Abstract
Cysteine residues can complicate the folding and storage of proteins due to improper formation of disulfide bonds or oxidation of residues that are natively reduced. Wild-type Rop is a homodimeric four-helix bundle protein and an important model for protein design in the understanding of protein stability, structure and folding kinetics. In the native state, Rop has two buried, reduced cysteine residues in its core, but these are prone to oxidation in destabilized variants, particularly upon extended storage. To circumvent this problem, we designed and characterized a Cys-free variant of Rop, including solving the 2.3 Å X-ray crystal structure. We show that the C38A C52V variant has similar structure, stability and in vivo activity to wild-type Rop, but that it has dramatically faster unfolding kinetics like virtually every other mutant of Rop that has been characterized. This cysteine-free Rop has already proven useful for studies on solution topology and on the relationship of core mutations to stability. It also suggests a general strategy for removal of pairs of Cys residues in proteins, both to make them more experimentally tractable and to improve their storage properties for therapeutic or industrial purposes.
Keywords: Rop, four-helix bundle, cysteine-free, unfolding kinetics
Introduction
Protein engineering for improved physical properties, such as stability, folding kinetics or solubility, remains a difficult challenge.1 A major focus has been the repacking of the hydrophobic core of proteins, because a number of lines of evidence suggest that hydrophobic core packing is the key parameter affecting the stability of most proteins. These residues tend to be the most conserved for a particular protein family, and the most intolerant of mutation.2,3 Remarkable successes in packing or repacking protein cores have come from rational and computational design, as well as directed evolution.4–8 But there is still no accurate predictive model for the thermodynamic or kinetic consequences of even a single point mutation in the core of a protein.
Engineering cysteine residues into or out of a protein has been a major area of investigation. Disulfide bonds can dramatically stabilize structures, and have been successfully engineered for this purpose.9–11 It is also frequently desirable to engineer cysteine-free variants of proteins for study. Cysteines complicate the folding and storage of proteins, both because disulfide bond formation tends to be a slow step in folding and because oxidation of cysteines (usually to adventitious disulfides) can result in misfolding and oligomerization. This is especially problematic for destabilized mutants of proteins. For example, a cysteine-free variant of T4 lysozyme was engineered by Matthews and colleagues as the basis for dozens of mutants designed to understand the structural and thermodynamic consequences of amino acid changes.12 A variant of human superoxide dismutase without free cysteines is resistant to thermal inactivation and has improved E. coli expression.13
An interesting question in engineering cysteines is, to what (or from what) residues should one make a cysteine mutation? In active sites, a reasonable choice is serine, although serine is less nucleophilic and less acidic than cysteine. It is often underappreciated that cysteine is quite hydrophobic and much larger than serine due to the oxygen-to-sulfur substitution. In fact, cysteine is intermediate between alanine and valine both in volume14 and in the Kyte and Doolittle hydropathy scale.15 Moreover, hydrogen bonds between the Cys thiol and backbone carbonyl oxygens' would be expected to be much weaker than those commonly observed from the Ser or Thr alcohol, due to the poorer pKA match of the thiol proton. The BLOSUM62 matrix16 shows that the highest probability substitution throughout evolution of Cys is for Ala (log-odds of zero), followed by Ile, Leu, Met, Ser, Thr, and Val (log-odds of −1).
Coiled coils and helical bundles have proven especially useful for protein engineering and in elucidating the effects of mutations on protein stability. For example, the four-helix bundle Rop is a homodimer of 63 amino acid monomers arranged in antiparallel fashion (Fig. 1).17 Extensive mutations have been made to the core and loop of Rop to probe their effects on stability, folding and structure.8,18–22 Recently, we have reported in vivo and in vitro screens for the folding and stability of Rop, with the goal of correlating the effects of many kinds of mutations with changes in thermodynamics.23,24 A proof-of-principle library of mutations in the central core of Rop (I15 T19 L41 A45 to all amino acids with the NNK residue) produced a library of active variants with a range of physical properties (TJM and Lynne Regan, manuscript in preparation). Wild-type Rop has reduced cysteine residues buried in the core at positions 38 and 52, and less-stable and molten globular variants were found to oxidatively dimerize upon storage. These proved difficult to reduce, even upon boiling in SDS-PAGE loading buffer containing dithiothreitol, presumably because once one disulfide forms between the monomers a second forms intramolecularly with little entropic penalty.
Figure 1.
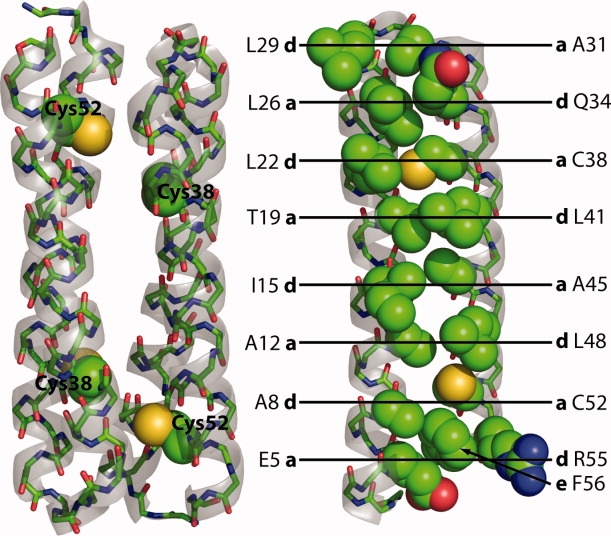
The structure of wild-type Rop. Rop is an antiparallel homodimer of 63 amino acid monomers, placing the reduced Cys38 and Cy52 residues near each other across the dimer interface. At right, in the monomer, residues in the hydrophobic core of Rop are shown as spheres and labeled with their position (a, d, or e) in the heptad repeat. Note that in the last repeat, R55 (d) is mostly exposed and F56 (e) instead packs into the core. Rendered from 1ROP with PyMOL (Delano Scientific).
