Abstract
In response to terminal differentiation signals that enable B cells to produce vast quantities of antibodies, a dramatic expansion of the secretory pathway and a corresponding increase in the molecular chaperones and folding enzymes that aid and monitor immunoglobulin synthesis occurs. Recent studies reveal that the unfolded protein response (UPR), which is normally activated by endoplasmic reticulum (ER) stress, plays a critical role in this process. Although B cells activate all three branches of the UPR in response to pharmacological inducers of the pathway, plasma cell differentiation elicits only a partial UPR in which components of the PKR-like ER kinase (PERK) branch are not expressed. This prompted us to further characterize UPR activation during plasma cell differentiation. We found that in response to lipopolysaccharides (LPS)-induced differentiation of the I.29 μ+ B cell line, Ire1 was activated early, which led to splicing of XBP-1. PERK was partially phosphorylated with similar kinetics, but this was not sufficient to activate its downstream target eIF-2α, which initiates translation arrest, or to induce other targets like CHOP or GADD34. Both of these events preceded increased Ig synthesis, arguing this is not the signal for activating these two transducers. Targets of activating transcription factor 6 (ATF6) were up-regulated considerably later, arguing that the ATF6 branch is activated by a distinct signal. Pretreatment with LPS inhibited activation of the PERK branch by pharmacological inducers of the UPR, suggesting that differentiation-induced signals specifically silence this branch. This unique ability to differentially regulate various branches of the UPR allows B cells to accomplish distinct outcomes via the same UPR machinery.
Keywords: Plasma cell, Differentiation, B cell, UPR, ER stress
Introduction
The terminal differentiation of a mature B cell to a plasma cell requires a dramatic change in both the cellular structure and function. During plasma cell differentiation, the production of immunoglobulin (Ig) heavy and light chain transcripts and proteins are significantly elevated to allow for the biosynthesis and secretion of vast amounts of antibody molecules. This coincides with a significant increase in the cytoplasmic to nuclear ratio of the cell, as well as a massive expansion of the rough endoplasmic reticulum (ER) where the nascent immunoglobulin chains are folded, assembled and, inspected by the ER quality control apparatus. These functions are achieved by resident ER chaperones and folding enzymes, which are also up-regulated during this differentiation process. Recent studies have identified two major master regulators of the plasma cell differentiation process: B lymphocyte induced maturation protein-1 (Blimp-1; Turner et al. 1994; Shapiro-Shelef et al. 2003) and x-box-binding protein-1 (XBP-1; Reimold et al. 2001; Iwakoshi et al. 2003). Both are required for plasma cell differentiation with Blimp-1 working upstream of XBP-1 (Shaffer et al. 2004). During plasma cell differentiation, Blimp-1 represses various B-cell-specific markers including Pax-5 (Lin et al. 2002), which has been shown in turn to repress the XBP-1 promoter (Shaffer et al. 2004). Thus, there is a direct link from Blimp-1 activation to XBP-1 transcriptional induction. More recently, plasma cell differentiation was linked to a signaling pathway induced during ER stress called the unfolded protein response (UPR) (Iwakoshi et al. 2003; Gass et al. 2002; van Anken et al. 2003) where XBP-1 is also a target.
The accumulation of unfolded or misfolded proteins in the ER serves as the activating signal of the UPR. This pathway has been largely characterized through the use of chemical reagents that disrupt global protein folding in the ER-like thapsigargin, which causes Ca++ deprivation in the ER lumen, tunicamycin, which blocks protein glycosylation in the ER, and DTT, which inhibits disulfide bond formation in the ER. The UPR can also be directly induced by over-expressing a malfolded ER protein (Kozutsumi et al. 1988) or by increased trafficking of proteins through the ER (Pahl and Baeuerle 1995). Three ER transmembrane proteins were identified that sense ER stress and transduce this signal from the lumen to the cytosol and nucleus: Ire1, activating transcription factor 6 (ATF6) and PKR-like ER kinase (PERK; reviewed in (Ma and Hendershot 2001)). All three proteins are kept inactive by binding to the ER resident chaperone BiP via their homologous luminal domains. Activation of the UPR causes BiP to dissociate from their luminal domains, allowing oligomerization and autophosphorylation of Ire1 and PERK (Bertolotti et al. 2000) and translocation of ATF6 from ER to Golgi, (Shen et al. 2002). For Ire1, autophosphorylation activates its cytosolic endonuclease activity, which specifically excises 26 bases from the XBP-1 transcript to produce a remodeled transcription factor, XBP-1(S), with both a DNA binding and transactivation domain (Yoshida et al. 2001; Calfon et al. 2002). Activated PERK phosphorylates eIF-2α, which in turn inhibits cap-dependent translation (Harding et al. 1999). The activation of the PERK-dependent branch of the UPR is required for the activation of targets like C/EBP homology protein (CHOP) and GADD34 (Harding et al. 2000a; Novoa et al. 2001), as well as down-regulation of cyclin D1 (Brewer and Diehl 2000) during ER stress. Thus, PERK activation ensures restricted levels of protein synthesis, cell cycle arrest and/or apoptosis in cells experiencing prolonged pathological ER stress. Once translocated to the Golgi, ATF6 is cleaved by S1P and S2P serving to release its cytosolic transcription factor domain (Ye et al. 2000). ATF6 can bind and transactivate the ERSE I, and to a lesser extent the ERSE II, site in the promoters of various UPR targets (Yoshida et al. 1998; Yamamoto et al. 2004), whereas XBP-1(S) appears to be more specific for UPRE elements (Yoshida et al. 2001; Yamamoto et al. 2004; Shen and Hendershot 2007).
XBP-1 was first identified by its binding to an x-box sequence in the promoter of HLA-DRα (Liou et al. 1990). XBP-1 is a ubiquitously expressed leucine zipper protein that belongs to the CRE-binding protein/ATF family of transcription factors. Disruption of the XBP-1 gene in mice dramatically impaired hepatocyte development, resulting in severe anemia and early embryonic death (Reimold et al. 2000). To assess the role of XBP-1 in B cell development, XBP-1-/- embryonic stem cells were injected into Rag-2 deficient blastocysts, which repopulated the B- and T-cell compartments of the resulting chimeric mice. These mice had normal numbers of B cells that could be stimulated to produce cytokines, but the XBP-1 null B cells were unable to secrete Ig or fully differentiate to plasma cells (Reimold et al. 2001). Additional studies suggest a role for XBP-1 in the specific translational control or ER insertion of IgM (Tirosh et al. 2005). Subsequently, XBP-1 was identified as the only known target of Ire1's endonuclease activity (Yoshida et al. 2001, Calfon et al. 2002), and its essential role in plasma cell differentiation was shown to be dependent on the splicing of its mRNA, presumably by Ire1. Over-expression of the unspliced form XBP-1 protein in XBP-1 null B cells did not rescue the plasma cell differentiation defect, whereas expression of XBP-1(S) did (Iwakoshi et al. 2003). Although enforced expression of XBP-1(S) in B cells is not sufficient to induce plasma cell differentiation, it is able to elevate production of the resident proteins of the secretory pathway and to expand ER membranes and other organelles (Shaffer et al. 2004).
