Abstract
Cullin-RING ubiquitin ligases promote the polyubiquitination and degradation of many important cellular proteins, which previous studies indicated can be targeted for degradation via interaction with BTB domain-containing subunits of this E3 ligase complex. PEST domains are known to promote the degradation of proteins that contain them. However, the molecular mechanism by which PEST sequences promote degradation of these proteins is not understood. Here we show that the PEST sequences of a short-lived protein called HSF2 interact with Cullin3, a subunit of a Cullin-RING E3 ubiquitin ligase, and that this interaction mediates the Cul3-dependent ubiquitination and degradation of HSF2. These results indicate how, at the molecular level, PEST sequences can promote the proteolysis of proteins that contain them. They also expand understanding of the mechanisms by which substrates can be recruited to Cullin-RING E3 ubiquitin ligases to include interactions between PEST sequences and Cul3.
Keywords: HSF2, Cul3, PEST, Ubiquitin, Protein turnover
Introduction
Cul3, a member of the Cullin family of proteins, is a subunit of a Cullin-RING ubiquitin E3 ligase complex that polyubiquitinates many important proteins, leading to their degradation by the proteasome (Bosu and Kipreos 2008; Hotton and Callis 2008; Petroski and Deshaies 2005; Sumara et al. 2008). Cul3-containing ubiquitin ligases have been found to play important roles in the regulation of mitosis, development, cytoskeletal proteins, and transcription factors (Pintard et al. 2004; Sumara et al. 2008; Willems et al. 2004).
Cul3 plays an important role in the organization of this complex, interacting not only with the RING protein Roc1 that associates with the E2 ubiquitinating enzyme but also with a large number of different BTB proteins that bind substrates and thereby recruit them to the complex. PEST domains, sequences enriched in proline (P), glutamate (E), serine (S), and threonine (T), are known to promote the degradation of proteins that contain them (Rechsteiner 1991; Rechsteiner and Rogers 1996; Rogers et al. 1986). However, the underlying molecular mechanism by which PEST sequences mediate increased protein turnover is not clear.
HSF2 is a DNA-binding protein that interacts with heat shock elements in the promoters of the hsp70 and other heat shock protein genes. One function of HSF2 is to mediate an epigenetic mechanism called gene bookmarking, in which compaction of the hsp70 promoter is inhibited during mitosis so that this gene can be transcribed as soon as G1 begins in the event of cell stress (Sarge and Park-Sarge 2005; Xing et al. 2005). HSF2 has a rapid rate of degradation by the proteasome, with a half-life of less than 60 min (Mathew et al. 2001; Mathew et al. 1998; Pirkkala et al. 1999). However, the underlying molecular mechanism that mediates the rapid turnover of HSF2 is not known.
The results of this study show that PEST sequences present within HSF2 contribute to the rapid turnover of this protein, and that they do so by interacting with the Cul3 subunit of the Cul3-RING E3 ubiquitin ligase, thereby promoting the ubiquitination of HSF2 and its subsequent degradation by the proteasome. Thus, these results provide an explanation for how PEST sequences can mediate the turnover of proteins that contain them. They also reveal a new mechanism by which substrates can be recruited to Cullin-RING E3 ubiquitin ligase complexes by showing that it can be mediated by PEST sequence interaction with Cul3, and not only by substrate interaction with BTB domain-containing subunits of these ubiquitin ligases.
Materials and methods
Yeast two-hybrid and β-galactosidase assay
An HSF2 “bait” construct consisting of full-length HSF2 inserted in-frame into the vector pGBD-C1 was transformed into yeast strain pJ69-4A. The resulting strain was then transformed with a mouse whole embryo cDNA library (Hollenberg et al. 1995). To confirm the interaction, the pGBD-HSF2 and pVP16 plasmid containing the partial Cul3 cDNA, referred to as pVP16-Cul3 (161-293), were transformed back into yeast and the ability of HSF2 and the Cul3 clone to interact was determined by growth on selective media lacking adenine or histidine. For assay of interaction strength using β-galactosidase activity yeast extracts were incubated with Z Buffer (60 mM Na2HPO4 and 40 mM NaH2PO4 (pH 7.0), 10 mM KCl, 1 mM MgSO4, 50 mM mercaptoethanol). After addition of 4 mg/ml o-nitrophenyl-β-d-galactoside substrate, samples were incubated at 30°C for 30 min and then the OD420 was measured.
