Abstract
TonB from Escherichia coli and its homologues are critical for the uptake of siderophores through the outer membrane of Gram-negative bacteria using chemiosmotic energy. When different models for the mechanism of TonB mediated energy transfer from the inner to the outer membrane are discussed, one of the key questions is whether TonB spans the periplasm. In this article, we use long range distance measurements by spin-label pulsed EPR (Double Electron–Electron Resonance, DEER) and CD spectroscopy to show that the proline-rich segment of TonB exists in a PPII-like conformation. The result implies that the proline-rich segment of TonB possesses a length of more than 15 nm, sufficient to span the periplasm of Gram-negative bacteria.
Keywords: TonB, outer membrane, active transport, EPR, DEER, polyproline II
Introduction
TonB from Escherichia coli and its homologues are critical for the uptake of siderophores and a number of other nutrients through the outer membrane of Gram-negative bacteria,1 and in E. coli for the docking of phages such as T1 (eponymous) and φ80.2 The TonB system is also critical for uptake of bacterial toxins like colicin Ia and B3 and certain antibiotics (albomycin, rifamycin, and microcin 254). In complex with the cytoplasmic membrane proteins ExbB and ExbD, and dependent on the proton motive force, TonB serves a large class of “TonB-dependent” outer membrane receptors, each responsible for the uptake of specific cargo molecules, including iron complexed by siderophores, heme,5 transferrin6 and lactoferrin, cobalt as cyanocobalamin, nickel, copper, thiamine, and carbohydrates.7 In the cell, these receptors outnumber TonB. For the FepA receptor and TonB, for example, a molar ratio of 12.5 has been estimated.8 This suggests that the transport mechanism involves a mobile sampling mechanism in which each TonB complex interacts transiently with many different receptors, recognizing those that are ligand loaded and in some way transducing energy derived from the proton motive force to them to effectuate transport.
Within an N-terminal stretch of 32 residues (in E. coli) a hydrophobic segment anchors TonB in the cytoplasmic membrane (CM). This is followed by a proline-rich segment of ∼100 residues that includes a stretch of 68 residues in which more than 1 in every 3 residues is a proline. A globular domain with affinity for TonB-dependent receptors forms the C-terminus of the protein. ExbB and ExbD, which exhibit distant homology with components of the flagellar motor,9 appear to form a complex with TonB.10,11 Suppressor mutation analysis has suggested that ExbB interacts with TonB through the transmembrane domain12 while an NMR study suggested that ExbD interacts weakly with the periplasmic domain.13
The crystal structures of several TonB-dependent receptors show a common architecture. 22-stranded β-barrels span the outer membrane, with the lumen filled almost completely by a cork domain formed by the N-terminal part of the receptor polypeptide. This leaves two pockets at the extracellular and the periplasmic openings of the β-barrel. Prior to TonB-mediated energy transduction, the cargo molecule is bound to a high-affinity site at the bottom of the extracellular pocket. Binding involves residues from the cork, the barrel and from the characteristic long loops connecting β-strands of the barrel on the extracellular side, which in some cases enclose the binding site like petals.14 The binding affinity is typically in the nanomolar range, and binding occurs whether or not the cell has a functional TonB system, indicating that energy transduction through the TonB-receptor complex is required to dissociate the cargo molecule from the binding site and to open an exit channel for its passage through the barrel and into the periplasm.
Because TonB had been found critical for energy transduction from the cytoplasmic membrane to the receptors, it was consistent that complexes of receptors with TonB were found to be stabilized upon binding of cargo molecules.15,16 It was also in accord with the observation of conformational transitions in the structure of some receptors on the periplasmic side after binding of cargo molecules.17,18 Crystal and NMR structures of the C-terminal globular domain of TonB alone15,19 and in complex with outer membrane receptors20,21 have been reported. In the complexes the domain was in contact with the so-called TonB-box of the receptors, a conserved stretch of ∼7 amino acids near the periplasmic N-terminal end of the cork domain.
Models have been proposed as to how TonB might transduce electrochemical energy to the receptors so as to effect cargo translocation from the receptor binding site to the periplasm.
In one model, TonB adopts an energy-rich conformation through interaction with ExbB/ExbD and the proton motive force. This “loaded-spring” dissociates from the cytoplasmic membrane, diffuses across the periplasmic space, binds to cargo-loaded receptors and discharges its energy so that by means of an unspecified mechanism the translocation of the cargo is effected (“shuttle model”).22 In another model, TonB molecules, which are anchored in the CM but touch a loaded receptor on the periplasmic side of the outer membrane, dimerize by means of their C-terminal domain. ExbB and ExbD by means of the proton motive force rotate the two TonB molecules. This torque leads to a twisting of the two TonB molecules pulling the C-terminal domains and the attached N-terminus of the receptor cork domain away from the outer membrane into the periplasmic space, thus unfolding the cork entirely or partially and abolishing the cargo binding site as well as creating the exit channel (“propeller model”).23 Gumbart et al.24 have studied the feasibility of partial unfolding or removal of the cork domain by a force pulling at the C-terminal TonB perpendicular to the membrane into the periplasm by a molecular dynamics simulation.
In accord with the shuttle model, the N-terminal domain of TonB, which would be in the cytoplasm if it remained in the CM, could be labeled with a reagent impermeable to the cytoplasmic membrane, and only when it was functional in siderophore transport.25 On the other hand, a study by Kaserer et al.26 using TonB fusions is at odds with the dissociation of TonB from the CM. For the propeller model, the strongest support for the generation of a rotational torque stems from the homology between the ExbB and ExbD and the MotA and MotB proteins of the flagellar motor. However, no evidence has been provided that TonB actually rotates and the observed 1:1 stochiometry of TonB and receptor in the structure of the complex is not in accord with this model without modifications.
Thus far, differentiating between the shuttle and propeller models is not possible, in spite of the large amount of information about protein–protein interactions in the TonB energy transduction system. One item of information which could address the feasibility of the models, however, would be knowledge of the physical length of TonB. The width of the periplasm is estimated to be 15–20 nm,27 so TonB should match this length in order to be able to deliver torque that is generated in the CM to a receptor in the outer membrane. TonB and its homologues share a central proline-rich segment of 68 residues of which 24 are prolines. Moreover, in its N-terminal half acidic residues but no basic residues are abundant whereas in its C-terminal half this pattern is reversed. Short distance information from NMR studies28,29 have already suggested that TonB adopts an extended structure allowing it to span the estimated 20 nm width of the periplasmic space. The anomalous migration in SDS-PAGE indicates rigidity and elongation as well.30 The high content of proline and the aqueous environment suggests that a polyproline helix II (PPII) would be a possible conformation, that is a left-handed helix with an axial translation of 0.31 nm per residue, rather than a right-handed polyproline I helix (PPI) with an axial translation of 0.19 nm per residue. Experiments showed that the PPII conformation is maintained in proline-rich polypeptides up to a certain content of other residues.31,32 The dihedral angles of a PPII conformation (φ = −75°, ψ = 145°, ω = 180°) resemble those of an antiparallel β-sheet. Both conformations are close to the most extended conformation which a polypeptide can adopt in the absence of steric constraints.
In this study, we present circular dichroism data as well as long range distance measurements for the proline-rich segment of TonB using pulsed EPR. EPR spectroscopy is a method for revealing structure and dynamics in disordered systems. EPR techniques can access distances between 0.5 and 8 nm measuring the dipole–dipole interaction between two spins.33 Diamagnetic systems can be studied using spin-labels.
The results show that the proline-rich segment of TonB exists in a PPII conformation with a length enabling it to span the periplasm of Gram-negative bacteria. On the basis of the conformational data obtained here, we propose a modification to the propeller model in which the mechanism of energy induces a torque-generated mutarotation of the PPII segment of TonB to a PPI conformation.