To circumvent this problem, we sought to engineer a cysteine-free Rop variant with native-like activity, structure and stability. To this end, we used our cell-based activity screen to examine combinations of Ser, Ala, and Val in positions 38 and 52. We found that a C38A C52V variant of Rop is stable and active, and the X-ray crystal structure demonstrates that there is little perturbation as a result of these mutations. However, Cys-free Rop unfolds much more rapidly than wild-type Rop, which has anomalously slow unfolding for a small protein lacking prolines or disulfide bonds. We have recently employed this useful variant as the basis for a comprehensive study of Rop core mutations using a dye-based screen for thermal stability we dubbed High-Throughput Thermal Scanning (HTTS).23
Results
Construction, activity, and stability
From the crystal structure of wild-type Rop (1ROP),17 a hydrogen bond between the thiol of Cys38 and the backbone carbonyl oxygen of Thr19 appears possible (3.4 Å), but it is mostly surrounded by hydrophobic residues and appears to be well-packed (Fig. 2). The thiol of Cys52 is slightly further from the backbone carbonyl oxygen of Leu48 (3.5 Å), and it appears to be somewhat less well packed. Both positions 38 and 52 are in the a position of the canonical heptad repeat, which are typically small amino acids; however, the next-to-last layers of the Rop hydrophobic core are reversed, and position 52 would be expected to be a large amino acid (Fig. 1).8 We decided to initially mutate Cys38 and Cys52 to Ser/Ser, Ala/Ala, and Val/Val. The genes for these variants were produced by PCR assembly of four ∼62-mer synthetic oligonucleotides. The genes were cloned both into pACT7lac24 to assess function and into pMR10125 for overexpression.
Figure 2.
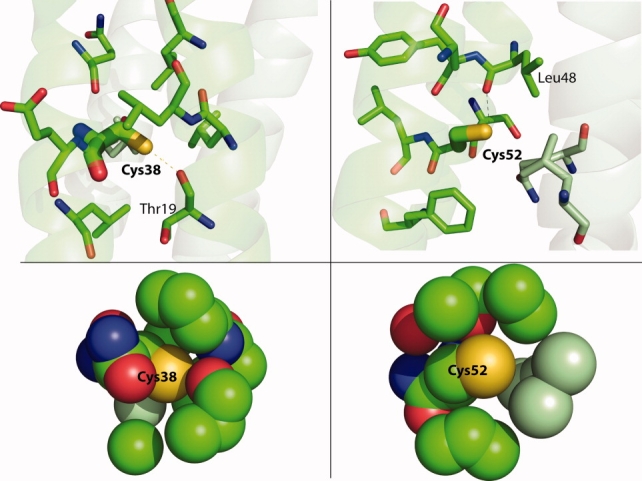
Packing and interactions of Cys38 and Cys52 in wild-type Rop. Residues within 5 Å are shown at the top, and atoms within 5 Å of the Cys sulfur atoms are shown below. Mint-colored carbons are from the other monomer. At left, Cys38 may make a hydrogen bond to the carbonyl oxygen of Thr19, but it is well-packed with mostly hydrophobic residues nearby. At right, Cys52 is slightly further from the carbonyl oxygen of Leu48 but appears to be much less tightly packed. Rendered from 1ROP with PyMOL.
Figure 3 shows the results of the in vivo functional assay. Rop controls the copy number of ColE1 plasmids in E. coli, and the basis of the screen is expression of green fluorescent protein driven from a ColE1 plasmid.24 In this implementation (the “positive screen”), active variants yield a high level of GFP expression, while an inactive variants shows a low level. The Ser/Ser mutant is not active. The other three constructs show a level of function equal to wild-type Rop. Analysis of ColE1 plasmid levels by miniprep and gel electrophoresis confirm this result (not shown).
Figure 3.
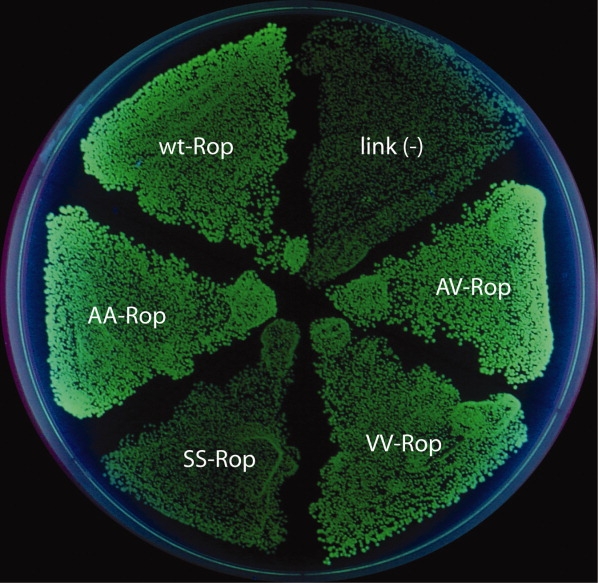
In vivo activity of Rop cysteine variants. The Ser/Ser variant is inactive, with low fluorescence like the negative control. The Ala/Ala, Val/Val, and Ala/Val variants are as active as wild-type Rop.
Although both the Ala/Ala and Val/Val variants are active, only the Ala/Ala variant appeared to be expressed at significant levels under control of a T7 promoter. The reason for the lack of expression of Val/Val is not clear. The Ala/Ala variant has a similar mean residue ellipticity to wild-type Rop at 222 nm, indicating similar secondary structure. However, it is considerably less thermally stable than the Cys/Cys wild-type (TM of 56°C vs. 70°C, Fig. 4).