Cell culture and transfection
HeLa cells were cultured at 37°C with 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum, and 50 µg/ml gentamycin until 80% confluent. HeLa cells were transfected with pEGFP-HSF2, pEGFP-HSFΔP1 (HSF2 lacking amino acids 241–309), pEGFP-HSFΔP2 (HSF2 lacking amino acids 479–499), or pGFP-HSFΔP12 (HSF2 lacking amino acids 241–309 and 479–499) along with empty pEGFP vector, using Effectene (Qiagen). Mouse embryonic Cul3+/− MEF cells (provided by Dr. Jeffrey Singer, Brown University, RI, USA) were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% FBS and antibiotics, 10 mM nonessential amino acids, 0.96 µl of β-mercaptoethanol/100 ml at 37°C with 5% CO2.
Measurement of HSF2 turnover
HeLa cells were transfected with pGFP-HSF2 wild type or pGFP-HSF2 ΔP12. One day after transfection, the cells were then treated with cycloheximide at a concentration of 10 μM for the indicated times. Cells were lysed using RIPA buffer (150 mM NaCl, 50 mM Tris–HCl pH 7.4, 1% NP-40, 0.2% SDS, 0.25% sodium deoxycholate, and 1 mM EDTA) with 1× protease inhibitor cocktail (Roche). The cell lysates were then separated by SDS-PAGE and subjected to Western blot assay using a 1:2,000 dilution of an anti-mouse monoclonal anti-GFP antibody (Clontech), and anti-mouse secondary antibodies (Zymed). Detection was done using enhanced chemiluminescence assay (Pierce), and quantitation of HSF2 band intensities was performed using ImageQuant software.
In vitro binding assay
This was done using our previously established procedures (Sarge and Park-Sarge 2005; Xing et al. 2005). For expression of recombinant proteins, pGEX-HSF2 and control pGEX-2T were transformed into BL21 (DE3) competent cells. The recombinant protein expression was induced by addition of 1 mM isopropyl-1-thio-β-d-glactopyranoside. The bacteria were lysed by sonication in PBS containing 0.5% Triton X-100 and complete mini protease inhibitor mixture (Roche Applied Science). GST-HSF2, GST-P12, or GST bound to glutathione-agarose beads were incubated with 35S-labeled in vitro translated Cul3 for 2 h at 4°C. After washing, bound proteins were analyzed by SDS-PAGE and autoradiography to detect the 35S-labeled Cul3 proteins. Amounts of GST and GST-fusion proteins bound to the beads were determined by SDS-PAGE followed by Western blot using goat polyclonal anti-GST antibody (Amersham).
Immunoprecipitation and western blot assay
This was done following our previously established procedures (Sarge and Park-Sarge 2005; Xing et al. 2005). For co-immunoprecipitation experiments, PBS-washed HeLa cells or HeLa cells transfected with the indicated expression vectors were lysed on ice with lysis buffer (25 mM Tris–HCl pH 8.0, 150 mM NaCl, 10% glycerol, 0.5% NP-40, 2 mM EDTA, 1 mM NaF, 10 mM β-glycerophosphate, 5 mM N-ethylmaleimide, and complete protease inhibitor (Roche Molecular Biochemicals)) for 15 min. Lysates were then cleared by centrifugation at 13,000 rpm for 10 min at 4°C. Soluble lysates were then precleared by incubation with non-specific (control) rabbit IgG and protein G-sepharose beads for 1 h at 4°C with gentle rotation. Precleared extracts were then incubated with a 1:200 dilution of rabbit anti-Cul3 antibodies (gift of Dr. Yue Xiong (Furukawa et al. 2003)) or control IgG (Sigma) and 20 µL of a 50% slurry of protein G-sepharose for 1 h at 4°C with rotation. After washing beads three times for 5 min each at 4°C with lysis buffer containing 300 mM NaCl, immunoprecipitated proteins were analyzed by SDS-PAGE followed by Western blot assay using a 1:200 dilution of anti-HSF2 rat monoclonal antibody (3E2, Neomarkers) or anti-GFP mouse monoclonal antibody (Clontech), and anti-rat or anti-mouse secondary antibodies, respectively (Zymed). Detection was done using enhanced chemiluminescence assay (Pierce), and quantitation of band intensities was performed using ImageQuant software.
