Abstract
Whereas the consequences of global warming at population or community levels are well documented, studies at the cellular level are still scarce. The study of the physiological or metabolic effects of such small increases in temperature (between +2°C and +6°C) is difficult because they are below the amplitude of the daily or seasonal thermal variations occurring in most environments. In contrast, subterranean biotopes are highly thermally buffered (±1°C within a year), and underground water organisms could thus be particularly well suited to characterise cellular responses of global warming. To this purpose, we studied genes encoding chaperone proteins of the HSP70 family in amphipod crustaceans belonging to the ubiquitous subterranean genus Niphargus. An HSP70 sequence was identified in eight populations of two complexes of species of the Niphargus genus (Niphargus rhenorhodanensis and Niphargus virei complexes). Expression profiles were determined for one of these by reverse transcription and quantitative polymerase chain reaction, confirming the inducible nature of this gene. An increase in temperature of 2°C seemed to be without effect on N. rhenorhodanensis physiology, whereas a heat shock of +6°C represented an important thermal stress for these individuals. Thus, this study shows that although Niphargus individuals do not undergo any daily or seasonal thermal variations in underground water, they display an inducible HSP70 heat shock response. This controlled laboratory-based physiological experiment constitutes a first step towards field investigations of the cellular consequences of global warming on subterranean organisms.
Keywords: Subterranean environment, Heat shock protein, Thermal stress, Climate change
Introduction
Global warming is now unequivocal, and specialists of the Intergovernmental Panel on Climate Change have predicted an average global warming between +2 and +6°C, depending on the scenarios, within the next 90 years (IPCC Report 2007). The consequences of this increase in temperature are now well documented on both the abundance and the geographic distribution of numerous taxa. In contrast, cellular or physiological effects for this range of temperature changes remain understudied. These temperature variations being well below the thermal stress endured daily or seasonally by most organisms, most would indeed expect little discriminative results for such temperature increases.
Organisms living in groundwater are not like most organisms since they inhabit a highly buffered biotope that does not undergo any daily or seasonal temperature variations (Ginet and Decou 1977). However, the temperatures of groundwater are directly correlated to the mean annual temperature of the surface. Thus, even if this peculiar environment lacks short-term variations, it will be affected by the consequences of global warming on mean annual temperature. Living in a highly thermally buffered environment, subterranean organisms might be particularly sensitive to temperature changes; they are thus well suited to study cellular responses to +2 or +6°C temperature increases.
Cellular defence mechanisms can occur in order to deal with thermal stress using chaperone proteins. These molecules prevent the aggregation of polypeptides and assist in the refolding of proteins damaged by thermal stress (Kregel 2002) but also play an essential role in unstressed cells by assisting in the initial folding processes (Hartl 1996). The most well-characterised and conserved chaperone protein family is the 70-kDa family of the heat shock proteins (HSP70; Feder and Hofmann 1999; Sorensen et al. 2003). Several studies have shown the physiological and ecological importance of the expression of these HSP70 genes in response to changing environments (Deane and Woo 2005; Hamdoun et al. 2003; Lejeusne et al. 2006). Moreover, the HSP70 family is the most abundant heat shock family in terms of number of isoforms found per organism (Sorensen et al. 2003), the most extensively studied as a biomarker of stress among HSP family (Boutet et al. 2003; Clark et al. 2008b; Ravaux et al. 2007) and is the primary family of heat shock proteins that is induced by thermal stress (Hartl 1996; Selvakumar and Geraldine 2005). For this purpose, inducible cytosolic HSP70 proteins could constitute a good molecular marker to quantify cellular stress due to thermal variations in subterranean organisms.
To test for cellular responses to +2 or +6°C in groundwater taxa, we thus investigated HSP70 genes in Niphargus rhenorhodanensis representatives. These crustaceans are particularly well suited as the ecophysiological and genetic background for this complex of species is abundant (Colson-Proch et al. 2009; Hervant et al. 1995; Issartel et al. 2005; Lefébure et al. 2007; Mathieu 1968). Furthermore, they are ubiquitous in Western Europe and colonise all types of aquifers at all altitudes (Dole-Olivier et al. 2009). Although no organisms here considered are living close to their short-term lethal thermal limit (e.g. 26°C, Mathieu 1968), a moderate increase in underground water temperature could have significant effects on individual fitness. This could further affect the whole population and then functioning and biodiversity of the subterranean ecosystem.
We characterised for the first time a gene encoding HSP70 stress proteins in the subterranean genus Niphargus and measured its expression pattern following heat shocks. The data are discussed within the context of the organisms’ evolutionary adaptation to life in an extreme environment and the applicability of HSP70 to monitor thermal stress in subterranean organisms in the context of global change.
Materials and methods
Identification of an HSP70 gene in the subterranean genus Niphargus
Animals
Organisms belonging to two species complexes of the crustacean genus Niphargus were studied. Taxonomic identifications at the species level were performed using both morphological (Ginet 1996) and molecular (Lefébure et al. 2007) identification. Six populations of the N. rhenorhodanensis complex were collected in the Jura mountains and were assigned to evolutionary significant unit DE (the populations of Volognat and Pissoir), FG (the populations of Froidières, Charabotte 2 and Alex) and I (the population of Chalamont) using molecular criteria (Lefébure et al. 2007; Colson-Proch et al., in preparation). These populations are located in France between 45°55′ and 46°12′ of latitude and 5°24′ and 5°37′ of longitude. Individuals from five of them were collected with a net placed at the emergence springs of different karstic systems: the populations of Volognat, Pissoir, Froidières, Charabotte 2 and Alex (Colson-Proch et al. 2009) and individuals from the population of Chalamont were collected using traps sunk into a porous system (Issartel et al. 2005).
Two populations of the Niphargus virei complex were collected: one at an emergence spring, the population of Source du Diable in the northeast part of France, and one in a well, the population of Dorpstratt in the eastern part of The Netherlands. These populations belong to evolutionary significant unit A for Source du Diable and C for Dorpstratt (Lefébure et al. 2006).
Primer design
HSP70 has been described from a wide range of organisms, but little is known for non-decapod crustaceans and no sequences are available within the taxonomic family of our focal taxa (Niphargidae). Thus, a large selection of cytosolic inducible HSP70 sequences was downloaded from the GenBank® database and was aligned by eyes using Seaview (Galtier et al. 1996). This multiple alignment included two bivalvia (Mytilus galloprovincialis GenBank number AY861684 and Mytilus edulis AF172607), 12 decapod crustaceans (Marsupenaeus japonicus EF091692, Penaeus monodon AF474375, Homarus americanus DQ173923, Fenneropenaeus chinensis AAW71958, Procambarus clarkii ABC01063, Macrobrachium rosenbergii AY466445, Rimiris exoculata ABF85673, Mirocaris fortunata DQ534064, Callinectes sapidus DQ663760, Litopenaeus vannamei AAT46566, Macrobrachium nipponense DQ660140, Pachygrapsus marmoratus AM410078), a crustacean amphipod (consensus sequence of Gammarus pulex EH276349, EH276222, EH276309, EH275268, EH273641) and an insect (Manduca sexta AAF09496). Primers for PCR amplifications on subterranean crustacean DNA were manually designed from this alignment.