The observation that Ire1 activation and XBP-1 splicing were required for plasma cell differentiation prompted further studies into the role of the UPR in this physiological differentiation process. It was shown that UPR sensors like Ire1 and ATF6 were activated, and UPR targets like XBP-1(S), BiP, GRP94, and ERdj3 were up-regulated during plasma cell differentiation (Gass et al. 2002; van Anken et al. 2003; Shen and Hendershot 2007). However, CHOP, a transcription factor whose induction during ER stress requires the activation of the PERK-dependent branch of the UPR (Harding et al. 2000a), did not appear to be significantly induced (Gass et al. 2002), and PERK null mice respond normally to lipopolysaccharides (LPS) (Gass et al. 2008). This raises the question of whether PERK, and its downstream pathway, is activated in the differentiation process, and if not, whether Ire1 and ATF6 are selectively activated or PERK selectively suppressed. The conventional UPR when induced by pathological stressors like thapsigargin, tunicamycin, DTT, glucose deprivation, or direct over-expression of malfolded proteins activates all three proximal sensors of the UPR simultaneously and no data has been obtained to suggest a mechanism by which the ER stress signal can distinguish between the three sensor proteins to selectively activate individual branches of the UPR.
To elucidate the difference between the conventional UPR and the UPR involved in plasma cell differentiation, we analyzed the activation of upstream sensors Ire1 and PERK, as well as downstream UPR targets of all three transducers in the I.29 μ+ cell line in response to both ER stress and LPS-induced differentiation. Our data reveal this B cell line is fully capable of activating all three branches of the UPR pathway if challenged with pharmacological stressors like thapsigargin or tunicamycin. However, in response to plasma cell differentiation signals, a modified UPR is initiated. Ire1 is activated early in the response leading to XBP-1 splicing. PERK shows signs of partial activation with kinetics similar to Ire1, but this is not sufficient to phosphorylate eIF-2α or to induce its downstream targets. These events occur prior to the up-regulation of Ig transcripts or proteins, arguing that this is not the signal for their activation. ATF6 appears to be activated considerably later and could conceivably be activated by increased processing of Ig proteins. Pretreatment of this B cell line with LPS specifically blocks the induction of PERK targets in response to conventional ER stressors. Together, our data demonstrate that plasma cell differentiation induces a modified UPR that initiates signals to specifically suppress the PERK-dependent branch of the UPR early in the differentiation process.
Materials and methods
Culture and stress treatment of NIH3T3, I.29 μ+, WT, and PERK null MEFs
Wild-type (WT) and PERK null MEFs were kind gifts from Dr. David Ron (Skirball Institute, NYU, New York, NY) and were cultured as previously described (Harding et al. 2000a). NIH3T3 cells were cultured in DMEM media (Mediatech, Herdon, VA) with 10% fetal bovine serum, 2 mM glutamine, 1% Fungizone , and 5% CO2. I.29 μ+ cells were generously provided by Dr. Janet Stavnezer (University of Massachusetts, Worcester, MA) and cultured in RPMI (Mediatech, Herdon, VA) with 20% fetal bovine serum, 2 mM glutamine, 10 μg/ml gentamicin, 1× non-essential amino acids, 1 mM sodium pyruvate, 55 μM mercaptoethanol (Invitrogen, Carlsbad, CA), and 8% CO2. To induce ER stress, cells were treated with 1 μM thapsigargin or 2.5 μg/ml tunicamycin for the indicated times, or with doses indicated in Fig. 5a and b. To induce plasma cell differentiation, I.29 μ+ cells were treated with 50 μg/ml LPS (Sigma, St. Louis, MO) for the indicated periods of time.
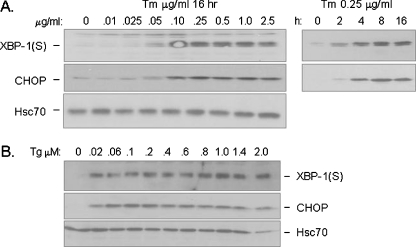
Fig. 5.
Ire1 and PERK are equally sensitive to ER stress induced by conventional stressors like Tm or Tg. a Cell lysates from I.29 μ+ B cells treated for 16 h with the indicated concentrations of Tm (left panel) or 0.25 ug/ml of Tm for the indicated periods of time (right panel) were harvested, separated on 10% SDS-polyacrylamide gel and transferred to a PVDF membrane. The membrane was then blotted for CHOP and XBP-1(S). Hsc70 was examined as a loading control. b Cell lysates were prepared from I.29 μ+ B cells that had been treated for 16 h with the indicated concentrations of Tg and examined as in a
Isolation and analysis of cytoplasmic RNA
Cytoplasmic RNA was harvested using the Qiagen RNeasy miniprep kit following the protocol provided by the supplier. Polymerase chain reaction (PCR) amplification and analysis of the spliced form of XBP-1 (XBP-1(S)) was performed as described previously (Ma and Hendershot 2003). Briefly, 1 μg of cytoplasmic RNA was used to generate 50 μl of cDNA using a poly(dT) primer (1 mM). PCR was then performed with the fail-safe PCR system (Epicenter, Madison, WI) using 2 μλ of cDNA, 0.25 mM of each spXBP-1 primer (5′-GCTGAGTCCGCAGCAGGTG and 5′-ACGAAAGAGACAGGCCTATGC), 2.5 units of Taq DNA polymerase, and 33 nM [α-32P]dCTP (Amersham Biosciences, Piscataway, NJ) for 18–20 cycles. The PCR products were run on a 4% TBE-acrylamide gel, dried and exposed to film. For Northern blotting, 20 μg of cytoplasmic mRNA from each experimental group was run on a Northern blot as described previously (Brewer et al. 1997). The Northern probes for BiP, GADD34, and glyceraldehyde-3 phosphate dehydrogenase (GAPDH) (Ma and Hendershot 2003), total XBP-1, Herp and CHOP (Ma and Hendershot 2004), and ERdj3 (Shen and Hendershot 2005) were described previously. Primer pairs 5′-CCAAAGAGTGTCCCCAAGAG and 5′-GGAAGAGTCTTGTAACCAGTC were used to PCR amplify the northern probe for mouse Blimp-1, pairs 5′-CCCGAGCGAGCGAGTGGAC and 5′-GGCAGATCCTCTCCTTCGAG were used for mouse p58IPK, pairs 5′-GCCCATTCCAGCTGTCGC and 5′-GTGGAGGGACTGGCAGCAC were used for mouse µ-heavy chain, and pairs 5′-CTCAGTACGAGAGGAACCGC and 5′-CGGATTCTGACTTAGAGGCG were used to amplify 28S rRNA.
Cell lysate preparation and Western blotting
For direct Western blotting, cells were lysed in Nonidet P-40 lysing buffer and equal amounts of total protein were electrophoresed under reducing conditions, transferred to PVDF membranes, and probed with the indicated primary antibodies: rabbit anti-CHOP as previously described (Ma et al. 2002), rabbit anti-XBP-1 (S), and goat anti-Hsc70, actin (Santa Cruz Biotechnology, Santa Cruz, CA), goat anti-μ heavy chain (Southern Biotechnology Associates, Birmingham, AL), rabbit anti-phosphorylated eIF-2α, and monoclonal mouse anti-pan eIF-2α (BIOSOURCE, Camarillo, CA). Rabbit anti-PERK and anti-Ire1 antisera were kind gifts from Dr. David Ron (Skirball Institute, NYU School of Medicine, New York, NY), the p58IPK antibody was generously provided by Dr. Michael G. Katze (NYU School of Medicine, New York, NY), and a rabbit anti-EDEM antibody was kindly provided by Dr. Dan Hebert (U. Mass, Amherst, MA). Loading equal amounts of protein for each time point provides a relative measure of induction of these proteins, as the concentration of protein per cell increases during differentiation.