Figure 4.

Thermal and urea denaturation of Rop cysteine variants observed by CD spectroscopy. The Ala/Val variant is slightly more thermally stable than wild-type Rop, while Ala/Ala is much less stable. The Ala/Val variant is slightly destabilized to urea denaturation compared to wild-type, but it is much more stable than Ala/Ala.
Because Cys52 appears to be less well packed in the structure of wild-type Rop, we decided to construct and examine an Ala/Val mutant (C38A C52V). This variant is active by the cell-based screen. Its activity is also evident from DNA miniprep to examine plasmid levels, and it expresses at high levels from a T7 promoter (6–7 mg L−1 culture, not shown). Essentially all of the protein appears to be in the soluble fraction of the lysate. It can be concentrated to very high levels (>10 mg mL−1) with no signs of precipitation upon storage at 4°C for months.
The Ala/Val variant has similar helical content to wild-type Rop as judged from CD, and it is slightly more thermally stable than wild-type Rop (TM of 74°C). While Ala/Val (D½ ∼4.1M) is considerably more stable to urea (Fig. 4) than Ala/Ala (D½ ∼2.3M), it is somewhat less stable than wild-type Rop (D½ ∼4.6M). Because Rop is a homodimer, its folding is bimolecular, but its unfolding is unimolecular. Consequently, we perform all CD experiments at a defined standard state of 50 μM protein (with respect to the monomer). Taking the molecularity into account by the method of Dalal et al.,26 we measure the stability of wild-type Rop to be 16 kcal mol−1 (compared to 14.6 kcal mol−1 reported by Dalal et al.) and C38V C52V to be 11 kcal mol−1. The m-value changes from 2.1 kcal mol−1 M−1 for wild-type (Dalal et al. report 2.3 kcal mol−1 M−1) to 1.4 kcal mol−1 M−1 for Ala/Val. (See Table I for a summary of these parameters.)
Table I.
Thermodynamic and Kinetic Parameters of Rop Variants
| M.R.E.a (deg cm2 dmol−1) | TM (°C) | D½ (M urea) | m-value (kcal mol−1 M−1) | ΔGH2O (kcal mol−1) | ku (s−1) | t½ (min) | |
|---|---|---|---|---|---|---|---|
| w.t. (Cys/Cys) | 21,000 | 70 | 4.6 | 2.1 | 16 | 5.1 × 10−6 | 2300 |
| Ala/Val | 20,000 | 74 | 4.1 | 1.3 | 11 | 7.3 × 10−4 | 16 |
| Ala/Ala | 20,000 | 56 | 2.3 | 0.9 | 7.7 | n.d. | n.d. |
Equilibrium parameters were determined from 2 to 3 trials. Unfolding rates were determined from 2 or 3 independent trials at 4–7 different urea concentrations (see Supporting Information).
M.R.E., mean residue ellipticity at 222 nm and 25°C; n.d., not determined.
We performed differential scanning calorimetry on Ala/Val Rop and compared the values to wild-type from the work of Munson et al.8,19 (Table II). The T½ values (temperature half-way through the calorimetric transition) at 0.3 mM are opposite those determined by CD spectroscopy at 50 μM (compare to Table I). Ala/Val Rop has a smaller enthalpic stabilization (ΔHcal), but its ΔCp value is within experimental error of the value for wild-type.
Table II.
Differential Scanning Calorimetry
| T½ (°C) | ΔH (kcal mol−1) | ΔCp (kcal mol−1 deg−1) | |
|---|---|---|---|
| w.t. (Cys/Cys)a | 73.9 ± 0.8 | 110 ± 6 | 0.6 ± 0.4 |
| Ala/Valb | 67.8 ± 0.8 | 65 ± 4 | 1.1 ± 0.4 |
Numbers are ± standard deviation of three trials.
From Munson et al.19 in 100 mM phosphate, pH 7, 200 mM NaCl.
50 mM phosphate, pH 6.3, 300 mM NaCl.
Unfolding kinetics
Wild-type Rop is particularly slow to unfold in chemical denaturant. Complete unfolding in the presence of 4M GdnHCl is reported to require more than a day.18 We examined the rate of unfolding of wild-type and Ala/Val Rop in urea. Even at high concentrations of urea, wild-type Rop requires more than 2 days to unfold, whereas C38A C52V requires less than an hour. By selecting several concentrations of urea where the proteins are more than 90% unfolded at equilibrium, it is possible to extrapolate the unfolding rates of the proteins in buffer (that is, no urea), assuming the logs of the rates are linearly related to the urea concentration over the full range (Fig. 5).18 The half-time (t½) of unfolding of the Ala/Val mutant is 16 min, whereas the t½ for wild-type Rop is about 38 h, or ∼1.5 days.
Figure 5.
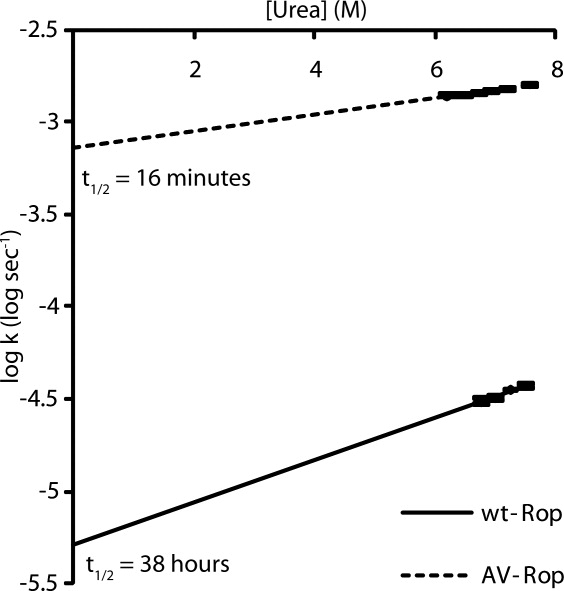
Unfolding kinetics of wild-type and Ala/Val Rop from urea observed by CD spectroscopy. Each log ku (where k is in s−1) is plotted as a function of urea concentration. Ala/Val Rop's unfolding rate constant is about 140-fold higher than wild-type in zero denaturant.