DNA templates and PCR amplification
For the amplification of HSP70 genes in the eight populations of Niphargus studied, genomic DNA was isolated from whole organisms using the DNeasy® tissue kit (Qiagen, Hilden, Germany). Amplifications were performed on 300 ng of DNA template in 50-μL volumes containing 1 μM of dNTP, 2X of BSA, 2.5 U of Taq polymerase (Biolabs, Ipswich, MA, USA), 0.4 μM of each primer and 1X of supplied PCR buffer (Biolabs) using an Eppendorf thermocycler (Eppendorf Mastercycler; Eppendorf, Hamburg, Germany). Partial HSP70 sequences were obtained using primer designed at the previous step (i.e. F324: 5′-GATCATCGCCAACGAYCAGGG-3′ and R960: 5′-CGCTTGAAYTCYTGGATGAAGT-3′). An alternative forward primer (F498: 5′-GACATGAARCAYTGGCCCTT-3′) was used when initial amplification was unsuccessful. Amplifications were conducted with the following settings: (1) one step of 2 min at 94°C, (2) 35 cycles of 30 s at 94°C, 30 s at 51°C and 30 s at 72°C; followed by (3) one step of 10 min at 72°C. PCR products were either purified in the laboratory using the NucleoSpin Extract II kit (Macherey-Nagel, Düren, Germany) and then sent to the local sequencing platform (DTAMB) of Lyon 1 University for sequencing or sent to GATC Biotech (Konstanz, Germany) for purification and sequencing.
Characterisation of the HSP70 gene in N. rhenorhodanensis
Phylogenetic reconstructions
In order to define the characteristics of HSP70 gene expression and HSP70 protein cell localisation in N. rhenorhodanensis, we downloaded from the GenBank database a selection of cytosolic HSP70 amino acid sequences from other crustaceans (Table 1) for which previous studies had proven the inducible or the constitutive character of the genes. Moreover, the cytosolic, reticulum and mitochondrial HSP70 sequences available in the GenBank database for Drosophila melanogaster, Caenorhabditis elegans, Saccharomyces cerevisiae, Crassostrea gigas, Danio rerio, Mus musculus and Homo sapiens (Table 1) were collected. The multiple alignment of all these sequences and of the deduced amino acid sequences of N. rhenorhodanensis and N. virei was carried out using ClustalX version 2.0.10 (Larkin et al. 2007) with default alignment parameters. To eliminate ambiguous positions in the alignment prior to phylogenetic reconstructions, GBlocks Server version 0.91b (http://molevol.cmima.csic.es/castresana/Gblocks_server.html) was used with the three options making the selection of blocks less stringent (smaller final blocks, gap positions within the blocks and less strict flanking positions; Castresana 2000). However, we manually reincluded two positions at the 5′ end of the alignment because they were unambiguously aligned and suppressed 15 positions within the alignment that we found ambiguous. Phylogenetic trees were constructed using a Bayesian inference under the CAT+Gamma model as implemented in Phylobayes (Lartillot and Philippe 2004). Two independent chains were run in parallel for 2,500,000 generations. Convergence of the chains was evaluated by monitoring the largest discrepancy observed across all bipartitions between the two independent chains (maxdiff < 0.1) considering trees sampled every 10 minus 806 (corresponding to a burn-in of 25%). In order to reduce phylogenetic artefact such as sequence saturation or long branch attraction, a second analysis was performed after exclusion of the reticulum and mitochondrial HSP70 sequences. The estimation of ambiguous sites and the phylogenetic analysis were conducted as previously described (21 sites were manually suppressed; 1,000,000 generations). Data sets are available upon request.
Table 1.
The HSP70 sequences retrieved from the public databases analysed in Fig. 2
| Species | Taxonomy | Accession number | Characterization | Abbreviations |
|---|---|---|---|---|
| Saccharomyces cerevisiae | Fungi; Ascomycota | M97225 | CYT, I | Scer_CYT_I |
| CAA31393 | CYT, C | Scer_CYT_C | ||
| M27229 | MT, C | Scer_MT_C | ||
| M31006 | ER, C | Scer_ER_C | ||
| Caenorhabditis elegans | Nematoda | Z80223 | CYT, C | Cele_CYT_C |
| Z83108 | CYT, I | Cele_CYT_I | ||
| M26604 | ER, C | Cele_ER_C | ||
| Drosophila melanogaster | Arthropoda; Insecta | AAB59186 | CYT, C | Dmel_CYT_C |
| P82910 | CYT, I | Dmel_CYT_I | ||
| LO1502 | MT, C | Dmel_MT_C | ||
| L01498 | ER, C | Dmel_ER_C | ||
| Gammarus locusta | Arthropoda; Crustacea; Amphipoda | CAQ60114 | CYT, I | Gloc_CYT_I |
| Artemia franciscana | Arthropoda; Crustacea; Branchiopoda | AAL27404 | CYT, I | Afra_CYT_I |
| Balanus amphitrite | Arthropoda; Crustacea; Maxillopoda | AAN74984 | CYT, I | Bamp_CYT_I |
| Rimicaris exoculata | Arthropoda; Crustacea; Malacostraca; Decapoda | ABF85673 | CYT, I | Rexo_CYT_I |
| Macrobrachium rosenbergii | AY466497 | CYT, I | Mros_CYT_I | |
| AAS45710 | CYT, C | Mros_CYT_C | ||
| Procambarus clarkii | ABC01063 | CYT, I | Pcla_CYT_I | |
| Macrobrachium nipponense | ABG45886 | CYT, C | Mnip_CYT_C | |
| Penaeus monodon | AAQ05768 | CYT, C | Pmon_CYT_C | |
| Homarus americanus | ABA02165 | CYT, C | Hame_CYT_C | |
| Callinectes sapidus | ABF83606 | CYT, C | Csap_CYT_C | |
| Pachygraptus marmoratus | ABA02164 | CYT, C | Pmar_CYT_C | |
| Crassostrea gigas | Mollusca; Bivalvia | AB122063 | CYT, I | Cgig_CYT_I |
| AF144646 | CYT, C | Cgig_CYT_C | ||
| AB122065 | ER, C | Cgig_ER_C | ||
| Mus musculus | Chordata; Craniata; Vertebrata; Rodentia | ABK96811 | CYT, I | Mmus_CYT_I |
| P63017 | CYT, C | Mmus_CYT_C | ||
| NP034611 | MT | Mmus_MT | ||
| Homo sapiens | Chordata; Craniata; Vertebrata; Primates | AF134726 | CYT, I | Hsap_CYT_I |
| Y00371 | CYT, C | Hsap_CYT_C | ||
| P38646 | MT | Hsap_MT | ||
| M19645 | ER, C | Hsap_ER_C | ||
| Danio rerio | Chordata; Craniata; Vertebrata; Teleostei | AB062116 | CYT, I | Drer_CYT_I |
| Y11413 | CYT, C | Drer_CYT_C | ||
| NM_213058 | ER, C | Drer_ER_C |
CYT cytoplasmic, ER endoplasmic reticulum, MT mitochondria, I inducible, C cognate
Gene expression levels following a heat shock
Animals Amphipod crustaceans (N. rhenorhodanensis) collected in Chalamont (Ain, France) were acclimated for 6 months in the dark at 10°C (±0.3°C). They were fed once a week with food for aquarium fish (TetraRubin; Tetra, Melle, Germany). Water was changed once a week, and pH, oxygen concentration and temperature were maintained constant during acclimation.