Results
We first investigated the structure of the proline-rich segment of TonB using a two-frequency, pulsed EPR method [double electron–electron resonance (DEER) or pulsed electron–electron double resonance (PELDOR)].33–37 The DEER method was chosen, because it allows the measurement of distances from 1.5 nm up to 8 nm in disordered materials.34
To obtain long-range information on the conformation of the central part of TonB, we expressed and purified the proline-rich segment between residues 56 and 126 of native E. coli TonB with a N-terminal Hexahis-tag (see Fig. S1 in Supporting Information). Distances were then measured between pairs of spin-labels introduced by site-specific cysteine mutagenesis and derivatized with the thiol reactive spin-label MTSL (1-oxy-2,2,5,5-tetramethylpyrroline-3-methyl-methanethiosulfonate). The following six double cysteine mutants derivatized with MTSL were investigated: TonB 59/69, TonB 59/76, TonB 69/76, TonB 69/84, TonB 88/106, and TonB 106/120 (The pairs of numbers indicate the two residues of native TonB which are replaced by the cysteines, see Figure S1 in Supporting Information).
The protein conformation in aqueous solution was trapped by shock-freezing, and the distance measurements were performed with the frozen solution. To minimize spin relaxation due to proton hyperfine interactions deuterated water was used as the solvent. Distances below 1.5 nm are accessible by analyzing the broadening of continuous wave (cw)-EPR spectra in frozen solution. In control experiments, no differences between cw-EPR spectra of singly and doubly labeled TonB mutants were obtained (data not shown). Therefore, intramolecular distances below 1.5 nm were excluded. Protein aggregation/dimerization under the conditions used was also ruled out by analysis of DEER traces of a singly labeled mutant which showed a homogeneous three-dimensional spin distribution (see Fig. S2 in Supporting Information).
The dipolar evolution curves obtained by DEER for the double-mutants are shown in Figure 1. Model free analysis revealed that the spin-label distance distributions could be well fitted by Gaussian distributions. To facilitate comparison we analyzed the DEER data assuming Gaussian distance distributions characterized by two parameters only. The experimental data could be fitted by this model (thick solid lines in Fig. 1). Table I lists the parameters of these distance distributions for all of the mutants. The widths of the distributions observed do not reflect the error of the method but the conformational variability of the protein itself and of the spin-label linkers. Assuming a linker length of the MTSL spin-label of ∼0.5 nm the findings suggest that the protein conformation is rather stiff but not completely rigid. To estimate the deviation from a linear backbone conformation a set of three double-mutants (TonB 59/69, TonB 69/76, and TonB 59/76) was designed to allow for triangulation. Adding the distances found individually for both sections (59–69 and 69–76) results in 5.4 nm in total which is about 20% longer than the distance of the 59–76 section measured directly, suggesting deviations from a linear conformation of the backbone. The assumption of a slightly flexible backbone is supported by the fact that the width of the distance distribution increases with the distance between the corresponding spin-label pair.
Figure 1.
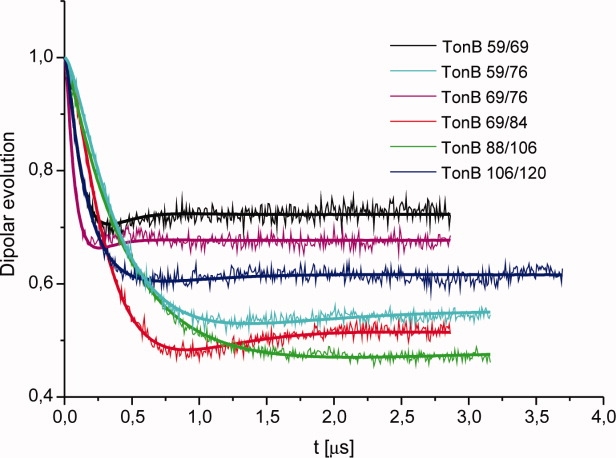
DEER traces. Background corrected dipolar evolution data from four-pulse DEER experiments for different double-labeled mutants of the proline-rich segment of TonB (thin solid lines). Thick solid lines correspond to the fit assuming a Gauss distribution, parameters shown in Table I.