In vivo ubiquitination assay
MEF cells were grown in DMEM and the cells were treated with 10 µM MG132 for 4 h to inhibit proteasome function. The cells were lysed in RIPA immunoprecipitation buffer (25 mM Tris–HCl (pH 8.0), 150 mM NaCl, 10% glycerol, 1% NP-40, 0.25% deoxycholic acid, 2 mM EDTA, 1 mM NaF, 10 mM β-glycerophosphate, with 5 mM N-ethylmaleimide). Lysates were cleared by centrifugation and immunoprecipitation was performed using HSF2 goat polyclonal antibody (Bethyl Laboratories), or anti-GFP antibodies for the experiments analyzing GFP-HSF2 proteins transfected into cells. The ubiquitination status of the HSF2 or GFP-HSF2 proteins was then analyzed by Western blot assay using a 1:1,000 dilution of a mouse monoclonal anti-ubiquitin antibody (Sigma), and anti-mouse secondary antibodies (Zymed). Detection was done using enhanced chemiluminescence assay (Pierce).
Statistical analysis
This was performed using the Student’s t test. A P value of ≤0.05 was considered a significant difference.
Results
Interaction between HSF2 and Cul3
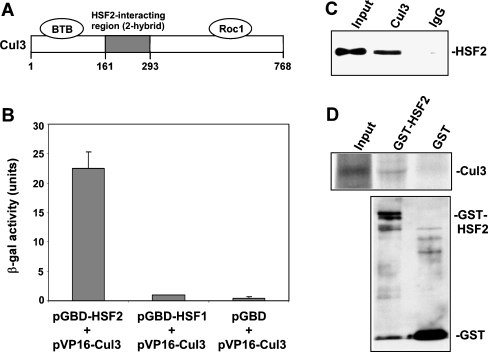
From a yeast two-hybrid screen for HSF2 partner proteins we identified an interaction between this factor and Cul3, with the interacting clone comprising amino acids 161–293 of the Cul3 protein (Fig. 1a). The interaction of Cul3 with HSF2 was confirmed using a β-galactosidase activity assay of yeast harboring the two plasmids (Fig. 1b). The results of this experiment also showed that this Cul3 clone does not interact with HSF1, a protein with high sequence-relatedness to HSF2 (Rabindran et al. 1991; Sarge et al. 1991; Schuetz et al. 1991), indicating the specificity of the interaction between HSF2 and Cul3. Co-immunoprecipitation analysis using extracts of HeLa cells demonstrates that endogenous HSF2 and Cul3 proteins interact (Fig. 1c). As an independent test of the interaction between HSF2 and Cul3, and also to determine whether the interaction between these two proteins is direct, in vitro binding experiments were performed in which 35S-labeled in vitro translated Cul3 were incubated with GST-HSF2 or GST bound to glutathione-agarose beads. The results of this experiment demonstrate that Cul3 is able to interact specifically and directly with HSF2 in vitro (Fig. 1d).
Fig. 1.
HSF2 interacts with Cul3. a The segment of Cul3 identified as an HSF2-interacting clone comprises amino acids 161–293 of this protein. Also shown are the Cul3 regions that interact with BTB domain proteins and Roc1. b β-galactosidase activity was measured in extracts of yeast strain pJ69-4A transformed with a pVP16 plasmid containing amino acids 161–293 of Cul3 and pGBD-HSF2, pGBD-HSF1, or empty pGBD-C1 plasmid. Data are presented as means ±SEM (n = 3). c Extracts of HeLa cells were subjected to immunoprecipitation with antibodies to Cul3 or non-specific IgG, followed by Western blot using HSF2 antibodies. d35S-labeled in vitro translated Cul3 was incubated with GST-HSF2 and GST bound to glutathione-agarose beads. After washing, the amount of bound 35S-labeled Cul3 was determined by SDS-PAGE and autoradiography (upper panel). The amounts of GST-HSF2 and GST bound to the beads were determined by anti-GST Western blot (lower panel)
Cul3 is involved in HSF2 turnover and ubiquitination
The data presented above showing interaction between HSF2 and Cul3, coupled with the known role of this cullin in protein turnover, suggested that Cul3 may be involved in the instability of the HSF2 protein. To test this hypothesis, we examined the effect of reducing levels of Cul3 on the amounts of HSF2 in cells. In one set of experiments we utilized Cul3+/+ vs. Cul3+/− MEF cells (McEvoy et al. 2007), and performed Western blot assays to determine whether HSF2 levels are lower in the Cul3+/+ vs. Cul3+/− MEF cells, as predicted by our hypothesis. We used Cul3+/− cells for this experiment because Cul3−/− MEF cells exhibit significant loss of viability (McEvoy et al. 2007). Consistent with our hypothesis, the results indicate that reduction of Cul3 is associated with increased HSF2 protein levels (Fig. 2a), suggesting a role for Cul3-dependent proteolysis in HSF2 turnover.