Heat shock of +15°C In order to study the effect of temperature on HSP70 gene expression, individuals were exposed to 25°C for 12 h and then returned to the control temperature (10°C) for a period of 48 h. Time course was performed at 0, 2, 6, 12, 24 or 48 h (N = 2 for each time) during the recovery period at 10°C. Organisms were rapidly frozen in liquid nitrogen and stored at −80°C until total RNA extraction.
Heat shock of +2°C and +6°C In order to study the potential effects of global warming, thermal stresses of +2°C and +6°C were performed on N. rhenorhodanensis. These temperatures were chosen according to the last IPCC Report (2007) which foresees a global warming in the next 90 years between 2°C and 6°C. Organisms were split into three groups: a control group (C; N = 30; 10°C), a +2°C heat-shocked group (HS+2°C; N = 30; 12°C) and a +6°C heat-shocked group (HS+6°C; N = 30; 16°C). Organisms were removed from water baths after 12 h (N = 6), 1 month (N = 6), 2 months (N = 6) or 3 months (N = 6) and immediately placed for 2 h at 10°C. They were then frozen in liquid nitrogen and stored at −80°C until total RNA extraction.
RNA extraction and purification Total RNAs were extracted by homogenisation in Tri Reagent LS (Euromedex, Souffelweyersheim, France) and chloroform was then added; these steps were performed successively twice in order to improve the quality of the extraction. Total RNAs were suspended in 50 μL DEPC water and then purified with Turbo DNA free reagent (Ambion Applied Biosystems, Foster City, CA, USA). Total RNA concentration and quality were determined by spectrophotometry (Nanodrop ND-1000; Thermo Scientific, Wilmington, DE, USA). The absence of genomic DNA contamination was controlled by amplification of total RNAs.
Calibrated reverse transcription Messenger RNAs contained in 1 μg of total RNAs were reverse-transcribed in single-stranded cDNAs using oligo_(dT15) primer and MMLV reverse transcriptase (Promega, Madison, WI, USA). Reactions were performed in a 40-μL volume containing 24 μL of total RNA at a concentration of 41.7 ng/μL, 0.5 μM dNTP mix (Promega), 80 U of RNAsin (Promega), 200 U of MMLV RNase H− (Promega), 2 μg of oligodT, 1X of MMLV RT reaction buffer (Promega) and 80 pg of a synthetic messenger poly(A)-RNA (SmRNA). This solution was incubated at 42°C for 1.5 h and 15 min at 70°C. After reverse transcription, aliquots of 5 μL of cDNA at 10 ng/μL were stored at −80°C until real-time PCR amplifications. The SmRNA included in our solution is a synthetic product designed to be non-homologous to any sequence found in living organisms (Morales and Bezin 2004; World Patent no. 2004/092414 A12004). It was used to normalise the reverse transcription reaction of mRNAs of biological samples (Morales et al. 2006). We chose this strategy over control by a housekeeping gene to avoid dealing with fluctuation in expression level of our control. Indeed, normalisation requires that expression level in control remain stable, and growing evidence attests that this is not the case for most organisms and housekeeping genes (Tricarico et al. 2002; Bustin et al. 2005; Huggett et al. 2005).
Quantitative real-time PCR analysis HSP70 gene expression data were quantified using quantitative polymerase chain reaction (qPCR) performed on a LightCycler® sequence analysis system (Roche Diagnostics, Basel, Switzerland) using the QuantiTect SYBR® Green PCR kit (Qiagen). qPCR amplification was performed using synthetic DNA-specific primers corresponding to the transcript of SmRNA that allowed us to determine the reverse transcription efficiency for each sample. qPCR amplification of the gene of interest, HSP70, was then performed using specific primers designed from the alignment of the eight sequences of HSP70 from Niphargus obtained in this study, forward primer F307-PCRQ 5′-GCTGCGATTGCTTACGG-3′ and reverse primer R408-PCRQ 5′-CGCCAGCAGTAGATTTCACCTC-3′. Amplifications of HSP70 were performed using 5 μL cDNA, 0.25 μmol of each primer and 1 X Mix (QuantiTect Master Mix; Qiagen) to a total volume of 20 μL under the following conditions: one denaturation cycle of 95°C for 15 min and 40 amplification cycles of 94°C for 15 s, 58°C for 30 s and 72°C for 6 s. At the end of each PCR, the absence of primer dimers was confirmed with the analysis of the melting curve obtained after one fusion cycle between 95°C and 68°C for 20 s and one cooling cycle of 40°C for 30 s. The specificity of the PCR products was confirmed by sequencing. Results obtained for HSP70 mRNA were finally normalised against the control (SmRNA).
Statistical analyses
Data were finally expressed as means ± standard deviation of the relative variation (fold induction) between each treatment and the control sample. All data tested were found to conform to assumptions of equal variance and normality. For the first heat shock experiment (+15°C), normalised transcript abundance for HSP70 amplified genes was subjected to a one-way ANOVA with time as main effect to test for significance, followed by a Newman–Keuls multiple comparison test. Data set from the second experiment of heat shocks (+2°C and +6°C) was analysed by a two-way ANOVA with time and heat shock temperature as the main effects, and post hoc comparisons were performed among the groups with the Newman–Keuls multiple range test. All statistical analyses were performed using Statistica v.7 (Statsoft, Tulsa, OK, USA), and differences were considered significant at P < 0.05.
Results
Identification of an HSP70 gene in the subterranean genus Niphargus
The F324, F498 and R960 primers used in this study allowed us to obtain eight DNA fragments from six populations of N. rhenorhodanensis and two populations of N. virei. Sequencing revealed a high level of similarity with known HSP70 gene sequences using BLASTN and BLASTP searches, thus demonstrating the specificity of the amplification product. Total length of the partial heat shock protein transcripts ranged between 386 and 580 nucleotides, and the eight sequences are deposited under GenBank accession nos. GQ324981 to GQ324988. Sequences were named according to the name of both the taxa and the geographic location (Fig. 1). The partial sequence of HSP70 from individuals belonging to the population of Chalamont, called Nrhe_Chalamont_HSP70, began 494 bp after the 5′ end and finished 1,552 bp before the 3′ end of the full Gammarus locusta nucleotide sequence (Crustacea amphipod; FM165078).
Fig. 1.