Table I.
Parameters Characterizing the Gaussian Distance Distributions Derived from DEER Measurements
| Spin-labeled residues | Distance (nm) | Width of the distribution (nm) |
|---|---|---|
| TonB 69/76 | 2.5 ± 0.1 | 0.8 ± 0.1 |
| TonB 59/69 | 2.9 ± 0.1 | 0.8 ± 0.1 |
| TonB 106/120 | 3.3 ± 0.15 | 1.4 ± 0.1 |
| TonB 69/84 | 4.0 ± 0.1 | 1.1 ± 0.1 |
| TonB 59/76 | 4.4 ± 0.1 | 1.6 ± 0.1 |
| TonB 88/106 | 4.6 ± 0.1 | 2.1 ± 0.15 |
The main value of the EPR distance measurements rests in the possibility of estimating the displacement per residue of the proline-rich segment of TonB by adding the lengths of the individually measured sections. This resulted in an average spin-label distance of 0.26 nm per residue and a full length of 14.4 nm (see Table I).
To calculate the distance projected onto the helix axis from the distance between spin-labels attached to a helical structure, one must take into account the distance due to the relative angular displacement of the two labeled residues. A rigid PPII helix can be modeled as a cylinder with a diameter D of about 1 nm38 (see Fig. S3 in Supporting Information). Owing to the threefold point symmetry component of the PPII helix the additional distance contribution is maximal when the relative angular displacement of the spin-labels corresponds to an angle of 120°. Using a linker length l of 0.5 nm the maximal additional distance contribution can be estimated. For all spin-label positions under investigation it is maximally 0.2 nm and on average 0.1 nm and therefore can be neglected. Relative angular displacement could affect the triangulation experiment as well. Here, the experimentally obtained difference between the sum of the distances of both sections and the distance in total is 1 nm. Relative angular displacement corresponding to an angle of 120° would have the largest effect, but would result in a difference of only 0.4 nm. In conclusion, the difference obtained in the triangulation experiment cannot be explained by angular displacement but indicates a slightly variable conformation.
The DEER results suggested that the TonB proline-rich segment has an extended structure and may in fact be in a PPII conformation. As an independent assessment of this conformation, the circular dichroism spectrum of the purified unmodified proline-rich segment of TonB was analyzed. The results in Figure 2 show a prominent dip at 205 nm in the spectrum that is characteristic for the PPII conformation as shown using model systems.
Figure 2.
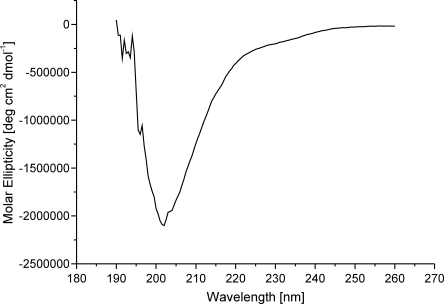
CD spectrum of the proline-rich segment of TonB. The CD spectrum of the TonB wild-type fragment containing residues 56–126 shows the characteristic feature of a polyproline-II-helical structure.
Discussion
Despite the wealth of information available concerning the protein–protein interactions of TonB with high affinity outer membrane receptors, the known requirement for an energized membrane for transport, the crystal structures of receptors, the C-terminal domain of TonB and the complex of both, the mechanism by which TonB acts is unknown. The shuttle and propeller models discussed in the literature entail radically different modes of action. Because there is no high resolution structure of TonB in complex with ExbB and ExbD available and because of the dynamic nature of the envisaged mechanisms, it is difficult to judge which of these models is reasonable. A central tenet of the propeller model however is that TonB must be able to reach the receptor in the outer membrane, that is that it can span the periplasm. The long range information concerning the structure of the proline-rich TonB segment obtained here via a spin-label approach indicates for the first time that TonB is long enough to span the periplasm. The total length of the different sections of the proline-rich segment as measured by DEER and listed in Table I indicates a length of ∼14.4 nm. In conjunction with an estimated 6.5 nm length for the C-terminal globular domain as measured on the crystal structure, a total length of more than 20 nm is conceivable (see Fig. 3).
Figure 3.
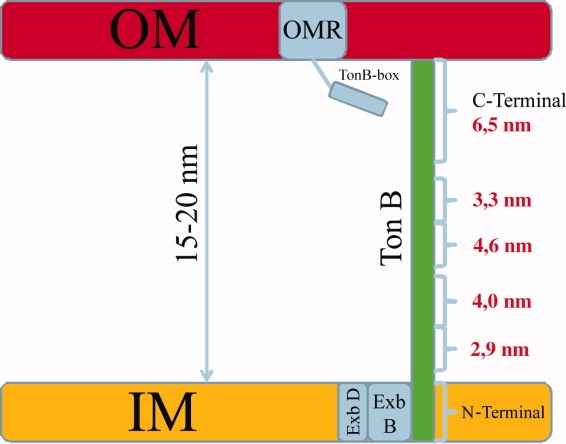
Scheme of the structure of TonB. Distances derived from DEER measurements are indicated in the diagram showing that the proline-rich segment of TonB spans the periplasm. [Color figure can be viewed in the online issue, which is available at http://www.interscience.wiley.com.]
The EPR data also indicated some flexibility of the protein conformation resulting in minor deflections from a stick-like conformation. Because of this backbone flexibility the individual spin-label distances of the TonB 59/69 and TonB 69/76 pairs were ∼20% larger than the distance obtained directly with the TonB 59/76 pair. The flexibility may facilitate the sampling by TonB of the OM for ligand loaded receptors. The averaged spin-label distance value was 0.26 nm per residue. Taking the backbone flexibility into account an averaged axial translation of 0.31 nm per residue results. This is significantly larger than the axial translation for an alpha-helical conformation (0.15 nm/residue), but is in full quantitative agreement with a PPII helix model. The possibility of a PPII conformation was also supported by the CD measurements which clearly indicated the signature dip at 205 nm. This is also in full agreement with recent NMR data28,29 which suggested based on short range informations an elongated structure of the proline-rich segment of TonB.
A PPII conformation is adopted by many proline-rich sequences, but other sequences that have few or even no prolines also adopt this structure. Charged residues, glutamine, and alanine have relatively high propensities for PPII conformation.31,32
The finding of a PPII conformation that provides for a length sufficient to span the periplasm may have implications for models of TonB function that go beyond the demonstration that it would not have to dissociate from the cytoplasmic membrane to contact receptors as suggested by the shuttle model. The results presented here are not in conflict with the propeller model, however, that model contradicts the 1:1 complex seen in crystal structures.
The tendency of PPII helices under certain conditions to perform a mutarotation to a PPI conformation39 that can result in a contraction of about 40% inspires us to propose the following modification to the propeller model: TonB is anchored in the cytoplasmic membrane flanked by the flagellum motor-like proteins ExbB and ExbD. These proteins exert a torque on TonB resulting in a mutarotation if TonB is bound to the TonB-box of the outer membrane receptor resulting in an angular change of ω from 180° (trans) to 0° (cis) in the peptide bonds of the proline-rich segment. The mutarotation leads to a partial or complete transition from the stable PPII to the unstable PPI conformation accompanied by a contraction which can amount up to 40% of the length of the proline-rich segment. This results in an inward force as is assumed for partial unfolding or removal of the cork from the receptor by Gumbart et al.,24 thereby mediating dissociation of the cargo molecule from its binding site and translocation to the periplasm. In future studies, we will examine whether or not the proline-rich segment of TonB can undergo a transition from a PPII to a PPI conformation.
Acknowledgments
The authors thank Bettina Nägele, Daniela Lehr, and Frederike Eggers for experimental contributions as well as Kay Diederichs for stimulating discussions.
References
- 1.Wang CC, Newton A. An additional step in the transport of iron defined by the TonB locus of Escherichia coli. J Biol Chem. 1971;246:2147–2151. [PubMed] [Google Scholar]
- 2.Hancock REW, Braun V. Nature of the energy requirement for the irreversible adsorption of bacteriophages T1 and φ80 to Escherichia coli. J Bacteriol. 1976;125:409–415. doi: 10.1128/jb.125.2.409-415.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hilsenbeck JL, Park HJ, Chen G, Youn B, Postle K, Kang CH. Crystal structure of th cytotoxic bacterial protein colicin B at 2.5 Å resolution. Mol Microbiol. 2004;51:711–720. doi: 10.1111/j.1365-2958.2003.03884.x. [DOI] [PubMed] [Google Scholar]
- 4.Salomón RA, Fariás RN. The peptide antibiotic microcin 25 is imported through the TonB pathway and the SbmA protein. J Bacteriol. 1995;177:3323–3325. doi: 10.1128/jb.177.11.3323-3325.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wandersman C, Stojiljkovic I. Bacterial heme sources: the role of heme, hemoprotein receptors and hemophores. Curr Opin Microbiol. 2000;3:215–220. doi: 10.1016/s1369-5274(00)00078-3. [DOI] [PubMed] [Google Scholar]
- 6.Cornelissen CN, Biswas GD, Tsai J, Paruchuri DK, Thompson SA, Sparling PF. Gonococcal transferrin-binding protein 1 is required for transferrin utilization and its homologous to TonB-dependent outer membrane receptors. J Bacteriol. 1992;174:5788–5797. doi: 10.1128/jb.174.18.5788-5797.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schauer K, Rodionov DA, de Reuse H. New substrates for TonB-dependent transport: do we see the ‘tip of the iceberg’. Trends Biochem Sci. 2008;33:330–338. doi: 10.1016/j.tibs.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 8.Higgs PI, Larsen RA, Postle K. Quantification of known components of the Escherichia coli TonB energy transduction system: TonB, ExbB and FepA. Mol Microbiol. 2002;44:271–281. doi: 10.1046/j.1365-2958.2002.02880.x. [DOI] [PubMed] [Google Scholar]
- 9.Cascales E, Lioubes R, Sturgis JN. The TolQ-TolR proteins energize TolA and share homologies with the flagellar motor proteins MotA-MotB. Mol Microbiol. 2001;42:795–807. doi: 10.1046/j.1365-2958.2001.02673.x. [DOI] [PubMed] [Google Scholar]
- 10.Braun V, Gaisser S, Herrmann C, Kampfenkel K, Killmann H, Traub I. Energy coupled transport across the outer membrane of Escherichia coli: ExbB binds ExbD and TonB in vitro, and leucine 132 in the periplasmic region and aspartate 25 in the transmembrane region are important for ExbD activity. J Bacteriol. 1996;178:2836–2284. doi: 10.1128/jb.178.10.2836-2845.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Higgs PI, Myers PS, Postle K. Interactions in the TonB-dependent energy transduction complex: ExbB and ExbD form heteromultimers. J Bacteriol. 1998;180:6031–6038. doi: 10.1128/jb.180.22.6031-6038.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Larson RA, Thomas MT, Wood GE, Postle K. Partial suppression of an Escherichia coli TonB transmembrane domain mutation (ΔV17) by a missense mutation in ExbB. Mol Microbiol. 1994;13:627–640. doi: 10.1111/j.1365-2958.1994.tb00457.x. [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Herrero A, Peacock RS, Howard SP, Vogel HJ. The solution structure of the periplasmic domain of the TonB system ExbD protein reveals and unexpected structural homology with siderophore-binding proteins. Mol Microbiol. 2007;66:872–889. doi: 10.1111/j.1365-2958.2007.05957.x. [DOI] [PubMed] [Google Scholar]
- 14.Ferguson AD, Chakraborty R, Smith BS, Esser L, van der Helm D, Deisenhofer J. Structural basis of gating by the outer membrane transporter FecA. Science. 2002;295:1715–1719. doi: 10.1126/science.1067313. [DOI] [PubMed] [Google Scholar]
- 15.Ködding J, Killig F, Polzer P, Howard PS, Diederichs K, Welte W. Crystal structure of a 92-residue C-terminal fragment of TonB from Escherichia coli reveals significant conformational changes compared to structures of smaller TonB fragments. J Biol Chem. 2005;280:3022–3028. doi: 10.1074/jbc.M411155200. [DOI] [PubMed] [Google Scholar]
- 16.Khursigara CM, De Crescenzo G, Pawelek PD, Coulton JW. Enhanced binding of TonB to a ligand-loaded outer membrane receptor: role of the oligomeric state of TonB in formation of a functional FhuA-TonB complex. J Biol Chem. 2003;279:7405–7412. doi: 10.1074/jbc.M311784200. [DOI] [PubMed] [Google Scholar]
- 17.Ferguson AD, Hofmann E, Coulton JW, Diederichs K, Welte W. Siderophore-mediated iron transport: crystal structure of FhuA with bound lipopolysaccharide. Science. 1998;282:2215–2220. doi: 10.1126/science.282.5397.2215. [DOI] [PubMed] [Google Scholar]
- 18.Chimento DP, Mohanty AK, Kadner RJ, Wiener MC. Substrate-induced transmembrane signaling in the cobalamin transporter BtuB. Nat Struct Biol. 2003;10:394–401. doi: 10.1038/nsb914. [DOI] [PubMed] [Google Scholar]
- 19.Peacock RS, Weljie AM, Howard SP, Price FD, Vogel HJ. The solution structure of the C-terminal domain of TonB and interaction studies with TonB box peptides. J Mol Biol. 2005;345:1185–1197. doi: 10.1016/j.jmb.2004.11.026. [DOI] [PubMed] [Google Scholar]
- 20.Pawelek PD, Croteau N, Ng-Thow-Hing C, Khursigara CM, Moiseeva N, Allaire M, Coulton JW. Structure of TonB in complex with FhuA, E. coli outer membrane receptor. Science. 2006;312:1399–1402. doi: 10.1126/science.1128057. [DOI] [PubMed] [Google Scholar]
- 21.Shultis DD, Purdy MD, Banchs CN, Wiener MC. Outer membrane active transport: structure of the BtuB:TonB complex. Science. 2006;312:1396–1399. doi: 10.1126/science.1127694. [DOI] [PubMed] [Google Scholar]
- 22.Larsen RA, Letain TE, Postle K. In vivo evidence of TonB shuttling between the cytoplasmic and outer membrane in Escherichia coli. Mol Microbiol. 2003;49:211–218. doi: 10.1046/j.1365-2958.2003.03579.x. [DOI] [PubMed] [Google Scholar]
- 23.Chang C, Mooser A, Plückthun A, Wlodawer A. Crystal structure of the dimer C-terminal domain of TonB reveals a novel fold. J Biol Chem. 2001;276:27535–27540. doi: 10.1074/jbc.M102778200. [DOI] [PubMed] [Google Scholar]
- 24.Gumbart J, Wiener MC, Tajkhorshid E. Mechanics of force propagation in TonB-dependent outer membrane transport. Biophys J. 2007;93:496–504. doi: 10.1529/biophysj.107.104158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larsen RA, Letain TE, Postle K. In vivo evidence of TonB shuttling between the cytoplasmic and outer membrane in Escherichia coli. Mol Microbiol. 2003;49:211–218. doi: 10.1046/j.1365-2958.2003.03579.x. [DOI] [PubMed] [Google Scholar]