Fig. 2.
HSF2 instability and ubiquitination are dependent on Cul3. a Extracts of Cul3+/+ vs. Cul3+/− MEF cells were subjected to Western blot using antibodies against HSF2, Cul3, and β-actin. b Cul3+/+ and Cul3+/− MEF cells were treated with MG132 for 4 h, and then HSF2 was immunoprecipitated from extracts of these cells followed by anti-ubiquitin Western blot assay. Positions of molecular weight markers are indicated to the left of the panel. c Extracts of cells transfected with scrambled (negative control) siRNA or Cul3 siRNA were subjected to Western blot using antibodies against HSF2, Cul3, and β-actin. d Cells transfected with scrambled (negative control) siRNA or Cul3 siRNA were treated with MG132 for 4 h, and then HSF2 was immunoprecipitated from extracts of these cells followed by anti-ubiquitin Western blot assay. Positions of molecular weight markers are indicated to the left of the panel
Cullin-dependent ubiquitin ligases target proteins to the 26S proteasome by catalyzing the formation of ubiquitin chains on these proteins. Therefore, we next tested whether the increase in HSF2 levels in Cul3+/− vs. Cul3+/+ cells is associated with decreased polyubiquitination of HSF2. Cul3+/+ and Cul3+/− MEF cells were treated with the proteasome inhibitor MG132 to allow accumulation of polyubiquitinated proteins, and then extracts of these cells were subjected to immunoprecipitation with HSF2 antibodies followed by anti-ubiquitin Western blot assay to determine the amounts of ubiquitinated HSF2. The results, shown in Fig. 2b, demonstrate that the amount of HSF2 polyubiquitination for Cul3+/− cells is significantly lower than that observed for Cul3+/+ cells. These results support the hypothesis that Cul3 is involved in HSF2 polyubiquitination and degradation.
As a complementary approach for testing the hypothesis that reduced Cul3 protein level results in higher HSF2 protein level, we also performed HSF2 Western blot assay of extracts of cells that had been transfected with scrambled negative control siRNA or Cul3 siRNA. The results, shown in Fig. 2c, indicate that HSF2 levels are indeed higher in the extracts of the Cul3 siRNA-treated cells compared to those of cells transfected with the scrambled control siRNA. The data in Fig. 2d show that, as predicted by our hypothesis, treatment of cells with the Cul3 siRNA results in decreased amounts of HSF2 polyubiquitination.
PEST sequences cause instability of HSF2
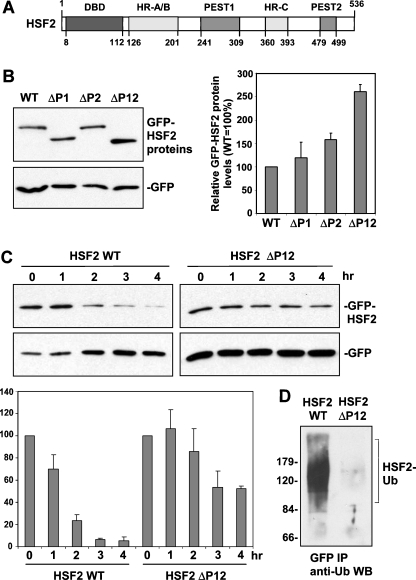
Analysis of the HSF2 sequence using the PESTFind algorithm reveals the presence of two putative PEST domains, comprising amino acids 241–309 (hereafter called PEST1) and 479–499 (PEST2) of this protein (Fig. 3a; Rechsteiner and Rogers 1996; https://emb1.bcc.univie.ac.at/toolbox/pestfind/). The PESTFind scores for the HSF2 PEST1 and PEST2 sequences are 6.52 and 9.58, respectively. Based on previous data indicating the role of PEST domains in promoting protein turnover (Rechsteiner and Rogers 1996; Rogers et al. 1986), we hypothesized that one or both of these PEST domains could be involved in the instability of HSF2. To test this hypothesis we transfected HeLa cells with expression plasmids encoding GFP-fusions of wild-type HSF2 and HSF2 in which PEST1, PEST2, or both PEST1 and PEST2, were deleted. The cells were also transfected with a construct encoding GFP alone to allow normalization for any potential differences in transfection efficiency. As shown in Fig. 3b, the results of this experiment indicate that deletion of either PEST domain is associated with a small increase in levels of the GFP-HSF2 proteins, and that deletion of both PEST domains results in a larger increase in HSF2 protein levels than that observed for deletion of either PEST domain alone. These results suggest that both PEST domains contribute to the instability of HSF2.