Metabolic labeling and quantitation of protein synthesis
I.29 μ+ cells were stimulated with LPS for varying times and then pulsed with 25 µCi of [35S] methionine/cysteine for 5 min. Cell lysates were prepared, and protein concentration was determined for each sample. A volume corresponding to 10 µg of total protein was loaded directly on a 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) in each case, and proteins were visualized by autoradiography. Additionally, a volume of lysate corresponding to 2 µg of protein was precipitated with 10% trichloroacetic acid and subjected to scintillation counting.
Isolation and LPS stimulation of splenic B cells
Mouse B cells were isolated from the spleen of 8- to 12-week-old female C57BL/6 mice by depleting non-B cells with surface markers (CD3, CD4, CD8, Mac1, GR1, and TER119) using autoMACS (Miltenyi Biotec, Auburn, USA). Plasma cell differentiation was induced by treating the purified splenic B cells with 50 μg/ml LPS (Sigma, St. Louis, MO) for the indicated times. Again, the concentration of total protein was quantitated for each sample and equal amounts of protein were added.
Results
Differentiation of I.29 μ+ B cells into plasma cells upon LPS stimulation involves activation of UPR targets
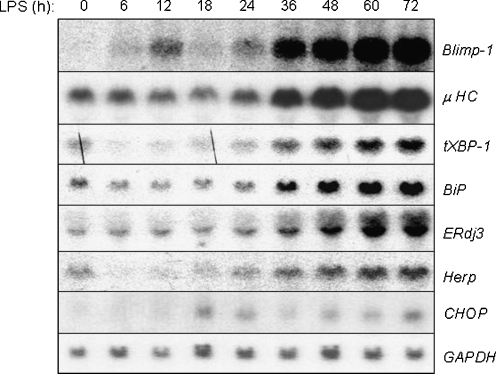
We chose the I.29 μ+ B cell line as a model to study UPR activation during plasma cell differentiation for several reasons. First, a recent proteomic study using this line revealed that LPS-induced plasma cell differentiation was accompanied by multiple waves of translation programs that occurred with distinct kinetics (van Anken et al. 2003) and differentiation is somewhat slower in this B cell line compared with primary splenic B cells (Gass et al. 2008), which suggested a clearer delineation of UPR activation might be easier to observe. Second, it is much easier to obtain large numbers of cells at early time points after LPS stimulation, and finally, these cells can be treated with conventional UPR inducers in the absence of other stimuli allowing us to determine the relative induction of various UPR targets during plasma cell differentiation compared with that achieved with a conventional UPR. We first extended the previous studies on this cell line by examining the timing of the transcriptional up-regulation of several B lineage genes and UPR targets during the differentiation process in this line. Consistent with published data, we found that up-regulation of the master plasma cell regulator gene Blimp-1 occurred with similar kinetics as the induction of Ig heavy chain transcripts (Fig. 1). We also detected transcriptional induction of a subset of UPR targets during the differentiation process (Fig. 1), although the kinetics were somewhat slower than that observed with another B cell line, CH12 (Gass et al. 2002), or in normal splenic B cells (Gass et al. 2008). Increased transcription of most UPR targets and Ig heavy chain occurred ∼36 h after LPS treatment. Increased heavy chain transcription did not precede that of UPR targets like XBP-1, BiP, ERdj3, or Herp. This is consistent with the proteomics study on this line (van Anken et al. 2003), which found that some changes in the translation program occurred well before detectable changes in Ig protein levels suggesting that increased synthesis through the secretory pathway may not be the trigger for the activation of the UPR. An initial very modest and transient induction of Blimp-1 mRNA was observed at 6 and 12 h of LPS treatment, which coincided with a transient transcriptional suppression of heavy chain and UPR targets like XBP-1, BiP and Herp, and may reflect the fact that it is negatively regulated in part. Consistent with published data on mouse splenic B cells (Gass et al. 2008), the UPR target CHOP was not significantly induced during LPS-induced differentiation when the signal was normalized to that of GAPDH (Fig. 1). CHOP is co-regulated by both the ATF6 and PERK/ATF4 branches of the UPR (Ma and Hendershot 2004), whereas targets like BiP and XBP-1 are regulated by ATF6 and Ire1. Thus, the absence of significant CHOP induction raised the question of how activation of only a subset of the UPR transducers might be accomplished.
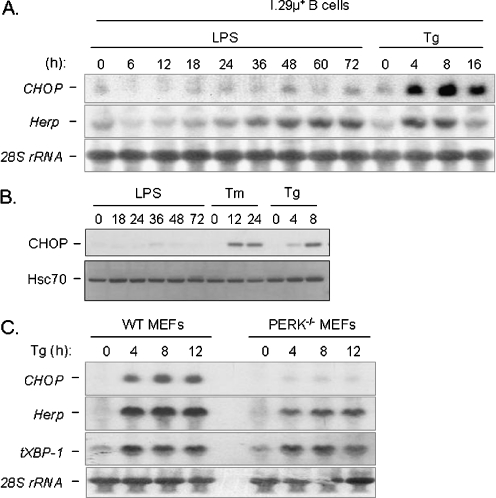
Fig. 1.
UPR targets are activated during the differentiation of I.29 μ+ B cells into plasma cells upon LPS stimulation. Cytosolic RNA from I.29 μ+ cells treated with LPS for the indicated periods of time were electrophoresed, transferred, and then probed with plasma cell markers like Blimp-1 and μ heavy chain (μ HC) or UPR targets like XBP-1, BiP, ERdj3, Herp, and CHOP. GAPDH was probed as control for loading
I.29 μ+ B cells are capable of activating all branches of the UPR
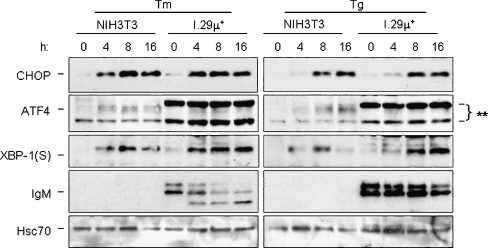
We first determined whether I.29 μ+ B cells were capable of activating all three branches of the UPR in response to conventional ER stressors like tunicamycin (Tm) and thapsigargin (Tg). To monitor the Ire1-ATF6 branches, we examined induction of the spliced form of XBP-1, XBP-1(S), which is dependent on both transcriptional up-regulation by ATF6 and cleavage by Ire1 (Yoshida et al. 2001). We found that XBP-1(S) protein levels were slightly higher in I.29 μ+ B cells than in NIH3T3 fibroblasts after treatment with either ER stressor (Fig. 2). The PERK-dependent target CHOP was examined to monitor this branch. We found that CHOP was induced to comparable levels in both fibroblasts and B cells upon treatment with either tunicamycin or thapsigargin, which argues that the absence of CHOP induction in response to LPS treatment is not due to an inability of this line to activate the PERK arm of the UPR. Thus, the I.29 μ+ B cells are capable of activating all branches of the UPR to similar levels as observed in a well characterized fibroblast line when they are treated with conventional ER stressors.