Structure
We solved the X-ray crystal structure of C38A C52V Rop to approximately 2.3 Å (Table III). The Ala/Val Rop crystallized in MES between pH 5.7–5.9 with 10% glycerol and 25–30% methanol as the precipitant. These conditions were suitably cryoprotective without additives. The crystal diffracted to 1.96 Å on Ohio State's rotating-anode home source, but 93.3% completeness was only seen to 2.32 Å. All of the data were used in the refinement. The phases were estimated by molecular replacement with wild-type Rop (1ROP). The structure was rebuilt and refined, applying TLS refinement and refinement with riding hydrogens. The only notable geometric distortions in the fully-refined structure are the somewhat long bonds between N and Cα in Gly1 and between Cα and Cβ in Val52.
Table III.
X-Ray Data Collection and Refinement Statistics
| Data collection wavelength (Å) | 1.54 |
| Resolution rangea (outer shell) (Å) | 29.63–2.32 (2.46–2.32) |
| Space group | C121 |
| Cell dimensions | |
| a, b, c (Å) | 44.7, 40.3, 31.7 |
| α, β, γ (°) | 90.0, 102.0, 90.0 |
| Mosaicity (°) | 0.82 |
| Rmerge | 0.030 (0.079) |
| I/σI | 47.4b (22.6) |
| Completeness (%) | 93.3% (74.5%) |
| Redundancy | 7.26b (7.04) |
| Refinement | |
| Reflections (workc/pairs/test) | 4,885/2,675/242 |
| Rwork/Rfree | 16.3/25.0 |
| Average B-factor (Å2) | 26.2 |
| Number of atoms | |
| Protein | 903 |
| Solvent | 69 |
| R.M.S. deviations | |
| Bond lengths (Å) | 0.013 |
| Bond angles (°) | 1.088 |
Reflections were observed to 1.96 Å, but the completeness beyond 2.32 Å is low. The data are 97.1% complete to 2.46 Å, and the overall completeness over the full resolution range is 66.1%.
Number corresponds to 29.63–1.96 Å.
Work reflections count the Fobs (+) and Fobs (−) reflections from the Bijveot pairs separately.
This variant is nearly superimposable with wild-type, with an all-atom RMSD of 0.41 Å (Fig. 6). As with 1ROP, there is no clear density for the last six residues, which compose a charged “tail” that is poorly resolved due to its flexibility. There are three regions with modest differences from 1ROP, the first two at the open end of the hairpin where the N- and C-termini meet. At the N-terminus, the Thr2 ψ angle is rotated nearly 180° between the two variants. This is likely mostly because 1ROP begins with the sequence MTK… and Ala/Val Rop begins GTK…, as it was generated from a TEV protease cleavage before Gly1. Secondly, there is some perturbation at the C-terminus around the C52V mutation, most notably forcing the phenyl group of Phe56 toward the open end of the hairpin. There also appears to be some small deviation of the backbone in this region, but we see density for residue 57 which was not seen in 1ROP, so it is difficult to say if this is mostly a result of minor refinement differences. Thirdly, near the closed end of the hairpin, there is a small displacement of the backbone in the vicinity of Leu23 toward helix 2, possibly compensating for the volume loss in the C38A mutation. The Cα and Cβ positions of Ala and Leu overlay with the corresponding positions in the wild-type, and one of the Cγ atoms points along the same vector as Cβ-S in the wild-type at position 52. Other than the slight displacement of Phe56, there are essentially no changes to the side chains around either mutation, and even the Phe56 shift is due to a slight movement of the backbone and not a dramatic change in the χ angles. Other side chain differences in the rotameric states of surface Arg, Glu, and His residues are likely to arise from differences in refinement and/or multiple conformations.
Figure 6.
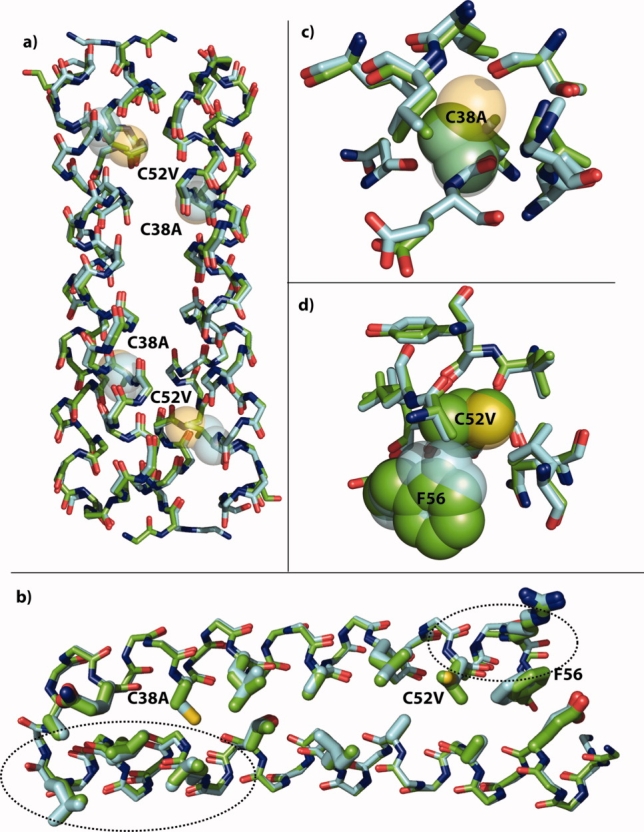
X-ray crystal structure of Ala/Val Rop. (a) Overlay of the backbones of the symmetry-derived dimers of wild-type Rop (cyan carbons) and Ala/Val Rop (green carbons). The positions of the C38A and C52V mutations are shown as 50% transparent spheres for the wild-type Cys side chains. There is little change in the overall structure. (b) Overlay of the monomer backbones with core residues rendered as thick sticks. The backbone in helix 1 opposite C38A has moved slightly toward position 38. Near C52V, a small movement of the backbone has displaced F56 toward the open end of the hairpin. (c) Resides with atoms within 5 Å of the Cys38 sulfur are nearly unchanged except for the slight backbone shift toward the site of mutation. (d) It is evident that Val38 would have clashed with position of Phe56 in the wild-type, and it is displaced slightly by movement of the backbone. In (c) and (d), C38, C52, and F56 are shown in 50% transparent spheres with cyan carbons in the wild-type, and A38, V52, and F56 are shown in solid spheres with green carbons in Ala/Val. Rendered with PyMOL from 1ROP and the structure solved here, 3K79.