Fig. 3.
Role of two PEST domains in HSF2 instability. a Schematic showing location of two PEST domains in HSF2 relative to the DNA-binding domain (DBD), Heptad Repeat oligomerization domain (HR-A/B), and Heptad Repeat C (HR-C). b Extracts of cells transfected with GFP fusion constructs of wild type, PEST domain 1-deleted (ΔP1), PEST domain 2-deleted (ΔP2), and PEST domain 1- and 2-deleted (ΔP12) HSF2, along with GFP alone expression plasmid (normalizing control) were subjected to Western blot using anti-GFP antibodies. For the graph shown to the right of this data, bands of the different GFP-HSF2 fusion proteins from replicates of this experiment were quantified, normalized to values of the GFP alone bands, and then graphed as percent relative to that of wild-type HSF2 (which was set to 100%). Data are presented as means ±SEM (n = 3). c HeLa cells were transfected with the GFP-HSF2 wild-type or the GFP-HSF2 construct lacking the PEST domains along with GFP alone expression plasmid (normalizing control), treated with cycloheximide to block translation, and then extracts of cells collected at the indicated time points after addition of cycloheximide were subjected to anti-GFP Western blot. For the graph shown below this data, bands of GFP-HSF2 fusion proteins from replicates of this experiment were quantified, normalized to values of GFP bands, and graphed as percent of wild-type HSF2 in the absence of cycloheximide (set to 100%). Data are presented as means ± SEM (n = 3). d Cells transfected with GFP-HSF2 wild-type and GFP-HSF2 lacking the PEST domains were treated with MG132 for 4 h, and then extracts of these cells were subjected to anti-GFP immunoprecipitation followed by anti-ubiquitin Western blot assay. Positions of molecular weight markers are indicated to the left of the panel
To support these conclusions implicating a role for these PEST sequence in HSF2 turnover, we transfected cells with the GFP-HSF2 wild-type construct or the GFP-HSF2 construct lacking the PEST sequences, added cycloheximide to prevent translation, collected cells at various time points, and then extracts of these cells were subjected to anti-GFP Western blot assay in order to compare the turnover rates of each protein. The results of this analysis confirm that the HSF2 protein lacking the PEST domains has a significantly lower turnover rate than the wild-type HSF2 protein (Fig. 3c).
Based on previous studies, we hypothesized that these PEST sequences contribute to HSF2 instability by promoting the polyubiquitination of this protein (Rechsteiner and Rogers 1996). We tested this by comparing the amount of polyubiquitination of GFP-HSF2 wild-type vs. GFP-HSF2 lacking the PEST domains, using anti-GFP immunoprecipitation of extracts of cells transfected with these constructs followed by anti-ubiquitin Western blot assay. The results of this experiment, shown in Fig. 3d, indicate that the amount of polyubiquitination of HSF2 lacking the PEST sequences is indeed reduced compared to wild-type HSF2.