Fig. 2.
The Ire1, ATF6, and PERK branches of the UPR are intact in I.29 μ+ cells. NIH3T3 fibroblasts or I.29 μ+ B cells were treated with the ER stressors tunicamycin (Tm) or thapsigargin (Tg) for the indicated period of time. Cell lysates were harvested and separated on 10% SDS-polyacrylamide gels and transferred for western blot analyses. The membrane was then blotted for the PERK targets CHOP and ATF4, the Ire1/ATF6 regulated target, XBP-1(S), and mouse μ heavy chain (which is only present in I.29 μ+ cells). Hsc70 was examined as a control for loading. The asterisks indicate two non-specific bands detected with the ATF4 antibody in I.29 μ+ cells
UPR pathway downstream of PERK is selectively excluded from the plasma cell differentiation process
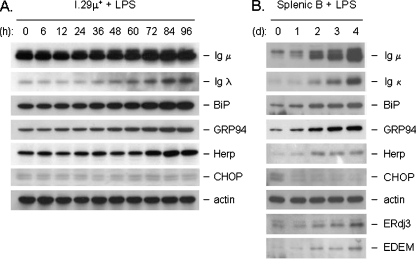
Somewhat surprisingly, unlike CHOP, Herp transcripts were induced during plasma cell differentiation (Fig. 1) even though we previously demonstrated that Herp and CHOP are similarly co-regulated by both the PERK/ATF4-dependent and the Ire1/ATF6-dependent branches of the UPR in fibroblast lines when they are treated with conventional ER stressors (Ma and Hendershot 2004). The differential induction of Herp and CHOP was further examined at the protein level in this cell line (Fig. 3a) and in splenic B cell stimulated with LPS (Fig. 3b). In both cases, the induction of Herp protein was observed, whereas there was no induction of CHOP protein in the case of I.29 μ+ B cells and an actual decrease in the slight expression of CHOP was observed in the LPS-stimulated splenic B cells. Induction of other UPR targets in the splenic B cells mirrored that seen in the I.29 μ+ B cells (Figs. 1 and 3a).
Fig. 3.
Splenic B cells show similar induction of UPR targets with LPS as I.29 μ+ cells as well as the differential induction of CHOP and Herp proteins. Cell lysates from I.29 µ+ B cells (a) or mouse splenic B cells (b) were treated with LPS for the indicated times and equal amounts of protein were loaded for each sample. After separation by SDS-PAGE and transfer, membranes were probed with the indicated antibodies. Actin served as a control for loading
We next compared the induction of these two genes in the I.29 μ+ B cell line in response to either LPS or thapsigargin treatment. This was not possible in the splenic B cells as they do not survive long in culture without the addition of mitogens or growth factors. We found that Herp transcripts were up-regulated to nearly similar levels with both LPS and thapsigargin treatment (Fig. 4a), and the kinetics of Herp induction with LPS was very similar to that observed for other UPR targets and Ig heavy chain (i.e., 36 h post-LPS treatment). In contrast, the induction of CHOP with LPS was dramatically impaired compared with that observed with thapsigargin treatment, a finding that was also observed at the protein level (Fig. 4b). To our knowledge, this is the first example of differential regulation of these two genes. Studies from the Ron lab and our group have suggested that a PERK-dependent signal other than ATF4 might be required for the activation of CHOP (Harding et al. 2000b; Ma et al. 2002). This requirement for an addition signal downstream of PERK activation was confirmed when we examined the induction of CHOP mRNA in WT and PERK null MEFs treated with thapsigargin (Fig. 4c). CHOP mRNA was not up-regulated in PERK null MEFs during UPR activation. However, Herp transcripts were still up-regulated in the PERK null MEFs, albeit to somewhat lower levels than in the WT cells. This demonstrated that although ATF6 and PERK/ATF4 both contribute to CHOP and Herp induction as we previously reported (Ma and Hendershot 2004), CHOP induction requires a signal downstream of PERK in addition to ATF4, whereas Herp does not. Thus Herp induction does not require PERK activation, whereas CHOP does. The differential induction of Herp and CHOP during plasma cell differentiation therefore suggests that the PERK-dependent branch of the UPR is selectively excluded during plasma cell differentiation.
Fig. 4.
UPR targets CHOP and Herp are regulated differently during B-cell differentiation. a Cytosolic RNA from I.29 µ+ B cells stimulated with either LPS or Tg for the indicated periods of time were harvested, separated, and then probed for CHOP and Herp. 28S rRNA was probed as a loading control. b Cell extracts were prepared from cells treated as indicated and CHOP protein expression was determined by Western blotting. Hsc70 protein served as a control for loading. c Cytosolic RNA from WT and PERK null MEFs treated with Tg for the indicated periods of time were harvested, separated, and then probed for CHOP, Herp, and total XBP-1 (tXBP-1) transcript levels. Again, 28S rRNA was probed as a loading control
PERK has a similar threshold for activation as Ire1 and ATF6
There are two possible scenarios that could explain the lack of PERK activation during plasma cell differentiation: the Ire1 and ATF6 branches of the UPR could be specifically activated without activating the PERK-dependent branch, or alternatively the signaling pathway downstream of PERK could be selectively silenced by a negative regulator during plasma cell differentiation. Given the fact that the signal for UPR induction during plasma cell differentiation is not known, we began by assessing whether the various transducers might have different sensitivity to minimal doses of conventional ER stressors. Cells were treated with decreasing amounts of tunicamycin and thapsigargin, and activation of the Ire1-ATF6-dependent target XBP-1(S) and the PERK-dependent target CHOP were monitored (Fig. 5). We found that the minimal doses of ER stressors required for the activation of XBP-1 and CHOP were very similar and that both pathways appeared to be exquisitely sensitive to these agents. CHOP and XBP-1(S) were induced by as little as 0.05 µg/ml of tunicamycin and 0.02 μM thapsigargin, which were the lowest doses examined and well below the concentration normally used (2.5 μg/ml and 1.0 μM respectively). Furthermore, the kinetics of XBP-1 and CHOP induction by a low dose of tunicamycin (i.e., 0.25 mg/ml, which is 1/10 the usual dose) were also very similar beginning as early as 2 h after treatment. Therefore, we conclude that there is no inherent difference in the sensitivity of the various UPR transducers to conventional ER stress signals and argue that this is not an obvious reason for the limited activation of PERK-dependent targets during LPS induced plasma cell differentiation. However, we cannot rule out the possibility that these UPR transducers have a different threshold to the signal generated by LPS.