Multiple alignment of newly characterised fragments and deduced amino acid sequence (at the first base of the codon) of the isolated heat shock protein 70 kDa. Dots indicate that residues were identical in reference to the top level sequence from the population of Chalamont (N. rhenorhodanensis), called Nrhe_Chalamont_Hsp70, and dashes stand for gaps. Boxed nucleotides represent non-synonymous substitutions, and substitutions in bold are in first or second position of codons. Nucleotides boxed in Nrhe_Chalamont_Hsp70 sequence represent the F307 and R408 primers used in quantitative PCR to quantify HSP70 mRNA. The nucleotides and amino acids are numbered along the right margin. The HSP70 characteristic motifs are underlined in bold, and the eukaryotic ATP-GTP binding site is in italic in the amino acid sequence
Sequences belonging to the same phylogenetic unit of N. rhenorhodanensis shared between 99.1% and 100% similarity, whereas sequences belonging to distinct phylogenetic units of the N. rhenorhodanensis complex shared between 96.6% and 98% similarity. Both sequences of N. virei (belonging to two distinct phylogenetic units; Lefébure et al. 2006) shared 98.9% similarity. The percentage of similarity between N. rhenorhodanensis and N. virei comprised between 89.8% and 92.5%. Among the 53 mutations present on this alignment, 47 (88.7%) were localised in the third position within codons and seven (13.2%) are non-synonymous substitutions.
The corresponding amino acid sequence of Nrhe_Chalamont_HSP70 contained three HSP70 family signatures of which one is in the prosite bank: TVPAYFND, NEPTAA and IFDLGGGTFDVSIL (prosite ID HSP70_2 PS00329; Karlin and Brocchieri 1998; Rensing and Maier 1994; Fig. 1). This fragment also included a sequence similar to the eukaryotic ATP-binding site described in the prosite bank (prosite ID PDOC00017), AEAYIGRK (Fig. 1).
On the other hand, this multiple alignment does not encompass important signatures such as: (1) a consensus pattern specific for the cytosolic isoforms of HSP70 situated approximately 1,230 bp after the 3′ end of our amplified fragment, (2) a characteristic motif of some but not all cognate proteins situated near the COOH ending (Boorstein et al. 1994) and (3) an insertion location for an intron reported in several constitutive proteins HSC70 (Boutet et al. 2003; Gunther and Walter 1994).
Characterisation of the Nrhe_Chalamont_Hsp70 gene
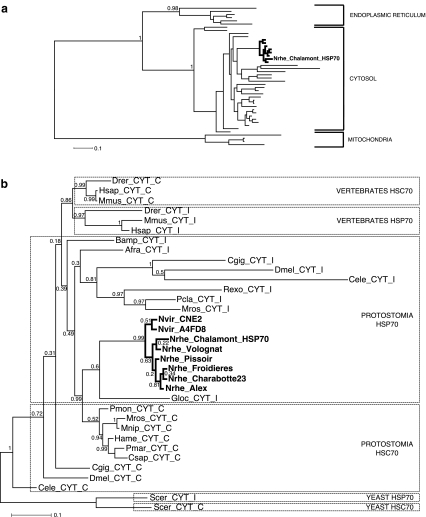
In order to determine the intracellular localisation of the protein encoded by the Nrhe_Chalamont_Hsp70 gene, a phylogenetic analysis was carried out (Fig. 2a). This analysis included sequences from cytosolic, endoplasmic reticulum and mitochondrial HSP70 amino acid sequences reported for crustaceans, yeast, the fruit fly, C. elegans, C. gigas and three vertebrates. As expected from previous studies (Boorstein et al. 1994; Rensing and Maier 1994), mitochondrial, endoplasmic reticulum and cytosolic genes were clearly separated into three groups (posterior probabilities of 0.98 and 1; Fig. 2a). All the sequences characterised in this study and in particular the sequence Nrhe_Chalamont_Hsp70 clustered within the cytosolic HSP70 clade with strong support (posterior probability of 1; Fig. 2a).
Fig. 2.
a Topology of the most probable phylogenetic tree showing the relationship of Niphargus HSP70 deduced amino acid sequences (branches in bold) to other HSP70 family members from different intracellular compartments (endoplasmic reticulum, cytosol and mitochondria). b Most probable phylogenetic tree showing the relationship of Niphargus HSP70 deduced amino acid sequences to other cytosolic HSP70 family members from a variety of species. Bold font indicates the sequences identified in this study (GenBank accession nos. GQ324981 to GQ324988). Boxes distinguish cytosolic constitutive (HSC70) and inducible (HSP70) sequences belonging to yeast, vertebrates and protostomia. The trees were reconstructed using Phylobayes programme under the CAT-Gamma model. Topology and posterior probabilities—given at each node—were evaluated after convergence of two independent chains (maxdiff < 0.1). GenBank accession nos. for amino acid sequences and abbreviations are reported in Table 1
Moreover, the cytosolic heat shock proteins comprised various inducible HSP70 and constitutive HSC70 from protostomes and vertebrates. Since the topology of the phylogenetic reconstructions for these cytosolic heat shock proteins can be biased by saturation or long branch attraction phenomena, we performed a second analysis with these cytosolic heat shock proteins only, excluding mitochondrial and reticulum HSP70 (Fig. 2b). It suggests (with a marginal support, posterior probability of 0.6) a relationship of Nrhe_Chalamont_Hsp70 sequence with an inducible HSP70 (G. locusta, another Crustacean amphipod; Fig. 2b). This trend is slightly reinforced by the fact that inducible and constitutive copies of vertebrates tend to cluster together and that constitutive cytosolic HSC70 of crustaceans are monophyletic (Fig. 2b), arguing for a relatively limited number of duplications. Our phylogenetic reconstructions thus ascertain the cellular localisation of the HSP70 genes amplified in this study, but they lack the necessary resolution to surely determine any specific expression regime for these genes.
Nevertheless, since the best criterion to distinguish between constitutive HSC70 and inducible HSP70 is the assessment of the gene expression level after an increase in temperature (Leignel et al. 2007), we performed a calibrated reverse transcription (cRT)-qPCR analysis after heat shock treatments. Primers designed for qPCR allowed the amplification of 99 bp corresponding to 32 amino acids (Fig. 1). Among them, 14 constitute an HSP70 signature which confirms that the primers used in qPCR are specific of HSP70 proteins. Considering all the thermal stress experiments, HSP70 expression was analysed for 26 control organisms kept at 10°C. The amount of Nrhe_Chalamont_Hsp70 mRNA in control organisms was 25,000 ± 1,480 copies per microgram of total RNA, and this value was considered as reference of onefold induction. The drastic thermal stress of +15°C induced a significant induction of the Nrhe_Chalamont_Hsp70 transcription between 23- and 28-fold (Fig. 3). Thus, the suggestion of our second phylogenetic analysis is highly corroborated by this expression measurement after a heat shock, and the Nrhe_Chalamont_HSP70 gene here characterised is a member of the inducible cytosolic HSP70 family.
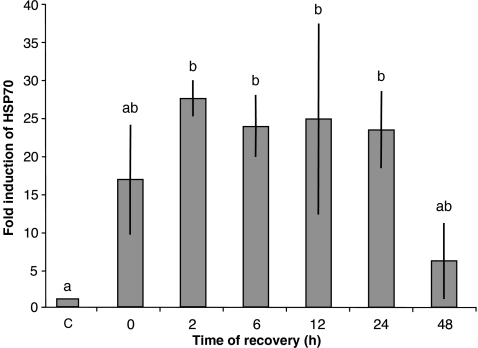
Fig. 3.