- 26.Kaserer WA, Jiang X, Xiao Q, Scott DC, Bauler M, Copeland D, Newton SMC, Klebba PE. Insight from TonB hybrid proteins into the mechanism of iron transport through the outer membrane. J Bacteriol. 2008;190:4001–4016. doi: 10.1128/JB.00135-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Graham LL, Beveridge TJ, Naninga N. Freeze-substitution of gram-negative eubacteria: general cell morphology and envelope profiles. J Bacteriol. 1991;173:1623–1633. doi: 10.1128/jb.173.5.1623-1633.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Evans JS, Levine BA, Trayer IP, Dorman CJ, Higgins CF. Sequence-imposed structural constraints in the TonB protein of E. coli. FEBS Lett. 1986;208:211–216. doi: 10.1016/0014-5793(86)81020-1. [DOI] [PubMed] [Google Scholar]
- 29.Brewer S, Tolley M, Trayer IP, Barr GC, Dorman CJ, Hannavy K, Higgins CF, Evans JS, Levine BA, Wormald MR. Structure and function of X-Pro dipeptide repeats in the TonB proteins of Salmonella typhimurium and Escherichia coli. J Mol Biol. 1990;216:883–895. doi: 10.1016/S0022-2836(99)80008-4. [DOI] [PubMed] [Google Scholar]
- 30.Postle K, Good RF. DNA sequence of the Escherichia coli TonB gene. Proc Natl Acad Sci USA. 1983;80:5235–5239. doi: 10.1073/pnas.80.17.5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kay BK, Williamson MP, Sudol M. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 2000;14:231–241. [PubMed] [Google Scholar]
- 32.Adzhubei AA, Sternberg MJE. Left-handed polyproline II helices commonly occur in globular proteins. J Mol Biol. 1993;229:472–493. doi: 10.1006/jmbi.1993.1047. [DOI] [PubMed] [Google Scholar]
- 33.Milov AD, Maryasov AG, Tsvetkov YD. Pulsed electron double resonance (PELDOR) and its applications in free-radicals research. Appl Magn Reson. 1998;15:107–143. [Google Scholar]
- 34.Jeschke G. Distance measurements in the nanometer range by pulse EPR. Chem Phys Chem. 2002;3:927–932. doi: 10.1002/1439-7641(20021115)3:11<927::AID-CPHC927>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 35.Larsen RG, Singel DJ. Double electron–electron resonance spin–echo modulation: spectroscopic measurement of electron spin pair separations in orientationally disordered solids. J Chem Phys. 1993;98:5134–5146. [Google Scholar]
- 36.Jeschke G, Polyhach Y. Distance measurements on spin-labelled biomacromolecules by pulsed electron paramagnetic resonance. Phys Chem Chem Phys. 2007;9:1895–1910. doi: 10.1039/b614920k. [DOI] [PubMed] [Google Scholar]
- 37.Schiemann O, Prisner TF. Long-range distance determinations in biomacromolecules by EPR spectroscopy. Q Rev Biophys. 2007;40:1–53. doi: 10.1017/S003358350700460X. [DOI] [PubMed] [Google Scholar]
- 38.Ladias JAA. Structural basis for polyproline recognition by the FE65 WW domain. J Mol Biol. 2007;372:970–980. doi: 10.1016/j.jmb.2007.06.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kakinoki S, Hirano Y, Oka M. On the stability of polyproline-I and II structures of proline oligopeptides. Polym Bull. 2005;53:109–115. [Google Scholar]