PEST sequences directly interact with Cul3
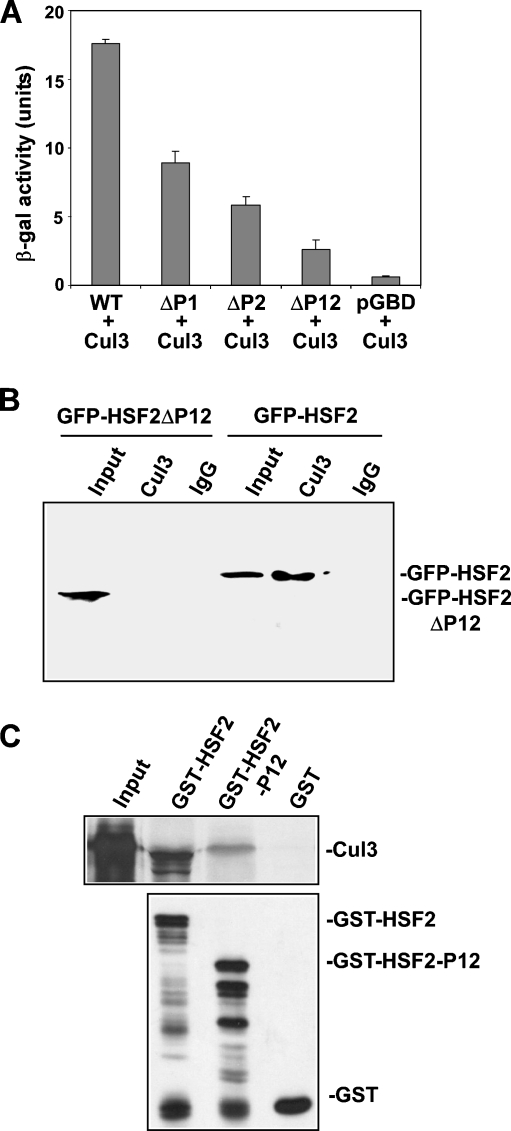
The results shown above indicate that HSF2 interacts with Cul3 and that it contains PEST domains that are involved in instability of the HSF2 protein. Based on this data, we hypothesized that HSF2 interacts with Cul3 via these PEST domains. As a first test of this hypothesis, we performed yeast two-hybrid assay to determine whether deleting the PEST domains affects the ability of HSF2 to interact with Cul3. The results indicate that deleting either PEST1 or PEST2 leads to a significant decrease in HSF2 interaction with Cul3, which is further decreased when both PEST domains are deleted (Fig. 4a). To test this hypothesis in the context of proteins expressed in mammalian cells, extracts of cells transfected with GFP-HSF2 wild-type or GFP-HSF2 lacking the PEST domains were subjected to immunoprecipitation using Cul3 antibodies followed by anti-GFP Western blot assay. The results of this experiment indicate that Cul3 interaction with HSF2 lacking the PEST sequences is significantly lower than with wild-type HSF2 (Fig. 4b).
Fig. 4.
PEST sequences interact with Cul3. a HSF2 interacts with Cul3 in yeast two-hybrid assay. β-galactosidase activity was measured in extracts of yeast carrying the pVP16-Cul3(161-293) plasmid and pGBD-HSF2 wild-type plasmid, pGBD-HSF2 in which one or both PEST domains were deleted, or empty pGBD-C1. Data are presented as means ± SEM (n = 3). b Extracts of cells transfected with GFP-HSF2 wild-type and GFP-HSF2 lacking the PEST domains were immunoprecipitated with anti-Cul3 or non-specific IgG antibodies followed by anti-GFP Western blot. c35S-labeled in vitro translated Cul3 was incubated with GST-HSF2, GST-HSF2-P12 (GST fused to HSF2 sequence containing PEST domains 1 and 2), or GST bound to glutathione-agarose beads. After washing, amounts of bound 35S-labeled Cul3 were determined by SDS-PAGE and autoradiography (upper panel). Amounts of GST-HSF2, GST-HSF2-P12, and GST bound to beads were measured by anti-GST Western blot (lower panel)
The results shown in Fig. 4a and b indicate that deletion of the PEST sequences is associated with loss of HSF2 interaction with Cul3, consistent with the hypothesis that Cul3 interacts with HSF2 via these PEST sequence. To strengthen support for this conclusion, as well as to test whether Cul3 binds directly to these PEST sequences, we performed an in vitro binding experiment to examine the interaction between in vitro translated Cul3 and GST-HSF2 as well as with a protein containing GST fused to the HSF2 sequence containing both PEST domains 1 and 2 (GST-HSF2-P12), comprising HSF2 amino acids 241–499. The results of this experiment, shown in Fig. 4c, indicate that Cul3 does interact specifically with GST-P12. The data shown in Fig. 4 suggest that Cul3 interacts with the HSF2 protein via its sequence containing PEST domains 1 and 2. Because the interaction of Cul3 with GST-HSF2-P12 was somewhat lower than that with full-length GST-HSF2, we cannot rule out the possibility that there may be other sequences in HSF2 that contribute to Cul3 binding, but this could also be a consequence of structural/folding differences due to the P12 region being removed from the context of the intact HSF2 structure.