Graphical representation of quantitative PCR results. Total RNA was extracted from control and heat-shocked crustaceans, and Nrhe_Chalamont_Hsp70 mRNA expression was analysed with specific probes. Control animals (C; fold induction = 1) were maintained at 10°C over the entire experimental period; treated animals were exposed to 25°C for 12 h and returned at 10°C for a period of post-stress recovery (0–48 h). Plotted data are expressed as fold induction with respect to sample maintained at 10°C (mean ± SD). Each bar represents the mean of two individuals. Different letters above the bars indicate values significantly different from each other (P < 0.05)
Moreover, this experiment showed that Nrhe_Chalamont_Hsp70 mRNA was over-expressed during the early phases of post-stress recovery, reaching a maximum after 2 h and decreasing after 24 h. Based on this experiment, we chose a 2-h recovery period at the control temperature (10°C) for subsequent treatments.
Heat shock experiments considering global change predictions
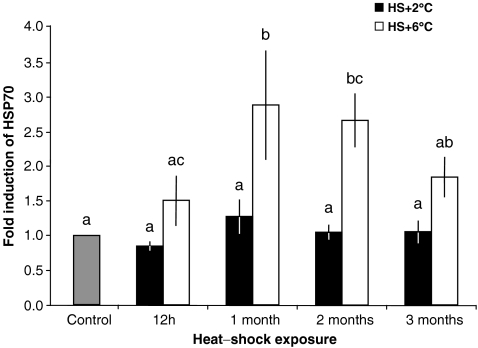
Thermal acclimation at 12°C (thermal shock of +2°C) did not produce a significant induction of Nrhe_Chalamont_Hsp70 gene transcription whatever the duration of the thermal stress (Fig. 4). A thermal shock of +6°C induced a significant approximately threefold induction of the amount of Nrhe_Chalamont_Hsp70 transcripts after 1 and 2 months of thermal stress. After 3 months, the quantity of Nrhe_Chalamont_Hsp70 transcripts reached a twofold induction that was no more significantly different from the controls. Two-way ANOVA for Nrhe_Chalamont_Hsp70 mRNA levels revealed a significant effect of time (P = 0.041) and heat shock temperature (P = 0), but no significant interaction (P = 0.159).
Fig. 4.
Graphical representation of quantitative PCR results. Total RNA was extracted from control and heat-shocked crustaceans, and Nrhe_Chalamont_Hsp70 mRNA expression was analysed with specific probes. Control animals (C; fold induction = 1) were maintained at 10°C over the entire experimental period; treated animals were exposed to 12°C (HS+2°C, black bars) or 16°C (HS+6°C, white bars) for 12 h, 1, 2 or 3 months and returned to 10°C for a 2-h period of post-stress recovery. Plotted data are expressed as fold induction with respect to a sample kept at 10°C (mean ± SD). Each bar represents the mean of six individuals. Different letters above the bars indicate values significantly different from each other (P < 0.05)
Discussion
This study reports the first characterisation of a HSP70 gene fragment in organisms inhabiting thermally buffered groundwaters (i.e. the crustaceans of N. rhenorhodanensis and N. virei complexes). The comparison of the amino acid sequences indicated that the Niphargus sequences partially amplified in this study are situated within the 44 kDa NH2 terminal ATPase domain of HSP70 proteins. The three HSP70 signatures found in the eight sequences are situated in highly conserved core blocks (Karlin and Brocchieri 1998) which are very strongly conserved across all known HSP70 sequences, suggesting that these blocks are functionally and/or structurally important. Nevertheless, the exact role of these signatures is not clearly known, except for the IFDLGGGTFDVSIL signature that is a PO−4 binding site (Modisakeng et al. 2004). Moreover, all the proteins that bind ATP share a conserved sequence that interacts with this nucleotide; a very similar sequence was identified in our sequences (AEAYIGRK) and was also found in other HSP70 sequences such as in the shrimps M. fortunata (AEAFLGGT) and Rimicaris exoculata (AESYLGKK; Ravaux et al. 2007). The DLFGGGTFD signature and the ATP-binding functional domain are characteristic of the HSP70 family in eukaryots. The gene sequenced in this study thus clearly belongs to the eukaryotic HSP70 family, hereby excluding prokaryotic contamination.
Structural analyses did not allow us to further characterise the cellular localisation nor the expression regime of the HSP70 amplified in Niphargus. Indeed, the amplified region did not encompass the C-terminal tail or the positions affected by an intron insertion in some constitutive HSC70 sequences. Yet the motifs characteristic of the cellular localisation and of the expression profiles of the HSP70 proteins are localised in these regions. Whilst a 5′ rapid amplification of cDNA ends (RACE) and 3′-RACE approach might have been developed, we chose an alternative approach implying both phylogenetic reconstruction and expression analysis by cRT-qPCR. This option was taken because structural analyses are at best indicative but never truly conclusive (Leignel et al. 2007), all these signatures being non-obligatory. Moreover, considering our aim to characterise cellular responses of subterranean crustaceans in a context of global change, obtaining a full-length sequence of the HSP70 was not our primary objective. All the sequences amplified in N. rhenorhodanensis and N. virei clustered together with the other sequences of HSP70 that are cytosolic, and percentages of similarity found are consistent with known phylogenies of N. rhenorhodanensis and N. virei (Lefébure et al. 2006, 2007). This strongly suggests that the sequences amplified in the different populations of Niphargus are not paralogs. The weak phylogenetic relation between Niphargus sequences and the inducible HSP70 of G. locusta tends to show that the gene amplified in this study encoded inducible 70 kDa heat shock chaperones. We concentrated our expression study on the N. rhenorhodanensis population from Chalamont (Ain, France) as individuals of this location have been extensively studied (Hervant et al. 1995; Issartel et al. 2005; Lefébure et al. 2007; Mathieu 1968). The inducibility of the Nrhe_Chalamont_HSP70 gene suggested by the phylogenetic analyses was then confirmed by its high sensitivity to heat shocks.
Nrhe_Chalamont_Hsp70 gene expression was time-dependent, with a maximum reached 2 h after a heat shock. The expression level of HSP70 returned to control levels after 48 h, thus clearly defining a stress-associated role for this HSP70. Such mRNA HSP70 expression was highly similar to the transcriptional profile of heat-induced response observed in one species of Pacific abalone (Cheng et al. 2007). This response might be considered atypical since it differed from several other studies in which mRNA levels were back to control levels much more rapidly, within 5–10 h after the heat shock (Bahrndorff et al. 2009; Franzellitti and Fabbri 2005). N. rhenorhodanensis live in a thermally buffered biotope (groundwater) such as Orchesella cincta (soil; Bahrndorff et al. 2009), whereas the Pacific abalone Haliotis discus hannai (Cheng et al. 2007) and the mollusc M. galloprovincialis (Franzellitti and Fabbri 2005) live in highly fluctuating environments represented by intertidal biotopes. Thus, there is apparently no clear links between environmental characteristics and the different patterns of HSP70 expression observed in these organisms.