PERK is partially activated along with other sensors of the UPR during plasma cell differentiation, although this is not sufficient to activate its downstream targets
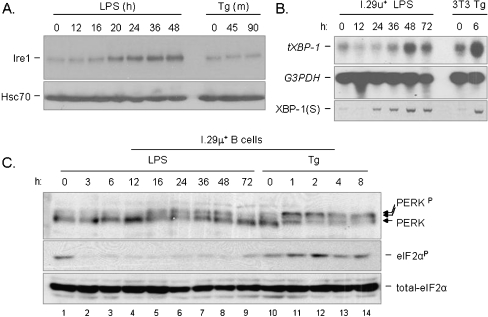
Next, we directly examined the activation state of Ire1 and PERK in I.29 μ+ B cells during LPS-induced differentiation. Although the mobility shift is quite modest in keeping with previous data on Ire1 activation (Bertolotti et al. 2000), our data revealed that phosphorylation of Ire1 could be detected between 16 to 20 h after LPS stimulation and within 45 min of tunicamycin treatment (Fig. 6a). Of note, Ire1 levels were also induced by LPS but not by thapsigargin, which is reminiscent of the increased expression of ATF6 in response to LPS observed in another study (Gunn et al. 2004). The kinetics of Ire1 phosphorylation during plasma cell differentiation coincided with the splicing of its downstream target, XBP-1 mRNA, demonstrating that this phosphorylation event was sufficient for the activation of Ire1’s nuclease activity (Fig. 6b). Interestingly, the up-regulation of total XBP-1 mRNA, which is regulated by ATF6 (Yoshida et al. 2003), did not begin until 36 h of LPS stimulation suggesting a kinetic delay in the activation of ATF6 as compared with that of Ire1.
Fig. 6.
Both Ire1 and PERK are activated to different extents during plasma cell differentiation, but eIF-2α phosphorylation downstream of PERK is blocked. a. Cell lysates from I.29 μ+ B cells treated with either LPS or Tg for the indicated period time was harvested and then separated on a 10% SDS-PAGE. The membrane was blotted for Ire1 and for Hsc70 as a loading control. b Cytosolic RNA from I.29 μ+ cells treated with LPS, or NIH3T3 cells treated with Tg for the indicated period of time was harvested and separated. Total XBP-1 (tXBP-1) and GAPDH mRNA levels were then probed, the latter as a loading control. RT-PCR was also performed with the same RNA samples using primers specific for the spliced form of XBP-1 (sXBP-1). c Cell lysates from I.29 μ+ cells treated with LPS or Tg for the indicated period of time were separated on a 8% SDS-PAGE which was then blotted for PERK, total and phosphorylated form of eIF-2α
PERK's eIF-2α kinase activity is activated through oligomerization and subsequent autophosphorylation in trans at multiple sites (Bertolotti et al. 2000; Marciniak et al. 2006). Some of these sites are required for its association with eIF-2α but not for its catalytic activity (Marciniak et al. 2006). We observed 3 major forms of PERK when I.29 μ+ B cells were stressed with thapsigargin (Fig. 6c, lane 10–14), which is in keeping with other published data (Marciniak et al. 2006; Zhang et al. 2005). The lower band represents unphosphorylated PERK and is the predominant form present before ER stress. The upper two bands represent phosphorylated forms of PERK, with the majority shifting to the slowest migrating, hyperphosphorylated state later during conventional ER stress. Coinciding with PERK hyperphosphorylation during thapsigargin treatment, we detected the phosphorylation of its downstream target eIF-2α, suggesting that this is the active form of PERK. However, when I.29 μ+ B cells were stimulated with LPS, a shift in PERK to the intermediate mobility form was detected between 16–48 h, but there was no evidence of the hyperphosphorylated form (Fig. 6c, lane 1–9). In keeping with this, there was no evidence of enhanced eIF-2α phosphorylation during LPS stimulation. Together, our data suggests that the intermediate migrating form of PERK observed during plasma cell differentiation corresponds to a partial activation state, whereas the slowest migrating, hyperphosphorylated form represents the completely activated state seen during a more conventional UPR. Second, it is noteworthy that the phosphorylation of PERK in response to LPS treatment was gradually reversed starting at 36 h after LPS stimulation and returned to its basal state 72 h after stimulation. This is similar to what is observed with PERK phosphorylation in response to thapsigargin treatment and suggests that a PERK phosphatase might be activated in both cases. Finally, the phosphorylation state of eIF-2α, a direct downstream target of PERK, was first elevated and then returned to basal levels during thapsigargin treatment, which is consistent with published data (Novoa et al. 2003; Ma and Hendershot 2003). In contrast, basal eIF-2α phosphorylation actually decreased upon LPS stimulation, which is consistent with a selective suppression of the pathway downstream of PERK during plasma cell differentiation (Fig. 6c).
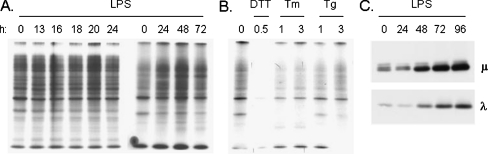
As an independent measure of PERK activation and eIF-2α suppression during plasma cell differentiation, we examined the effects on total protein synthesis. Cell were stimulated with LPS or incubated with conventional UPR inducers for the indicated times and then pulse labeled with [35S] methionine/cysteine. Cell lysates were prepared and either analyzed by SDS-PAGE (Fig. 7) or subjected to trichloroacetic acid precipitation and scintillation counting to measure changes in both the synthesis of specific proteins as well as global changes in synthesis. We saw no indication of inhibition of protein synthesis during the time period where hypophosphorylation of PERK was observed (16–48 h), and in fact total protein synthesis increased at 48 h as detected on the gels (Fig. 7a) and when measuring TCA precipitated counts (data not shown). In contrast, total protein synthesis was inhibited most dramatically in response to DTT treatment and to a lesser but significant amount when cells were treated with either tunicamycin or thapsigargin (Fig. 7b). In keeping with northern blot data (Fig. 1), the activation of Ire1 and partial activation of PERK occurred before an increase in Ig heavy or light chains could be detected (Fig. 7c). Thus, the hypophosphorylation of PERK observed at early time points after LPS stimulation does not represent an active form that can phosphorylate eIF-2α, leading to an inhibition of protein synthesis or induction of CHOP.
Fig. 7.
LPS induced differentiation does not induce an arrest in protein synthesis. I.29 μ+ B cells were treated with LPS (a) or conventional UPR inducers (b) for the indicated times and then pulse-labeled with [35S] methionine and cysteine for 5 min. Cell lysates were analyzed directly by SDS-PAGE and proteins were visualized by autoradiography. c An aliquot of cell lysates from the indicated times after LPS induction were examined by Western blotting for Ig heavy and light chain expression
Plasma cell differentiation induces p58IPK, a negative regulator of the PERK-dependent branch of the UPR, but not GADD34
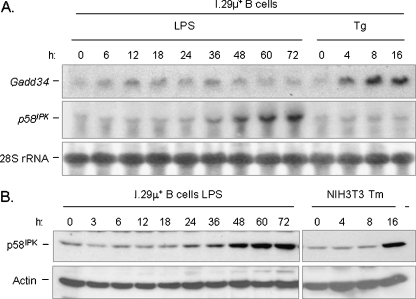
We next attempted to identify the negative regulating factors that are responsible for the suppression of the PERK-dependent branch. Two negative regulators of the branch have been identified: p58IPK and GADD34. The former has been reported to inhibit PERK kinase activity (Yan et al. 2002), although this activity was recently questioned (Rutkowski et al. 2007), while the latter facilitates the PP-1 phosphatase in reversing eIF-2α phosphorylation (Novoa et al. 2001). Therefore, induction of either factor could explain the absence of eIF-2α phosphorylation in differentiating plasma cells, although only activation of p58IPK would be expected to affect the phosphorylation status of PERK. We first examined the mRNA level of both GADD34 and p58IPK in differentiating I.29 μ+ B cells (Fig. 8). We were unable to detect an increase in GADD34 mRNA at any point during plasma cell differentiation, which further corroborates our data suggesting that the pathway downstream of PERK is not activated in differentiating plasma cells, as GADD34 is a direct target of eIF-2α phosphorylation and the subsequent induction of ATF4, which transactivates GADD34 (Ma and Hendershot 2003). In contrast, we detected an increase in p58IPK transcripts and protein beginning at ∼36 h of LPS stimulation. The induction of p58IPK relatively late in differentiation of this cell line is in keeping with previous data on splenic B cells (Gass et al. 2008), which further argues the validity of this cell lines as a model for UPR activation during plasma cell differentiation and also strongly suggests that p58IPK is not responsible for suppressing the PERK branch.
Fig. 8.
p58IPK, but not GADD34, is induced during the differentiation of I.29 μ+ B cells into plasma cells. a I.29 μ+ B cells were treated with LPS or Tg for the indicated periods of time and RNA was isolated and prepared for Northern blotting. The membrane was probed for GADD34 and p58IPK. 28S rRNA levels were also examined to control for loading. b Cell lysates were prepared from I.29 μ+ B cells that had been treated with LPS or from NIH3T3 fibroblasts treated with Tm for the indicated periods of time. Samples were examined on a 10% SDS-PAGE and transferred. The membrane was then blotted for p58IPK protein and actin as a loading control
LPS stimulation suppresses the ability of differentiating plasma cells to activate PERK targets in response to conventional UPR activators
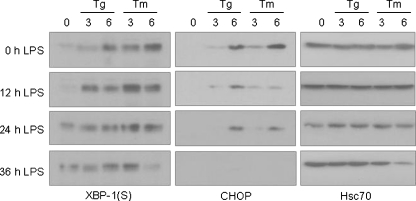
To further confirm the presence of negative regulators of the PERK pathway during plasma cell differentiation, we tested our hypothesis from a different angle. We measured the levels of XBP-1(S) and CHOP protein in I.29 μ+ B cells that were pre-stimulated with LPS for various periods of time before being treated with conventional ER stressors like tunicamycin and thapsigargin (Fig. 9). We found that up-regulation of XBP-1(S) protein could be readily detected 3 h after either tunicamycin or thapsigargin treatment, and the level of XBP-1(S) induction was actually higher when cells were pretreated with LPS. Cells that were incubated with LPS for longer periods of time expressed XBP-1(S) prior to treatment with the conventional ER stressors but still managed to increase these levels further after treatment with the conventional UPR inducers (Fig. 9, left panel). The apparent decrease in XBP-1(S) expression observed after 36 h of LPS coupled with 6 h of tunicamycin is due to the loss of cells, which was consistently seen with this combination of treatment (see loading control in right panel). As tunicamycin is a stronger inducer of the UPR than thapsigargin in this cell line (see 0 h LPS panels), it is likely that the inability to activate the PERK branch after LPS treatment has greater consequences on cell survival when tunicamycin is used as the UPR inducer. In contrast, LPS pretreatment for as short as 12 h significantly decreased the induction of CHOP protein by tunicamycin or thapsigargin, and the longer the pretreatment, the more significant the suppression of CHOP induction by conventional stressors (Fig. 9, middle vertical panel). Together, these data further support our earlier conclusion that although PERK is at least partially activated during plasma cell differentiation, its downstream pathway is actively suppressed.
Fig. 9.
LPS pretreatment of I.29 μ+ B cells augment XBP-1(S) levels but inhibits CHOP induction by pharmacological ER stressors. I.29 μ+ B cells were pretreated with LPS for 0, 12, 24, or 36 h and then further cultured with Tm or Tg for 0, 3 or 6 h. Cell lysates were harvested for each experimental group and analyzed by SDS-PAGE. After transferring, the membranes were blotted for XBP-1(S) (left panel), CHOP (middle panel) or Hsc70 (right panel) as a control for loading
Discussion
Plasma cell differentiation involves the significant expansion of the endoplasmic reticulum to support the elevated production of immunoglobulins through the secretory pathway (Wiest et al. 1990; Hendershot and Sitia 2004). This is achieved through the activation of a modified UPR in which some key targets of the UPR pathway, including ER chaperones like BiP, GRP94, and ERdj3, are induced, while others like CHOP, a target of the PERK branch of the UPR, are not. Because activation of the PERK branch and the attendant global suppression of protein synthesis would seem to be at odds with producing a cell designed to synthesize vast quantities of protein, it would make sense if indeed the PERK-dependent branch of the UPR was not activated during this differentiation process. However, it was unclear that in fact this was true and if so, how it was achieved. Although several other studies have been conducted on cell lines (Gass et al. 2002) and normal splenic B cells (Gass et al. 2008), which are compatible with the finding presented in this paper, the fact that I.29 μ+ B cells have slower kinetics of differentiation and can be treated with UPR inducers in the absence of LPS allowed us to more fully delineate the activation of various UPR signal transducers during plasma cell differentiation.
We first examined the activation status of the Ire1 and PERK-dependent branches of the UPR during LPS induced plasma cell differentiation. Our data demonstrate that PERK initially appears to be phosphorylated upon LPS treatment, although to a lesser extent than that observed with conventional ER stressors like tunicamycin and thapsigargin. The kinetics of PERK phosphorylation paralleled that of Ire1, which began around 16 h after LPS treatment, suggesting that both might be activated by the same upstream signal, which is yet to be identified. The partial phosphorylation of PERK induced by LPS did not result in eIF-2α phosphorylation or any of the downstream effects including inhibition of protein synthesis or transcription of the CHOP and GADD34 genes. This is in keeping with a recent study conducted on PERK phosphorylation site mutants, which found that full phosphorylation of PERK was required for recruiting eIF-2α and phosphorylating it (Marciniak et al. 2006).
Two putative negative regulators of the PERK-dependent pathway have been described that act above or at the level of eIF-2α phosphorylation: GADD34 and p58IPK. GADD34 binds to and activates PP-1 phosphatase, which then dephosphorylates eIF-2α. The GADD34 promoter is directly transactivated by ATF4, which is dependent on PERK activation during the UPR (Ma and Hendershot 2003). Therefore, our data demonstrating that GADD34 transcription is not elevated during plasma cell differentiation not only excludes it as a candidate for silencing the PERK branch, but further substantiates our conclusion that signaling pathway downstream of PERK is selectively shut down. Over-expression and recombinant pull-down assays demonstrated that p58IPK can bind directly to the kinase domain of eIF-2α kinases like PKR and PERK and inhibit their activities in vitro (Yan et al. 2002). However, recent data have shown that the original reported orientation of p58IPK in the ER membrane is incorrect and that this protein is actually inside the ER lumen where it appears to inhibit PERK phosphorylation indirectly by preventing protein misfolding during ER stress (Rutkowski et al. 2007). Thus, it is unlikely that p58IPK is responsible for inhibiting this branch, particularly given the late kinetics for its induction.
Importantly, our data revealed that whatever the differentiation-induced negative regulator(s) of PERK is, it also blocked the subsequent induction of PERK-dependent targets by pharmacological ER stressors but not Ire1 dependent targets (Fig. 9). This is somewhat contrary to data obtained with splenic B cells (Gass et al. 2008), which showed evidence of an ability to modestly induce GADD34 protein in response to tunicamycin after 3 days of LPS stimulation. However, it is important to note that in this case the cells were not treated with tunicamycin in the absence of LPS stimulation to measure relative activation of the PERK branch, since splenic B cells do not survive long in culture without stimulation and respond poorly to additional ER stress. More in keeping with our data, the induction of p58IPK protein, which is downstream of Ire1/XBP-1(S) was quite robust with the combination of tunicamycin and LPS in another study on splenic B cells (Rutkowski et al. 2007).
The splicing of XBP-1 mRNA, which is directly downstream of Ire1 activation, and the accumulation of XBP-1(S) protein occurred as early as 24 h after LPS treatment (Figs. 6b and 9), while transcription of most ERSE-dependent ATF6 targets including BiP, Herp, and total XBP-1 mRNA were not induced until 36 h after LPS treatment (Fig. 1). This apparent lapse of time between induction of Ire1 and ATF6 targets suggests that all three proximal transducers of the UPR may not be activated by the same signal during plasma cell differentiation, although it remains unclear exactly what triggers the activation of these proximal transducers during the differentiation process. When fibroblasts like NIH3T3 and MEF are activated by pharmacological stressors all three proximal sensors are activated at the same time. Our data in I.29 μ+ B cells revealed that B cells are no different from fibroblasts in their capacity to activate all branches of the UPR when stimulated with conventional pharmacological ER stressors (Fig. 2), nor is there an apparent difference among the three proximal sensors in their sensitivity to these pharmacological stressors (Fig. 5). The increased synthesis of Ig, as well as other ER resident proteins like BiP and ERdj3, during plasma cell differentiation occurred later than the phosphorylation of either Ire1 or PERK. Together, our data suggests that the initial trigger for their phosphorylation is unlikely to be the increased synthesis of ER proteins as in a conventional UPR, but this increased synthesis may well play a role in activating ATF6 and amplifying the response later during differentiation. This is in keeping with data showing that μH-chain deficient splenic B cells from the B1-8f/+ mouse are still able to induce XBP-1 splicing, albeit at a reduced level (Iwakoshi et al. 2003). Since both Ire1 and PERK are modifided by phosphorylation, it is possible that this occurs during the initial phases of plasma cell differentiation by either the activation of a kinase or inhibition of a phosphatase. This could also explain the non-native migration of PERK at this time. The increased synthesis of Ig later in the response could trigger a more conventional UPR that further amplifies the pathway, but is unlikely to significantly include the PERK branch, as a negative regulator(s) for this branch appears to already be in put into place. The differential regulation of individual branches of the UPR seen here is not entirely unprecedented. It has also been observed in other physiological processes like the synthesis of thyroglobulin by thyrocytes. Hormone stimulated thyroglobulin production in thyrocytes activates both the PERK and ATF6-dependent branches of the UPR, but not the splicing of XBP-1 mRNA (Sargsyan et al. 2004). Whether or not Ire1 is activated at all during the hormone stimulation was not tested.
In conclusion, our study revealed that all three proximal sensors of the UPR were activated to varying degrees and with different kinetics during plasma cell differentiation. The lack of expression of PERK downstream targets was not due to a failure to initiate activation of the PERK branch, but instead by a specific inhibition of this transducer. The varying kinetics of activation suggests that all three transducers are not likely to be activated by the same mechanism. We propose that by differentially regulating individual branches of the UPR, it is possible to achieve a variety of outcomes in different cell types that are tailored to the specific need of different tissue and cell types via the same UPR machinery.
Acknowledgements
This work was supported by NIH Grant GM54068 (LMH), the Cancer Center CORE Grant CA21765, and the American Lebanese Syrian Associated Charities of St. Jude Children's Research Hospital.
References
- Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol. 2000;2:326–332. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- Brewer JW, Diehl JA. PERK mediates cell-cycle exit during the mammalian unfolded protein response. Proc Natl Acad Sci USA. 2000;97:12625–12630. doi: 10.1073/pnas.220247197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer JW, Cleveland JL, Hendershot LM. A pathway distinct from the mammalian unfolded protein response regulates expression of endoplasmic reticulum chaperones in non-stressed cells. EMBO J. 1997;16:7207–7216. doi: 10.1093/emboj/16.23.7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clask SG, Ron D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- Gass JN, Gifford NM, Brewer JW. Activation of an unfolded protein response during differentiation of antibody-secreting B cells. J Biol Chem. 2002;277:49047–49054. doi: 10.1074/jbc.M205011200. [DOI] [PubMed] [Google Scholar]
- Gass JN, Jiang HY, Wek RC, Brewer JW. The unfolded protein response of B-lymphocytes: PERK-independent development of antibody-secreting cells. Mol Immunol. 2008;45:1035–1043. doi: 10.1016/j.molimm.2007.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunn KE, Gifford NM, Mori K, Brewer JW. A role for the unfolded protein response in optimizing antibody secretion. Mol Immunol. 2004;41:919–927. doi: 10.1016/j.molimm.2004.04.023. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6:1099–1108. doi: 10.1016/S1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Bertolotti A, Zeng H, Ron D. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol Cell. 2000;5:897–904. doi: 10.1016/S1097-2765(00)80330-5. [DOI] [PubMed] [Google Scholar]
- Hendershot LM, Sitia R. Antibody synthesis and assembly. In: Alt FW, Honjo T, Neuberger MS, editors. Molecular biology of B cells. New York: Elsevier; 2004. pp. 261–273. [Google Scholar]
- Iwakoshi NN, Lee AH, Vallabhajosyula P, Otipoby KL, Rajewsky K, Glimcher LH. Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1. Nat Immunol. 2003;4:321–329. doi: 10.1038/ni907. [DOI] [PubMed] [Google Scholar]
- Kozutsumi Y, Segal M, Normington K, Gething MJ, Sambrook J. The presence of malfolded proteins in the endoplasmic reticulum signals the induction of glucose-regulated proteins. Nature. 1988;332:462–464. doi: 10.1038/332462a0. [DOI] [PubMed] [Google Scholar]
- Lin KI, Angelin-Duclos C, Kuo TC, Calame K. Blimp-1-dependent repression of Pax-5 is required for differentiation of B cells to immunoglobulin M-secreting plasma cells. Mol Cell Biol. 2002;22:4771–4780. doi: 10.1128/MCB.22.13.4771-4780.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou HC, Boothby MR, Finn PW, Davidon R, Nabavi N, Zeleznik-Le NJ, Ting JP, Glimcher LH. A new member of the leucine zipper class of proteins that binds to the HLA DR alpha promoter. Science. 1990;247:1581–1584. doi: 10.1126/science.2321018. [DOI] [PubMed] [Google Scholar]
- Ma Y, Hendershot LM. The unfolding tale of the unfolded protein response. Cell. 2001;107:827–830. doi: 10.1016/S0092-8674(01)00623-7. [DOI] [PubMed] [Google Scholar]
- Ma Y, Hendershot LM. Delineation of the negative feedback regulatory loop that controls protein translation during ER stress. J Biol Chem. 2003;278:34864–34873. doi: 10.1074/jbc.M301107200. [DOI] [PubMed] [Google Scholar]
- Ma Y, Hendershot LM. Herp is dually regulated by both the endoplasmic reticulum stress-specific branch of the unfolded protein response and a branch that is shared with other cellular stress pathways. J Biol Chem. 2004;279:13792–13799. doi: 10.1074/jbc.M313724200. [DOI] [PubMed] [Google Scholar]
- Ma Y, Brewer JW, Diehl JA, Hendershot LM. Two distinct stress signaling pathways converge upon the CHOP promoter during the mammalian unfolded protein response. J Mol Biol. 2002;318:1351–1365. doi: 10.1016/S0022-2836(02)00234-6. [DOI] [PubMed] [Google Scholar]
- Marciniak SJ, Garcia-Bonilla L, Hu J, Harding HP, Ron D. Activation-dependent substrate recruitment by the eukaryotic translation initiation factor 2 kinase PERK. J Cell Biol. 2006;172:201–209. doi: 10.1083/jcb.200508099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoa I, Zeng H, Harding HP, Ron D. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2alpha. J Cell Biol. 2001;153:1011–1022. doi: 10.1083/jcb.153.5.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoa I, Zhang Y, Zeng H, Jungreis R, Harding HP, Ron D. Stress-induced gene expression requires programmed recovery from translational repression. EMBO J. 2003;22:1180–1187. doi: 10.1093/emboj/cdg112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pahl HL, Baeuerle PA. A novel signal transduction pathway from the endoplasmic reticulum to the nucleus is mediated by transcription factor NF-kappa B. EMBO J. 1995;14:2580–2588. doi: 10.1002/j.1460-2075.1995.tb07256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimold AM, Etkin A, Clauss I, Perkins A, Friend DS, Zhang J, Horton HF, Scott A, Orkin SH, Byrne MC, Grusby MJ, Glimcher LH. An essential role in liver development for transcription factor XBP-1. Genes Dev. 2000;14:152–157. [PMC free article] [PubMed] [Google Scholar]
- Reimold AM, Iwakoshi NN, Manis J, Vallabhajosyula P, Szomolanyi-Tsuda E, Gravallese EM, Friend D, Grusby MJ, Alt F, Glimcher LH. Plasma cell differentiation requires the transcription factor XBP-1. Nature. 2001;412:300–307. doi: 10.1038/35085509. [DOI] [PubMed] [Google Scholar]
- Rutkowski DT, Kang SW, Goodman AG, Garrison JL, Taunton J, Katze MG, Kaufman RJ, Hegde RS. The role of p58IPK in protecting the stressed endoplasmic reticulum. Mol Biol Cell. 2007;18:3681–3691. doi: 10.1091/mbc.E07-03-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargsyan E, Baryshev M, Mkrtchian S. The physiological unfolded protein response in the thyroid epithelial cells. Biochem Biophys Res Commun. 2004;322:570–576. doi: 10.1016/j.bbrc.2004.07.155. [DOI] [PubMed] [Google Scholar]
- Shaffer AL, Shapiro-Shelef M, Iwakoshi NN, Lee AH, Qian SB, Zhao H, Yu X, Yang L, Tan BK, Rosenwald A, Hurt EM, Petroulakis E, Sonenberg N, Yewdell JW, Calame K, Glimcher LH, Staudt LM. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity. 2004;21:81–93. doi: 10.1016/j.immuni.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Shapiro-Shelef M, Lin KI, McHeyzer-Williams LJ, Liao J, McHeyzer-Williams MG, Calame K. Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity. 2003;19:607–620. doi: 10.1016/S1074-7613(03)00267-X. [DOI] [PubMed] [Google Scholar]
- Shen Y, Hendershot LM. ERdj3, a stress-inducible endoplasmic reticulum DnaJ homologue, serves as a cofactor for BiP's interactions with unfolded substrates. Mol Biol Cell. 2005;16:40–50. doi: 10.1091/mbc.E04-05-0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Hendershot LM. XBP-1 regulates the OBF-1/BOB-1/OCA-B transcription coactivator, which controls Ig heavy chain expression during plasma cell differentiation. J Immunol. 2007;179:2969–2978. doi: 10.4049/jimmunol.179.5.2969. [DOI] [PubMed] [Google Scholar]
- Shen J, Chen X, Hendershot L, Prywes R. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev Cell. 2002;3:99–111. doi: 10.1016/S1534-5807(02)00203-4. [DOI] [PubMed] [Google Scholar]
- Tirosh B, Iwakoshi NN, Glimcher LH, Ploegh HL. XBP-1 specifically promotes IgM synthesis and secretion, but is dispensable for degradation of glycoproteins in primary B cells. J Exp Med. 2005;202:505–516. doi: 10.1084/jem.20050575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner CA, Mack DH, Davis MM. Blimp-1, a novel zinc finger-containing protein that can drive the maturation of B lymphocytes into immunoglobulin-secreting cells. Cell. 1994;77:297–306. doi: 10.1016/0092-8674(94)90321-2. [DOI] [PubMed] [Google Scholar]
- Anken E, Romijn EP, Maggioni C, Mezghrani A, Sitia R, Braakman I, Heck AJ. Sequential waves of functionally related proteins are expressed when B cells prepare for antibody secretion. Immunity. 2003;18:243–253. doi: 10.1016/S1074-7613(03)00024-4. [DOI] [PubMed] [Google Scholar]
- Wiest DL, Burkhardt JK, Hester S, Hortsch M, Meyer DI, Argon Y. Membrane biogenesis during B cell differentiation: most endoplasmic reticulum proteins are expressed coordinately. J Cell Biol. 1990;110:1501–1511. doi: 10.1083/jcb.110.5.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Yoshida H, Kokame K, Kaufman RJ, Mori K. Differential contributions of ATF6 and XBP1 to the activation of endoplasmic reticulum stress-responsive cis-acting elements ERSE, UPRE and ERSE-II. J Biochem (Tokyo) 2004;136:343–350. doi: 10.1093/jb/mvh122. [DOI] [PubMed] [Google Scholar]
- Yan W, Frank CL, Korth MJ, Sopher BL, Novoa I, Ron D, Katze MG. Control of PERK eIF2alpha kinase activity by the endoplasmic reticulum stress-induced molecular chaperone P58IPK. Proc Natl Acad Sci USA. 2002;99:15920–15925. doi: 10.1073/pnas.252341799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Rawson RB, Komuro R, Chen X, Dave UP, Prywes R, Brown MS, Goldstein JL. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell. 2000;6:1355–1364. doi: 10.1016/S1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Haze K, Yanagi H, Yura T, Mori K. Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins. Involvement of basic leucine zipper transcription factors. J Biol Chem. 1998;273:33741–33749. doi: 10.1074/jbc.273.50.33741. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/S0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Matsui T, Hosokawa N, Kaufman RJ, Nagata K, Mori K. A time-dependent phase shift in the mammalian unfolded protein response. Dev Cell. 2003;4:265–271. doi: 10.1016/S1534-5807(03)00022-4. [DOI] [PubMed] [Google Scholar]
- Zhang K, Wong HN, Song B, Miller CN, Scheuner D, Kaufman RJ. The unfolded protein response sensor IRE1alpha is required at 2 distinct steps in B cell lymphopoiesis. J Clin Invest. 2005;115:268–281. doi: 10.1172/JCI21848. [DOI] [PMC free article] [PubMed] [Google Scholar]