Discussion
Rop has proven to be an important model protein for understanding the effects of mutations on structure, stability and folding kinetics, especially in the core and loop. As a homodimeric four-helix bundle, Rop is sufficiently simple to make rational mutations, and a great deal is known about the association of helices as coiled-coils. Still, Rop has afforded many surprises that could not easily be predicted, such as the topological inversion of a core variant repacked with Ala and Ile,27 or the topological rearrangement of the A31P mutant to a “bisecting U.”28 Recently, we introduced a means of measuring the melting temperatures of 96 variants of a protein at once (High-Throughput Thermal Scanning, or HTTS), and we demonstrated its use with Rop core variants.23 However, the buried Cys residues in the core at positions 38 and 52 complicate the mutagenesis of Rop, because destabilized mutants are prone to the formation of adventitious disulfides. This affects the reversibility of thermal denaturation and reduces the storage time of the protein. Consequently, we sought to generate a Cys-free variant with very similar structure and thermodynamic properties for further mutagenesis to understand the effects of mutation on protein stability. Cys-free variants of proteins such as T4 lysozyme and human superoxide dismutase have proven very useful in related studies.
Here we have shown that the C38A C52V variant of Rop is active, has wild-type like thermal stability, exhibits a cooperative thermal denaturation, and has similar although slightly reduced stability to urea denaturation. In contrast, the Ser/Ser variant was not active, the Val/Val variant did not express in appreciable quantities, and the Ala/Ala variant was substantially thermally destabilized. The decrease in the calorimetrically measured ΔH for folding suggests that slight imperfections in packing and bond angles, for example, have been introduced relative to wild-type Rop, and this is in accord with the slight perturbations observed around the two mutation sites in the crystal structure. The similar thermodynamic stability of wild-type Rop and the Ala/Val variant coupled with the lower ΔH suggest that the Ala/Val variant has considerably more favorable entropy of folding.
One way in which the Ala/Val variant does differ dramatically from wild-type Rop is in its much faster unfolding kinetics. The reason for Rop's very slow unfolding is not clear; it contains no disulfide bonds or proline residues, it is a very small protein, and its variants with much faster unfolding kinetics (such as Ala/Val) are active, can be purified in large amounts, and do not aggregate. By way of comparison, several other core variants of Rop have extremely different folding kinetics. Rop can be considered to have eight 4-residue layers in its hydrophobic core. For wild-type, or in mutants wherein the central two or four layers are replaced with Ala2Leu2, the unfolding rates were found to be around 10−4 s−1 (extrapolated from GdnHCl denaturation); when six or eight layers are repacked, the rates increase to 10−2–100 s−1.18 Cys38 is replaced in the four-layer variants and beyond, but Cys52 is not replaced until the six-layer variants. The effects of mutations on Cys52 alone may be worth exploration. Recently, Dalal et al. examined several homologs of Rop from other bacteria.26 Intriguingly, Cys38 is conserved among five identified homologs of Rop, but Cys52 is mutated to Ile or Leu in four of the five homologs. The two homologs examined experimentally both unfold more rapidly than Rop, one about 10-fold faster and one about 104-fold faster. Both are C52L.
The Ala2Leu2-6 (AL6) and Ala2Ile2-6 (AI6) variants of Rop are both stable, but the AI6 variant does not bind RNA.8 When AI6 was crystallized, it was found in a parallel (or syn) topology, the opposite of that observed for wild-type Rop (anti, Fig. 7).27 The much faster unfolding of AL6 compared to wild-type Rop led Levy et al. to speculate that fast-unfolding variants might prefer the syn topology.29 In fact, we found that AI6 variant unfolds much faster than wild-type, but AL6 unfolds even faster, in the dead time of the CD experiment (JJL and TJM, unpublished). In collaboration with Deniz and Onuchic, we recently demonstrated by single-molecule FRET that at zero denaturant, both AL6 and AI6 adopt syn topology, while wild-type adopts anti.30 The AL6 variant can uniquely form both syn and anti at low denaturant. Both of these variants repack the Cys38 and Cys52 positions, and it will be interesting to examine the solution topological preferences of Ala/Val itself. It also remains to be seen if the differences in folding enthalpy and entropy are themselves related to the folding rates and solution topological preferences. It is notable that the Ala/Val variant was used as a labeling control in the smFRET experiments.
Figure 7.