Despite the quasi-absence of thermal variations in their biotope, subterranean crustaceans such as N. rhenorhodanensis exhibited the capacity to induce HSP70 gene over-expression in response to a heat stress. This observation is somewhat consistent with a recent study that found similar physiological response in highly stenothermal organisms, Antarctic marine molluscs (Clark et al. 2008a). In contrast, previous studies showed the lack of a HSP70 heat shock response in Antarctic fishes, sea stars and crustaceans (Clark et al. 2008b; Hofmann et al. 2000). As N. rhenorhodanensis is subjected to a maximum temperature variation of 1°C or 2°C within a year (Ginet and Decou 1977), the question arises as to how this species living in a highly stable biotope still possesses the ability to up-regulate its HSP70 genes. HSP70 is often referred as a ubiquitous response to stress (Feder and Hofmann 1999). Thus the over-expression observed in N. rhenorhodanensis could protect them from alternative stresses such as hypoxia (Sorensen et al. 2003) as this stress can be encountered in subterranean environments (Malard and Hervant 1999). Another explanation could be a vestigial attribute from a past phylogenetic history. Previous studies (Colson-Proch et al. 2009; Issartel et al. 2005; Lefébure et al. 2007) have indeed suggested that N. rhenorhodanensis complex, at least some populations or evolutionary significant unit, survived the last glacial event within or close to areas covered by ice, in groundwater at very low temperatures. As exposure to cold can induce HSP70 expression (Ali et al. 2003; Feder and Hofmann 1999), the ability to up-regulate HSP70 genes could be a remnant capacity from an ancestor exposed to very low temperatures, until 10,000 years ago. Finally, a third hypothesis is that N. rhenorhodanensis could be a recent subterranean species displaying heat shock responses inherited from a recent epigean ancestor. However, multiple facts suggest that N. rhenorhodanensis colonised groundwater several millions of years ago. The genus Niphargus Schiödte comprised over 300 described species and subspecies, and except 12 species, almost all of these taxa inhabit subterranean waters (Väinölä et al. 2008). Moreover, N. rhenorhodanensis displays a much longer life cycle than its closest epigean relatives (Ginet and Decou 1977), high faculties to survive to long-term starvation (Hervant et al. 1997) and hypoxic conditions (Hervant et al. 1995) and several other adaptations to life in groundwaters, suggesting that the time spent in subterranean waters is relatively long. Thus, these characteristics suggest that the colonisation of the subterranean environment by Niphargus ancestors is very old.
At the cellular level, N. rhenorhodanensis seemed to be insensible to an increase in temperature of 2°C even after 3 months at 12°C. This is congruent with an ecophysiological study performed by Issartel et al. (2005) on the same population of N. rhenorhodanensis that showed no significant variations in oxygen consumption, locomotory and ventilatory activities between 10°C and 12°C. A heat shock of +6°C over 12 h did not induce a significant accumulation of Nrhe_Chalamont_Hsp70 mRNA either, but it is interesting to note that inter-individual variability was high. Thus, N. rhenorhodanensis seemed to be able to live in water at 16°C during several hours and in water at 12°C without up-regulating the expression of this particular HSP70 gene. Background HSP70 expression or constitutive HSC70 proteins can be sufficient for this kind of thermal stress (Ali et al. 2003; Fangue et al. 2006). An alternative would be that other HSP70 genes not studied here can be upregulated. Results from multiple studies have indeed shown that heat stress induced several members of the HSP70 gene family, called isoforms (Clark et al. 2008b; Deane and Woo 2005; Fangue et al. 2006). One inducible form of HSP70 is surveyed in this study of N. rhenorhodanensis, but preliminary sequence data indicated that other isoforms could be present (Colson-Proch, unpublished data). Investigations on these other HSP70 genes are currently underway. Moreover, other chaperone proteins such as HSP90 or HSP60 could also be involved in dealing with this short-term and small-scale stress (Fangue et al. 2006; Smith et al. 1999).
In contrast, a 6°C increase in temperature induced a threefold induction of the HSP70 transcription after 1 month of acclimation, but after 3 months, the amount of HSP70 mRNA returned to a level that no longer differed significantly from the control level. At least three hypotheses can be invoked to explain this transient expression pattern:
Organisms can be acclimated at the elevated temperature of 16°C after 2 months, thanks to the accumulation of new thermostable protein isoforms as suggested by Berger and Emlet (2007), and other inducible HSP70 isoforms or heat shock proteins can be sufficient to tolerate this temperature. Interestingly, this alternative is supported by Franzellitti and Fabbri (2005) suggestion that constitutive HSC70 are engaged in long-term protection, whereas HSP70s represent a short-term cytoprotective mechanism.
Due to high physiological stress, organisms can be unable to maintain a large amount of HSP70 mRNA in their cells. Indeed, the energetic cost associated with the production of heat shock proteins is very high (Feder and Hofmann 1999; Sorensen et al. 2003). A decrease in HSP70 mRNA amount could be representative of the global reduction of their metabolism, leading to the death of the individuals. We can note that inter-individual variability in gene expression after 12 h and 1 month was clearly higher at 16°C (coefficients of variation of 51 and 60, respectively) than it was at 12°C (coefficients of variation of 18 and 42 after 12 h and 1 month, respectively), whereas there was no difference after 3 months (coefficients of variation of 32 at 12°C and 31 at 16°C). This suggests that a heat shock of +6°C represented a high thermal stress for individuals at least after 12 h and 1 month, whereas a +2C heat shock did not. The similar inter-individual variability found in the two groups after 3 months could be explained by the selection of the most resistant individuals who responded similarly to the heat exposure. The mortality observed during the experiment of heat acclimation at 16°C was in agreement with this explanation. Despite the difficulty to replicate survival experiments, a preliminary study of survival at 17°C for individuals naturally living at 11°C suggested that a +6°C heat treatment caused irreversible deleterious effects leading to the death of 50% of the individuals after 3 months, as observed in our study (Colson-Proch, unpublished data).
A third hypothesis could be that the turnover of mRNA and HSP70 proteins can change at elevated temperature and a high level of HSP70 proteins can be maintained for a long time (over 3 months) with the concomitant decrease in quantity of HSP70 mRNA. Indeed, our study examined mRNA levels, but as a decoupling of transcription and translation for HSP70 can exist, mRNA levels are not necessarily predictive of the behaviour of the protein pool (Bahrndorff et al. 2009; Hamdoun et al. 2003; Smith et al. 1999). Changes in the rate of HSP70 protein synthesis and catabolism may be important regulatory components of the long-term modulation in control HSP70 level (Hamdoun et al. 2003). Bahrndorff et al. (2009) showed that HSP70 mRNA reached a peak after 2 h of recovery after a 1-h heat shock of +20°C, whereas protein levels reached the highest level 49 h after heat hardening. Such prolonged stable response was also found in some fish species that maintained a high level of HSP70 proteins for at least 5 days after a 2-h heat shock (Bierkens 2000). Interestingly, Krebs and Feder (1998) predicted that individuals living in environments where thermal stress is not a common issue may produce HSP70 proteins more slowly than individuals living in stressful biotopes. We can thus expect that N. rhenorhodanesis display a high decoupling between the up-regulation of the transcription and the accumulation of newly synthesised HSP70 proteins.
This controlled laboratory-based physiological experiment allowed the first characterisation of an HSP70 gene in N. rhenorhodanensis and was essential as a first step towards field investigations of heat shock response in subterranean organisms. It demonstrated that HSP70 expression and induction have been maintained in subterranean organisms despite a lack of environmental temperature stress over several thousands of years.
Acknowledgements
This research was supported by funds from University Claude Bernard–Lyon 1 from the IFR41 of the National Centre of French Scientific Research (CNRS) and from the Institut Universitaire de France. The authors greatly thank Laurent Bezin for delivery of SmRNA aliquots and the local platform of Développement des Techniques d’Analyse Moléculaire de la Biodiversité (DTAMB) of the University of Lyon 1 for sequencing. We also wish to thank C. Richardson and an anonymous reviewer who helped improve this article.
References
- Ali KS, Dorgai L, Abraham M, Hermesz E. Tissue- and stressor-specific differential expression of two hsc70 genes in carp. Biochem Bioph Res Co. 2003;307:503–509. doi: 10.1016/S0006-291X(03)01206-3. [DOI] [PubMed] [Google Scholar]
- Bahrndorff S, Mariën J, Loeschcke V, Ellers J. Dynamics of heat-induced thermal stress resistance and Hsp70 expression in the springtail, Orchesella cincta. Funct Ecol. 2009;23:233–239. doi: 10.1111/j.1365-2435.2009.01541.x. [DOI] [Google Scholar]
- Berger MS, Emlet RB. Heat-shock response of the upper intertidal barnacle Balanus glandula: thermal stress and acclimation. Biol Bull. 2007;212:232–241. doi: 10.2307/25066605. [DOI] [PubMed] [Google Scholar]
- Bierkens JGEA. Applications and pitfalls of stress-proteins in biomonitoring. Toxicology. 2000;153:61–72. doi: 10.1016/S0300-483X(00)00304-8. [DOI] [PubMed] [Google Scholar]
- Boorstein WR, Ziegelhoffer T, Craig EA. Molecular evolution of the HSP70 multigene family. J Mol Evol. 1994;38:1–17. doi: 10.1007/BF00175490. [DOI] [PubMed] [Google Scholar]
- Boutet I, Tanguy A, Moraga D. Organization and nucleotide sequence of the European flat oyster Ostrea edulis heat shock cognate 70 (hsc70) and heat shock protein 70 (hsp70) genes. Aquat Toxicol. 2003;65:221–225. doi: 10.1016/S0166-445X(03)00137-1. [DOI] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Nolan T, Pfaffl MW. Quantitative real-time RT-PCR—a perspective. J Mol Endocrinol. 2005;34:597–601. doi: 10.1677/jme.1.01755. [DOI] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Cheng P, Liu X, Zhang G, He J. Cloning and expression analysis of a HSP70 gene from Pacific abalone (Haliotis discus hannai) Fish Shellfish Immun. 2007;22:77–87. doi: 10.1016/j.fsi.2006.03.014. [DOI] [PubMed] [Google Scholar]
- Clark MS, Fraser KPP, Peck LS. Antarctic marine molluscs do have an HSP70 heat shock response. Cell Stress Chaperon. 2008;13:39–49. doi: 10.1007/s12192-008-0014-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MS, Fraser KPP, Peck LS. Lack of an HSP70 heat shock response in two Antarctic marine invertebrates. Polar Biol. 2008;31:1059–1065. doi: 10.1007/s00300-008-0447-7. [DOI] [Google Scholar]
- Colson-Proch C, Renault D, Gravot A, Douady CJ, Hervant F. Do current environmental conditions explain physiological and metabolic responses of subterranean crustaceans to cold? J Exp Biol. 2009;112:1859–1868. doi: 10.1242/jeb.027987. [DOI] [PubMed] [Google Scholar]
- Deane EE, Woo NYS. Cloning and characterization of the hsp70 multigene family from silver sea bream: modulated gene expression between warm and cold temperature acclimation. Biochem Bioph Res Co. 2005;330:776–783. doi: 10.1016/j.bbrc.2005.03.039. [DOI] [PubMed] [Google Scholar]
- Dole-Olivier MJ, Malard F, Martin D, Lefébure T, Gibert J. Relationship between environmental variables and groundwater biodiversity at the regional scale. Freshwat Biol. 2009;54:797–813. doi: 10.1111/j.1365-2427.2009.02184.x. [DOI] [Google Scholar]
- Fangue NA, Hofmeister M, Schulte PM. Intraspecific variation in thermal tolerance and heat shock protein gene expression in common killifish, Fundulus heteroclitus. J Exp Biol. 2006;209:2859–2872. doi: 10.1242/jeb.02260. [DOI] [PubMed] [Google Scholar]
- Feder ME, Hofmann GE. Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu Rev Physiol. 1999;61:243–282. doi: 10.1146/annurev.physiol.61.1.243. [DOI] [PubMed] [Google Scholar]
- Franzellitti S, Fabbri E. Differential HSP70 gene expression in the Mediterranean mussel exposed to various stressors. Biochem Bioph Res Co. 2005;336:1157–1163. doi: 10.1016/j.bbrc.2005.08.244. [DOI] [PubMed] [Google Scholar]
- Galtier N, Gouy M, Gautier C. SeaView and Phylo_win, two graphic tools for sequence alignment and molecular phylogeny. CABIOS. 1996;12:543–548. doi: 10.1093/bioinformatics/12.6.543. [DOI] [PubMed] [Google Scholar]
- Ginet R. Bilan systématique du genre Niphargus en France. Lyon: Société Linéenne de Lyon and Hydrobiologie et Ecologie Souterraines; 1996. [Google Scholar]
- Ginet R, Decou V. Initiation à la biologie et à l’écologie souterraines. Paris: Delarge; 1977. [Google Scholar]
- Gunther E, Walter L. Genetic aspects of the hsp70 multi-family in vertebrates. Experientia. 1994;50:987–1001. doi: 10.1007/BF01923453. [DOI] [PubMed] [Google Scholar]
- Hamdoun AM, Cheney DP, Cherr GN. Phenotypic plasticity of HSP70 and HSP70 gene expression in the Pacific Oyster (Crassostrea gigas): implications for thermal limits and induction of thermal tolerance. Biol Bull. 2003;205:160–169. doi: 10.2307/1543236. [DOI] [PubMed] [Google Scholar]
- Hartl FU. Molecular chaperones in cellular protein folding. Nature. 1996;381:571–579. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- Hervant F, Mathieu J, Garin D, Freminet A. Behavioral, ventilatory and metabolic responses to severe hypoxia and subsequent recovery of the hypogean Niphargus rhenorhodanensis and the epigean Gammarus fossarum (Crustacea: Amphipoda) Physiol Biochem Zool. 1995;68:223–244. [Google Scholar]
- Hervant F, Mathieu J, Barre H, Simon K, Pinon C. Comparative study on the behavioral, ventilatory and respiratory responses of hypogean and epigean crustaceans to long-term starvation and subsequent feeding. Comp Biochem Physiol A—Mol Integr Physiol. 1997;118:1277–1283. doi: 10.1016/S0300-9629(97)00047-9. [DOI] [Google Scholar]
- Hofmann GE, Buckley BA, Airaksinen S, Keen JE, Somero GN. Heat-shock protein expression is absent in the Antarctic fish Trematomus bernacchii family Nototheniidae. J Exp Biol. 2000;203:2331–2339. doi: 10.1242/jeb.203.15.2331. [DOI] [PubMed] [Google Scholar]
- Huggett J, Dheda K, Bustin S, Zumla A. Real-time PCR normalisation; strategies and considerations. Genes Immun. 2005;6:279–284. doi: 10.1038/sj.gene.6364190. [DOI] [PubMed] [Google Scholar]
- IPCC (2007) Climate change 2007. Contribution of Working Groups I, II and III to the fourth assessment report of the Intergovernmental Panel on Climate Change (edited by Pachauri RK, Reisinger A), Switzerland
- Issartel J, Hervant F, Voituron Y, Renault D, Vernon P. Behavioural, ventilatory and respiratory responses of epigean and hypogean crustaceans to different temperatures. Comp Biochem Physiol A—Mol Integr Physiol. 2005;141:1–7. doi: 10.1016/j.cbpb.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Karlin S, Brocchieri L. Heat Shock protein 70 family: multiple sequence comparisons, function, and evolution. J Mol Evol. 1998;47:565–577. doi: 10.1007/PL00006413. [DOI] [PubMed] [Google Scholar]
- Krebs RA, Feder ME. Hsp70 and larval thermotolerance in Drosophila melanogaster: how much is enough and when is more too much? J Insect Physiol. 1998;44:1091–1101. doi: 10.1016/S0022-1910(98)00059-6. [DOI] [PubMed] [Google Scholar]
- Kregel KCJ. Heat shock proteins: modifying factors in physiological stress responses and acquired thermotolerance. J Appl Physiol. 2002;92:2177–2186. doi: 10.1152/japplphysiol.01267.2001. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Philippe H. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol Biol Evol. 2004;21:1095–1109. doi: 10.1093/molbev/msh112. [DOI] [PubMed] [Google Scholar]
- Lefébure T, Douady CJ, Gouy M, Trontelj P, Briolay J, Gibert J. Phylogeography of a subterranean amphipod reveals cryptic diversity and dynamic evolution in extreme environments. Mol Ecol. 2006;15:1797–1806. doi: 10.1111/j.1365-294X.2006.02888.x. [DOI] [PubMed] [Google Scholar]
- Lefébure T, Douady CJ, Malard F, Gibert J. Testing dispersal and cryptic diversity in a widely distributed groundwater amphipod (Niphargus rhenorhodanensis) Mol Phylogenet Evol. 2007;42:676–686. doi: 10.1016/j.ympev.2006.08.020. [DOI] [PubMed] [Google Scholar]
- Leignel V, Cibois M, Moreau B, Chénais B. Identification of new subgroup of HSP70 in Bythograeidae (hydrothermal crabs) and Xanthidae. Gene. 2007;396:84–92. doi: 10.1016/j.gene.2007.02.025. [DOI] [PubMed] [Google Scholar]
- Lejeusne C, Pérez T, Sarrazin V, Chevaldonné P. Baseline expression of heat-shock proteins (HSPs) of a “thermotolerant” Mediterranean marine species largely influenced by natural temperature fluctuations. Can J Fish Aquat Sci. 2006;63:2028–2037. doi: 10.1139/F06-102. [DOI] [Google Scholar]
- Malard F, Hervant F. Oxygen supply and the adaptations of animals in groundwater. Freshw Biol. 1999;41:1–30. doi: 10.1046/j.1365-2427.1999.00379.x. [DOI] [Google Scholar]
- Mathieu J. Lethal temperatures and thermic acclimatization in Niphargus longicaudatus (amphipode, gammaridé) Bull Soc Zool France. 1968;93:595–603. [Google Scholar]
- Modisakeng KW, Dorrington RA, Blatch GL. Isolation of genes encoding heat shock protein 70 (hsp70s) from the coelacanth, Latimeria chalumnae. S Afr J Sci. 2004;100:683–686. [Google Scholar]
- Morales A, Bezin L (2004) Method of calibration of reverse transcription using a synthetic messenger RNA (SmRNA). Patent WO2004092414
- Morales A, Bonnet C, Bourgoin N, Touvier T, Nadam J, Laglaine A, Navarro F, Moulin C, Georges B, Pequignot JM, Bezin L. Unexpected expression of orexin-B in basal conditions and increased levels in the adult rat hippocampus during pilocarpine-induced epileptogenesis. Brain Res. 2006;1109:164–175. doi: 10.1016/j.brainres.2006.06.075. [DOI] [PubMed] [Google Scholar]
- Ravaux J, Toullec JY, Léger N, Lopez P, Gaill F, Shillito B. First hsp70 from two hydrothermal vent shrimps, Mirocaris fortunata and Rimicaris exoculata: characterization and sequence analysis. Gene. 2007;386:162–172. doi: 10.1016/j.gene.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Rensing SA, Maier UG. Phylogenetic analysis of the stress-70 protein family. J Mol Evol. 1994;39:80–86. doi: 10.1007/BF00178252. [DOI] [PubMed] [Google Scholar]
- Selvakumar S, Geraldine P. Heat shock protein induction in the freshwater prawn Macrobrachium malcolmsonii : acclimation-influenced variations in the induction temperatures for Hsp 70. Comp Biochem Physiol A—Mol Integr Physiol. 2005;140:209–215. doi: 10.1016/j.cbpb.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Smith TR, Tremblay GC, Bradley TM. Charaterization of the heat shock protein response of Atlantic salmon (Salmo salar) Fish Physiol Biochem. 1999;20:279–292. doi: 10.1023/A:1007743329892. [DOI] [Google Scholar]
- Sorensen JG, Kristensen TN, Loeschcke V. The evolutionary and ecological role of heat shock proteins. Ecol Lett. 2003;6:1025–1037. doi: 10.1046/j.1461-0248.2003.00528.x. [DOI] [Google Scholar]
- Tricarico C, Pinzani P, Bianchi S, Paglierani M, Distante V, Pazzagli M, Bustin SA, Orlando C. Quantitative real-time reverse transcription polymerase chain reaction: normalization to rRNA or single housekeeping genes is inappropriate for human tissue biopsies. Anal Biochem. 2002;309:293–300. doi: 10.1016/S0003-2697(02)00311-1. [DOI] [PubMed] [Google Scholar]
- Väinölä R, Witt JDS, Grabowski M, Bradbury JH, Jazdzewski K, Sket B. Global diversity of amphipods (Amphipoda; Crustacea) in freshwater. Hydrobiologia. 2008;595:241–255. doi: 10.1007/s10750-007-9020-6. [DOI] [Google Scholar]