Discussion
The results presented in this paper indicate that the Cul3 subunit of the Cul3-RING E3 ubiquitin ligase complex interacts with the PEST sequences within the HSF2 protein, and that this contributes to the ubiquitination and proteasomal degradation of this protein. These results elucidate a mechanism that contributes to the short half-life of the HSF2 protein. More importantly, they provide an explanation for how PEST sequences can promote the degradation of proteins that contain them, namely, by interacting with Cul3 to recruit these proteins to Cul3-RING E3 ubiquitin ligases.
The results also reveal a new mechanism by which substrates can be recruited to Cullin-RING E3 ubiquitin ligases. It has previously been shown that this can be mediated by interaction between substrate proteins and BTB domain-containing subunits of the ubiquitin ligase complexes (Bosu and Kipreos 2008; Hotton and Callis 2008; Petroski and Deshaies 2005; Sumara et al. 2008). However, our results indicate that recruitment can also occur by interaction between PEST sequences in a substrate protein, in this case HSF2, and the Cul3 subunit of the Cul3-RING ligase complexes. Related to this point, since the location of the HSF2-interacting region of Cul3 found in the yeast two-hybrid screen is distinct from the Cul3 region that interacts with BTB domain substrate adapter proteins, we note the possibility that a BTB protein(s) may also interact with HSF2 and thereby contribute to HSF2 recruitment to the ubiquitin ligase complex. However, our results do indicate that the interaction between PEST sequences in HSF2 and Cul3 plays a role in the instability of the HSF2 protein.
In summary, the results of this study have revealed a mechanism by which PEST domains can promote protein degradation as well as expanded understanding of the mechanisms by which Cullin-RING E3 ubiquitin ligases recruit substrates. One interesting question for future studies is whether there are cases where the interaction between HSF2 and Cul3 is regulated as a means for controlling HSF2 protein levels. For example, HSF2 levels are regulated during development in both a temporal and cell type-dependent manner (Akerfelt et al. 2007; Brown and Gozes 1998; Morange et al. 1998; Pirkkala et al. 2001). Thus, future studies could address whether regulation of HSF2 turnover via alterations in its Cul3 interactions may contribute, at least to some extent, to some of the cases of developmental changes in HSF2 levels.
Acknowledgments
We are grateful to Dr. Jeffrey Singer for his gift of the Cul3+/+ and Cul3+/− MEF cells, to Dr. Yue Xiong for providing anti-Cul3 antibodies, and to other members of the laboratory for comments. This research was supported by NIH grant GM64606 to K.D.S.
References
- Akerfelt M, Trouillet D, Mezger V, Sistonen L. Heat shock factors at a crossroad between stress and development. Ann N Y Acad Sci. 2007;1113:15–27. doi: 10.1196/annals.1391.005. [DOI] [PubMed] [Google Scholar]
- Bosu DR, Kipreos ET. Cullin-RING ubiquitin ligases: global regulation and activation cycles. Cell Div. 2008;3:7–19. doi: 10.1186/1747-1028-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown IR, Gozes I. Stress genes in the nervous system during development and aging diseases. Ann N Y Acad Sci. 1998;851:123–128. doi: 10.1111/j.1749-6632.1998.tb08985.x. [DOI] [PubMed] [Google Scholar]
- Furukawa M, He YJ, Borchers C, Xiong Y. Targeting of protein ubiquitination by BTB-Cullin 3-Roc1 ubiquitin ligases. Nat Cell Biol. 2003;5:1001–1007. doi: 10.1038/ncb1056. [DOI] [PubMed] [Google Scholar]
- Hollenberg SM, Sternglanz R, Cheng PF, Weintraub H. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol Cell Biol. 1995;15:3813–3822. doi: 10.1128/mcb.15.7.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotton SK, Callis J. Regulation of cullin RING ligases. Annu Rev Plant Biol. 2008;59:467–489. doi: 10.1146/annurev.arplant.58.032806.104011. [DOI] [PubMed] [Google Scholar]
- Mathew A, Mathur SK, Morimoto RI. Heat shock response and protein degradation: regulation of HSF2 by the ubiquitin-proteasome pathway. Mol Cell Biol. 1998;18:5091–5098. doi: 10.1128/mcb.18.9.5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew A, Mathur SK, Jolly C, Fox SG, Kim S, Morimoto RI. Stress-specific activation and repression of heat shock factors 1 and 2. Mol Cell Biol. 2001;21:7163–7171. doi: 10.1128/MCB.21.21.7163-7171.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEvoy JD, Kossatz U, Malek N, Singer JD. Constitutive turnover of cyclin E by Cul3 maintains quiescence. Mol Cell Biol. 2007;27:3651–3666. doi: 10.1128/MCB.00720-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morange M, Favet N, Loones MT, Manuel M, Mezger V, Michel E, Rallu M, Sage J. Heat-shock genes and development. Ann N Y Acad Sci. 1998;851:117–122. doi: 10.1111/j.1749-6632.1998.tb08984.x. [DOI] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- Pintard L, Willems A, Peter M. Cullin-based ubiquitin ligases: Cul3-BTB complexes join the family. EMBO J. 2004;23:1681–1687. doi: 10.1038/sj.emboj.7600186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirkkala L, Alastalo TP, Nykanen P, Seppa L, Sistonen L. Differentiation lineage-specific expression of human heat shock transcription factor 2. FASEB J. 1999;13:1089–1098. doi: 10.1096/fasebj.13.9.1089. [DOI] [PubMed] [Google Scholar]
- Pirkkala L, Nykanen P, Sistonen L. Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. FASEB J. 2001;15:1118–1131. doi: 10.1096/fj00-0294rev. [DOI] [PubMed] [Google Scholar]
- Rabindran SK, Giorgi G, Clos J, Wu C. Molecular cloning and expression of a human heat shock factor, HSF1. Proc Natl Acad Sci U S A. 1991;88:6906–6910. doi: 10.1073/pnas.88.16.6906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechsteiner M. Natural substrates of the ubiquitin proteolytic pathway. Cell. 1991;66:615–618. doi: 10.1016/0092-8674(91)90104-7. [DOI] [PubMed] [Google Scholar]
- Rechsteiner M, Rogers SW. PEST sequences and regulation by proteolysis. Trends Biochem Sci. 1996;21:267–271. [PubMed] [Google Scholar]
- Rogers S, Wells R, Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science. 1986;234:364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- Sarge KD, Park-Sarge OK. Gene bookmarking: keeping the pages open. Trends Biochem Sci. 2005;30:605–610. doi: 10.1016/j.tibs.2005.09.004. [DOI] [PubMed] [Google Scholar]
- Sarge KD, Zimarino V, Holm K, Wu C, Morimoto RI. Cloning and characterization of two mouse heat shock factors with distinct inducible and constitutive DNA-binding ability. Genes Dev. 1991;5:1902–1911. doi: 10.1101/gad.5.10.1902. [DOI] [PubMed] [Google Scholar]
- Schuetz TJ, Gallo GJ, Sheldon L, Tempst P, Kingston RE. Isolation of a cDNA for HSF2: evidence for two heat shock factor genes in humans. Proc Natl Acad Sci U S A. 1991;88:6911–6915. doi: 10.1073/pnas.88.16.6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumara I, Maerki S, Peter M. E3 ubiquitin ligases and mitosis: embracing the complexity. Trends Cell Biol. 2008;18:84–94. doi: 10.1016/j.tcb.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Willems AR, Schwab M, Tyers M. A hitchhiker’s guide to the cullin ubiquitin ligases: SCF and its kin. Biochim Biophys Acta. 2004;1695:133–170. doi: 10.1016/j.bbamcr.2004.09.027. [DOI] [PubMed] [Google Scholar]
- Xing H, Wilkerson DC, Mayhew CN, Lubert EJ, Skaggs HS, Goodson ML, Hong Y, Park-Sarge OK, Sarge KD. Mechanism of hsp70i gene bookmarking. Science. 2005;307:421–423. doi: 10.1126/science.1106478. [DOI] [PubMed] [Google Scholar]