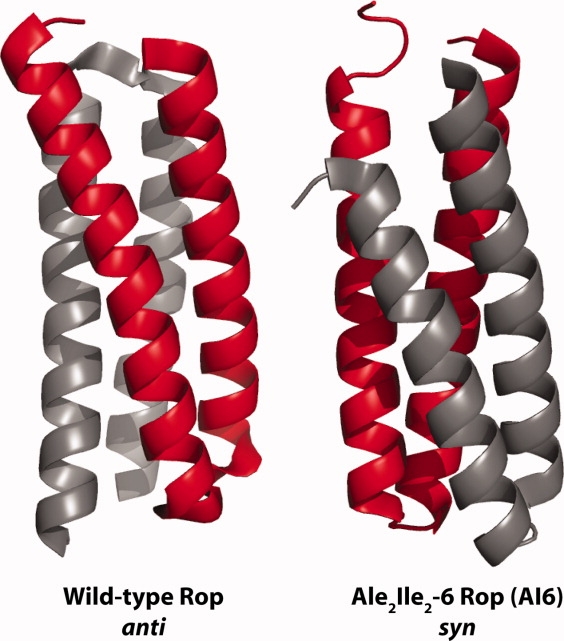
Topologies of Rop variants. Wild-type Rop (1ROP, anti) and AI6 Rop (1F4M, syn) were rendered with PyMOL. [Color figure can be viewed in the online issue, which is available at http://www.interscience.wiley.com.]
The unfolding rate we measured here for wild-type Rop is about an order of magnitude slower than Munson et al. measured, and the TM is about 6°C higher. This is probably mostly because of slight differences in the conditions: we used 50 μM protein, 50 mM phosphate, pH 6.3, 300 mM NaCl, and the previous study used 20 μM protein, 100 mM phosphate, pH 7, 200 mM NaCl. It is also possible that extrapolation back to zero denaturant from urea gives a different result from GdnHCl, perhaps because of the increasing ionic strength of the GdnHCl solutions.
Ala/Val Rop crystallized as an antiparallel homodimer, consistent with its activity. Overall, there is very little change to the structure as compared to wild-type. There is essentially no change in the positions of the residues known to be required for RNA binding: Asn10, Phe14, Gln18, or Lys25.31 Lys3 is weakly implicated in binding and is in a different rotameric state, but there was apparently not extensive density for this residue in 1ROP since the Cɛ and ɛ-amino groups are not shown. Very minimal perturbations are seen in the structure, principally in the backbone opposite C38A and with a small displacement of Phe56 next to C52V due to some backbone shifting.
Since buried cysteine residues are quite hydrophobic and have a side-chain volume that is intermediate between Ala and Val, we suggest that replacement of pairs of Cys residues with one Ala and one Val may be a good general strategy for cysteine mutagenesis. For example, we have even been able to replace a disulfide bond in another protein with Ala/Val, where either of the individual Cys → Ala mutants results in significant loss of activity (V. Shete & TJM, manuscript in preparation). This also suggests that it may be possible to introduce new disulfide bonds into proteins by mutating adjacent Ala and Val residues. The C38A C52V mutant of Rop has already proven extremely useful to us in development of a screen for protein stability. Since the unfolding kinetics of Ala/Val are more like virtually every core variant we have engineered and other small proteins lacking prolines and disulfides, we suggest this makes Ala/Val Rop a more useful reference state than wild-type for biophysical analysis. This Cys mutagenesis strategy may prove useful in many other contexts as well, such as removal of Cys residues to improve E. coli expression or to improve the storage properties of therapeutic proteins.
Materials and Methods
Cloning
Oligonucleotides (Sigma-Genosys) were used to assemble genes with point mutations at positions 38 and 52 of wild-type Rop (see Supporting Information Table S1). The mutation combinations were Ala/Ala, Ser/Ser, Val/Val, and Ala/Val. Each gene was amplified with two sets of primers to form “screening” genes that were used for the in vivo functional assay, and “expression” genes that contained N-terminal His6 and TEV sites. Screening genes (without His6 or TEV sites) were digested with NcoI and BamHI (New England Biolabs) and spin-column purified (Qiagen). Screening vector pACT7lac was digested with the same and gel-purified (Qiagen). Expression genes were digested with BanI and NdeI and spin-column purified. Expression vector pMR101 was digested with the same and gel-purified. The digested genes were ligated [T4 DNA ligase (NEB)] with their respective vectors at 16°C, spin-column purified, and transformed into DH10B. Plasmids used for expression were transformed into BL21 (DE3) for protein purification.
Screening
The cell-based assay for Rop activity was performed as described.24 Briefly, genes in the screening vector were transformed into electrocompetent DH10B already containing the screening plasmid pUCBAD-GFPuv. Cultures were plated onto LB agar medium containing 100 μg mL−1 ampicillin, 35 μg mL−1 kanamycin, 0.1 mM IPTG, and 0.0005% arabinose (w/v), and incubated overnight at 42°C. Cellular fluorescence was visualized under hand-held UV illumination.
Protein purification
Rop variants in pMR vectors were overexpressed in BL21(DE3) in 1 L 2YT media. The culture was grown to log phase (OD600 ∼0.7) and induced with 0.1 mM IPTG. The cells were harvested after growing at 30°C overnight or 37°C for 3 h. Frozen cell pellets were resuspended in 25 mL lysis buffer (50 mM Tris-HCl pH 8, 300 mM NaCl, 10 mM imidazole), mixed with 5 mM MgCl2, 0.5 mM CaCl2, 0.17 μg mL−1 DNase I, 0.17 μg mL−1 RNase A, 0.1% Triton X-100, and 0.3–1 mg mL−1 HEW lysozyme, and allowed to incubate on ice for 30 min. The solution was then sonicated at 300 W for 3× 30 s with 2 min on ice in between pulses. After centrifugation, the cleared lysate was mixed on ice with 1 mL Ni-NTA agarose (Qiagen) for 1 h. The slurry was poured into a prefritted column (Bio-Rad), washed with wash buffer (lysis buffer with 20 mM imidazole), and the protein was eluted with elution buffer (lysis buffer with 250 mM imidazole). TEV protease (0.5 mg) was added twice to the protein and incubated each time overnight at RT or 3 h at 30°C. The solution was desalted with PD-10 columns (GE Amersham) and mixed with 1 mL Ni-NTA. After incubating on ice for 1 h, the solution was added to a column, and the flow-through was collected to yield cleaved protein. This protein was concentrated through a YM-3 filter (Millipore) and exchanged into appropriate buffer for subsequent analysis.
CD spectroscopy
Data were collected on an Aviv 202 Circular Dichroism Spectrometer. Rop variants were analyzed at 50 μM monomer, as measured by UV absorption at 280 nm (using a calculated extinction coefficient of 1490M−1 cm−1), in CD buffer (50 mM sodium phosphate, pH 6.3, 300 mM NaCl). Thermal denaturations were acquired at 1°C min−1, 25–90°C, at 222 nm. Urea denaturations were performed in the same conditions but using varying concentrations of urea, with spectra acquired after equilibrating 24–48 h at RT, at 222 nm.
DSC
DSC measurements were made on a Microcal VP-DSC. Ala/Val Rop samples were in 500 μL with 2 mg mL−1 (0.3 mM) protein in CD buffer. Denaturation was observed from 10 to 95°C at 1.5°C min−1. Curves were fit to the data using Origin assuming a progressive baseline and a dissociative dimeric model (see below).
Data analysis
To calculate the ΔGH2O, the thermodynamic stability the protein in buffer (i.e., zero denaturant), we followed the method of Dalal et al., assuming an equilibrium between the folded dimer and unfolded monomer.
| (1) |
The equilibrium constants for each urea concentration D were determined from fraction unfolded (FU), fraction folded (FN), and total protein concentration (CT) using:
| (2) |
These were converted to ΔG values from –RT ln KD and the ΔGH20 and m-values were determined by fitting a line:
| (3) |
The rate constants for unfolding at each concentration of denaturant were determined by fitting single exponential functions (A0e-kt) to the time traces (Supporting Information Fig. S1). The rate constant for unfolding in water, ku, was linearly extrapolated from the rate constants for folding at given concentrations of denaturant, D:
| (4) |
Crystal growth and data collection
Rop was purified as above, followed by gel filtration purification using a Superdex 75 10/300 GL column (GE Life Sciences). The protein was then exchanged into a mild buffer (10 mM PIPES pH 6.3, 50 mM NaCl) and concentrated, if necessary, to approximately 2 mg mL−1 as determined by Bradford assay. Sitting drop and hanging drop trials were performed. Plates were obtained with both ammonium sulfate and 2-methyl-2,4-pentanediol (MPD) precipitant. Rhombohedral prisms were obtained from hanging drop wells using 1 mL reservoirs containing 25–30% methanol, 100 mM MES, pH 5.7–5.9, 300 mM NaCl, and 10% glycerol. Crystals appeared overnight at 22°C and grew for 3–5 days. Diffraction data were collected on a Rigaku R-AXIS IV++ at −160°C and a detector distance of 200 mm. Images were collected at 0.5° increments with exposure times of 2 min. The images were processed, scaled, and reduced using the program d*TREK.32
Structure determination and refinement
Molecular replacement modeling was performed with Phaser,33 using 1ROP as the model, and ARP/wARP34 was used to rebuild the structure. Refinement was performed using phenix.refine.35 The structure was hand-corrected with Coot.36 Later refinement included TLS refinement (using TLSMD37 parameters), followed by a final refinement using hydrogens added with Molprobity.38
Acknowledgments
SBH was intern of a Department of Biochemistry Summer Undergraduate Research Program and a Mayers Summer Research Intern. JJL was an NIH Chemistry-Biology Interface Program fellow and a predoctoral fellow of the Great Rivers American Heart Association. TJM is grateful to all the instructors in the 2007 Cold Spring Harbor Lab course in X-ray crystallography, particularly Jim Pflugrath, Alex McPherson, Paul Emsley, David and Jane Richardson, and Randy Read, who made this article possible. Coordinates for Rop (C38A C52V) have been deposited in the Protein Data Bank (PDB) under the accession code 3K79.
References
- 1.Roodveldt C, Aharoni A, Tawfik DS. Directed evolution of proteins for heterologous expression and stability. Curr Opin Struct Biol. 2005;15:50–56. doi: 10.1016/j.sbi.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 2.Bashford D, Chothia C, Lesk AM. Determinants of a protein fold. Unique features of the globin amino acid sequences. J Mol Biol. 1987;196:199–216. doi: 10.1016/0022-2836(87)90521-3. [DOI] [PubMed] [Google Scholar]
- 3.Bowie JU, Reidhaar-Olson JF, Lim WA, Sauer RT. Deciphering the message in protein sequences: tolerance to amino acid substitutions. Science. 1990;247:1306–1310. doi: 10.1126/science.2315699. [DOI] [PubMed] [Google Scholar]
- 4.Axe DD, Foster NW, Fersht AR. Active barnase variants with completely random hydrophobic cores. Proc Natl Acad Sci USA. 1996;93:5590–5594. doi: 10.1073/pnas.93.11.5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dahiyat BI, Mayo SL. Protein design automation. Protein Sci. 1996;5:895–903. doi: 10.1002/pro.5560050511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Desjarlais JR, Handel TM. De novo design of the hydrophobic cores of proteins. Protein Sci. 1995;4:2006–2018. doi: 10.1002/pro.5560041006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuhlman B, Dantas G, Ireton GC, Varani G, Stoddard BL, Baker D. Design of a novel globular protein fold with atomic-level accuracy. Science. 2003;302:1364–1368. doi: 10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]
- 8.Munson M, Balasubramanian S, Fleming KG, Nagi AD, O'Brien R, Sturtevant JM, Regan L. What makes a protein a protein? Hydrophobic core designs that specify stability and structural properties. Protein Sci. 1996;5:1584–1593. doi: 10.1002/pro.5560050813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martensson LG, Karlsson M, Carlsson U. Dramatic stabilization of the native state of human carbonic anhydrase II by an engineered disulfide bond. Biochemistry. 2002;41:15867–15875. doi: 10.1021/bi020433+. [DOI] [PubMed] [Google Scholar]
- 10.Matsumura M, Becktel WJ, Levitt M, Matthews BW. Stabilization of phage T4 lysozyme by engineered disulfide bonds. Proc Natl Acad Sci USA. 1989;86:6562–6566. doi: 10.1073/pnas.86.17.6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robinson CR, Sauer RT. Striking stabilization of Arc repressor by an engineered disulfide bond. Biochemistry. 2000;39:12494–12502. doi: 10.1021/bi001484e. [DOI] [PubMed] [Google Scholar]
- 12.Matsumura M, Matthews BW. Control of enzyme activity by an engineered disulfide bond. Science. 1989;243:792–794. doi: 10.1126/science.2916125. [DOI] [PubMed] [Google Scholar]
- 13.Lepock JR, Frey HE, Hallewell RA. Contribution of conformational stability and reversibility of unfolding to the increased thermostability of human and bovine superoxide dismutase mutated at free cysteines. J Biol Chem. 1990;265:21612–21618. [PubMed] [Google Scholar]
- 14.Tsai J, Taylor R, Chothia C, Gerstein M. The packing density in proteins: standard radii and volumes. J Mol Biol. 1999;290:253–266. doi: 10.1006/jmbi.1999.2829. [DOI] [PubMed] [Google Scholar]
- 15.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 16.Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Banner DW, Kokkinidis M, Tsernoglou D. Structure of the ColE1 rop protein at 1.7 A resolution. J Mol Biol. 1987;196:657–675. doi: 10.1016/0022-2836(87)90039-8. [DOI] [PubMed] [Google Scholar]
- 18.Munson M, Anderson KS, Regan L. Speeding up protein folding: mutations that increase the rate at which Rop folds and unfolds by over four orders of magnitude. Fold Des. 1997;2:77–87. doi: 10.1016/S1359-0278(97)00008-4. [DOI] [PubMed] [Google Scholar]
- 19.Munson M, O'Brien R, Sturtevant JM, Regan L. Redesigning the hydrophobic core of a four-helix-bundle protein. Protein Sci. 1994;3:2015–2022. doi: 10.1002/pro.5560031114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagi AD, Anderson KS, Regan L. Using loop length variants to dissect the folding pathway of a four-helix-bundle protein. J Mol Biol. 1999;286:257–265. doi: 10.1006/jmbi.1998.2474. [DOI] [PubMed] [Google Scholar]
- 21.Nagi AD, Regan L. An inverse correlation between loop length and stability in a four-helix-bundle protein. Fold Des. 1997;2:67–75. doi: 10.1016/S1359-0278(97)00007-2. [DOI] [PubMed] [Google Scholar]
- 22.Predki PF, Agrawal V, Brunger AT, Regan L. Amino-acid substitutions in a surface turn modulate protein stability. Nat Struct Biol. 1996;3:54–58. doi: 10.1038/nsb0196-54. [DOI] [PubMed] [Google Scholar]
- 23.Lavinder JJ, Hari SB, Sullivan BJ, Magliery TJ. High-throughput thermal scanning: a general, rapid dye-binding thermal shift screen for protein engineering. J Am Chem Soc. 2009;131:3794–3795. doi: 10.1021/ja8049063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Magliery TJ, Regan L. A cell-based screen for Rop function: a new tool for combinatorial experiments in biophysics. Protein Eng Des Sel. 2004;17:77–83. doi: 10.1093/protein/gzh010. [DOI] [PubMed] [Google Scholar]
- 25.Munson M, Predki PF, Regan L. ColE1-compatible vectors for high-level expression of cloned DNAs from the T7 promoter. Gene. 1994;144:59–62. doi: 10.1016/0378-1119(94)90203-8. [DOI] [PubMed] [Google Scholar]
- 26.Dalal S, Canet D, Kaiser SE, Dobson CM, Regan L. Conservation of mechanism, variation of rate: folding kinetics of three homologous four-helix bundle proteins. Protein Eng Des Sel. 2008;21:197–206. doi: 10.1093/protein/gzm088. [DOI] [PubMed] [Google Scholar]
- 27.Willis MA, Bishop B, Regan L, Brunger AT. Dramatic structural and thermodynamic consequences of repacking a protein's hydrophobic core. Structure. 2000;8:1319–1328. doi: 10.1016/s0969-2126(00)00544-x. [DOI] [PubMed] [Google Scholar]
- 28.Glykos NM, Cesareni G, Kokkinidis M. Protein plasticity to the extreme: changing the topology of a 4-alpha-helical bundle with a single amino acid substitution. Structure. 1999;7:597–603. doi: 10.1016/s0969-2126(99)80081-1. [DOI] [PubMed] [Google Scholar]
- 29.Levy Y, Cho SS, Shen T, Onuchic JN, Wolynes PG. Symmetry and frustration in protein energy landscapes: a near degeneracy resolves the Rop dimer-folding mystery. Proc Natl Acad Sci USA. 2005;102:2373–2378. doi: 10.1073/pnas.0409572102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gambin Y, Schug A, Lemke EA, Lavinder JJ, Ferreon AC, Magliery TJ, