313–33324. doi: 10.1074/jbc.M605250200. [DOI] [PubMed] [Google Scholar]
- Kosaka Y, Inoue H, Ohmachi T, Yokoe T, Matsumoto T, Minori K, Tanaka F, Watanabe M, Mori M. Tripartite motif-containing 29 (TRIM29) is a novel marker for lymph node metastasis in gastric cancer. Ann Surg Oncol. 2007;14:2543–2549. doi: 10.1245/s10434-007-9461-1. [DOI] [PubMed] [Google Scholar]
- Lamphere L, Fiore F, Xu X, Brizuela L, Keezer S, Sardet C, Draetta GF, Gyuris J. Interaction between Cdc37 and Cdk4 in human cells. Oncogene. 1997;14:1999–2004. doi: 10.1038/sj.onc.1201036. [DOI] [PubMed] [Google Scholar]
- Lotz GP, Brychzy A, Heinz S, Obermann WM. J Cell Sci. 2008;121:717–723. doi: 10.1242/jcs.015610. [DOI] [PubMed] [Google Scholar]
- McClellan AJ, Xia Y, Deutschbauer AM, Davis RW, Gerstein M, Frydman J. Diverse cellular functions of the Hsp90 molecular chaperone uncovered using systems approaches. Cell. 2007;131:121–135. doi: 10.1016/j.cell.2007.07.036. [DOI] [PubMed] [Google Scholar]
- Millson SH, Truman AW, King V, Prodromou C, Pearl LH, Piper PW. A two-hybrid screen of the yeast proteome for Hsp90 interactors uncovers a novel Hsp90 chaperone requirement in the activity of a stress-activated mitogen-activated protein kinase, Slt2p (Mpk1p) Eukar Cell. 2005;4:849–860. doi: 10.1128/EC.4.5.849-860.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modi S, Stopeck AT, Gordon MS, Mendelson D, Solit DB, Bagatell R, Ma W, Wheler J, Rosen N, Norton L, Cropp GF, Johnson RG, Hannah AL, Hudis CA. Combination of trastuzumab and tanespimycin (17-AAG, KOS-953) is safe and active in trastuzumab-refractory HER-2 overexpressing breast cancer: a phase I dose-escalation study. J Clin Oncol. 2007;25(Modi, S., Stopeck, A.T., Gordon, M.S., Mendelson, D., Solit, D.B., Bagatell, R., Ma, W., Wheler, J., Rosen, N., Norton, L., Cropp, G.F., Johnson, R.G., Hannah, A.L., and Hudis, C.A):5410–5417. doi: 10.1200/JCO.2007.11.7960. [DOI] [PubMed] [Google Scholar]
- Nisole S, Stoye JP, Saib A. TRIM family proteins: retroviral restriction and antiviral defense. Nat Rev Microbiol. 2005;3:799–808. doi: 10.1038/nrmicro1248. [DOI] [PubMed] [Google Scholar]
- Pearl LH, Prodromou C, Workman P. The Hsp90 molecular chaperone: an open and shut case for treatment. Biochem J. 2008;410:439–453. doi: 10.1042/BJ20071640. [DOI] [PubMed] [Google Scholar]
- Pratt WB, Toft DO. Regulation of signaling protein function and trafficking by Hsp90/Hsp70-based chaperone machinery. Exp Biol Med. 2003;228:11–133. doi: 10.1177/153537020322800201. [DOI] [PubMed] [Google Scholar]
- Prince T, Matts RL. Definition of protein kinase sequence that trigger high affinity binding of Hsp90 and Cdc37. J Biol Chem. 2004;279:39975–39981. doi: 10.1074/jbc.M406882200. [DOI] [PubMed] [Google Scholar]
- Prince T, Sun L, Matts RL. Cdk2: a genuine protein kinase client of Hsp90 and Cdc37. Biochem. 2006;44:15287–15295. doi: 10.1021/bi051423m. [DOI] [PubMed] [Google Scholar]
- Ramalingam SS, Egorin MG, Ramanathan RK, Remick SC, Sikorski RP, Lagattuta TF, Chatta GS, Friedland dM, Stoller RG, Potter DM, Ivy SP, Belani CP. A phase I study of 17-allylamino-17-demethoxygeldanamycin combined with paclitaxel in patients with advanced solid malignancies. Clin Cancer Res. 2008;14:3456–3461. doi: 10.1158/1078-0432.CCR-07-5088. [DOI] [PubMed] [Google Scholar]
- Richter K, Muschler P, Hainzl O, Reinstein J, Buchner J. Sti1 is a non-competitive inhibitor of the Hsp90 ATPase. J Biol Chem. 2003;278:10328–10333. doi: 10.1074/jbc.M213094200. [DOI] [PubMed] [Google Scholar]
- Roe SM, Ali MMU, Meyer P, Vaughan CK, Panaretou B, Piper PW, Prodromou C, Pearl L. The mechanism of Hsp90 regulation by the protein kinase-specific cochaperone p50cdc37. Cell. 2004;116:87–98. doi: 10.1016/S0092-8674(03)01027-4. [DOI] [PubMed] [Google Scholar]
- Rüdiger S, Freund SM, Verprintsev DB, Fersht AR. CRINEPT-TROSY NMR reveals p53 core domain bound in an unfolded form to the chaperone Hsp90. Proc Natl Acad Sci U S A. 2002;99:11085–11090. doi: 10.1073/pnas.132393699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlatter H, Langer T, Rosmus S, Onneken ML, Fasold H. A novel function for the Hsp90 kDa heat-shock protein (Hsp90): facilitating nuclear export of 60 S ribosomal subunits. Biochem J. 2002;362:675–684. doi: 10.1042/0264-6021:3620675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegel T, Mirus O, Haeseler A, Schleiff E. The tetratricopeptide repeats of receptors involved in protein translocation across membranes. Mol Biol Evol. 2007;24:2763–2774. doi: 10.1093/molbev/msm211. [DOI] [PubMed] [Google Scholar]
- Schumacher JA, Crockett DK, Elenitoba-Johnson KSJ, Lim MS. Proteome-wide changes induced by the Hsp90 inhibitor, geldanamycin in anaplastic large cell lymphoma cells. Proteomics. 2007;7:2603–2616. doi: 10.1002/pmic.200700108. [DOI] [PubMed] [Google Scholar]
- Shiau AK, Harris SF, Southworth DR, Agard DA. Structural analysis of E. coli hsp90 reveals dramatic nucleotide-dependent conformational rearrangements. Cell. 2006;127:329–340. doi: 10.1016/j.cell.2006.09.027. [DOI] [PubMed] [Google Scholar]
- Sreedhar AS, Kalmar E, Csermely P, Shen YF. Hsp90 isoforms: functions, expression and clinical importance. FEBS Lett. 2004;562:11–15. doi: 10.1016/S0014-5793(04)00229-7. [DOI] [PubMed] [Google Scholar]
- Stebbins CE, Russo AA, Schneider C, Rosen N, Hartl FU, Pavletich NP. Crystal structure of an Hsp90-geldanamycin complex: targeting of a protein chaperone by an antitumor agent. Cell. 1997;89:239–250. doi: 10.1016/S0092-8674(00)80203-2. [DOI] [PubMed] [Google Scholar]
- Stepanova L, Leng X, Parker SB, Harper JW. Mammalian p50cdc37 is a protein kinase-targeting subunit of Hsp90 that binds and stabilizes Cdk4. Gen Develop. 1996;10:1491–1502. doi: 10.1101/gad.10.12.1491. [DOI] [PubMed] [Google Scholar]
- Te J, Jia L, Rogers J, Miller A, Hartson SD. Novel subunits of the mammalian Hsp90 signal transduction chaperone. J Prot Res. 2007;6:1963–1973. doi: 10.1021/pr060595i. [DOI] [PubMed] [Google Scholar]
- Terasawa K, Yoshimatsu K, Iemura S, Natsume T, Tanaka K, Minami Y. Cdc37 interacts with the glycine-rich loop of Hsp90 client kinases. Mol Cell Biol. 2006;26:3378–3389. doi: 10.1128/MCB.26.9.3378-3389.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan CK, Gohlke U, Sobott F, Good VM, Ali MMU, Prodromou C, Robinson CV, Saibil HR, Pearl LH. Structure of an Hsp90–Cdc37–Cdk4 complex. Mol Cell. 2006;23:697–707. doi: 10.1016/j.molcel.2006.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein IB, Joe AK. Mechanisms of disease: oncogene addiction—a rationale for molecular targeting in cancer therapy. Nat Clin Pract Oncol. 2006;3:448–457. doi: 10.1038/ncponc0558. [DOI] [PubMed] [Google Scholar]
- Picard D (2008) Hsp90 interactors. http://www.picard.ch/downloads/Hsp90interactors.pdf
- Yang CS, Weiner H. Yeast two-hybrid screening identifies binding partners of human Tom34 that have ATPase activity and form a complex with Tom34 in the cytosol. Arch Biochem Biophys. 2002;400:105–110. doi: 10.1006/abbi.2002.2778. [DOI] [PubMed] [Google Scholar]
- Ye X, Zhu C, Harper JW. A premature-termination mutation in the Mus musculus cyclin-dependent kinase 3 gene. Proc Natl Acad Sci U S A. 2001;98:1682–1686. doi: 10.1073/pnas.041596198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JC, Obermann WMJ, Hartl FU. Specific binding of tetratricopeptide repeat proteins to the C-terminal 12-kDa domain of Hsp90. J Biol Chem. 1998;273:18007–18010. doi: 10.1074/jbc.273.29.18007. [DOI] [PubMed] [Google Scholar]
- Young JC, Hoogenraad NJ, Hartl FU. Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell. 2003;112:41–50. doi: 10.1016/S0092-8674(02)01250-3. [DOI] [PubMed] [Google Scholar]
- Zhao R, Davey M, Hsu Y, Kaplanek P, Tong A, Parsons AB, Krogan N, Cagney G, Mai D, Greenblatt J, Boone C, Emili A, Houry WA. Navigating the chaperone network: an integrative map of physical and genetic interactions mediated by the Hsp90 chaperone. Cell. 2005;120:715–727. doi: 10.1016/j.cell.2004.12.024. [DOI] [PubMed] [Google Scholar]
- Zhao R, Kakihara Y, Gribun A, Huen J, Yang G, Khanna M, Costanzo M, Brost RL, Boone C, Hughes TR, Yip CM, Houry WA. Molecular chaperone Hsp90 stabilizes Pih1/Nop17 to maintain R2TP complex activity that regulates snoRNA accumulation. J Cell Biol. 2008;180:563–578. doi: 10.1083/jcb.200709061. [DOI] [PMC free article] [PubMed] [Google Scholar]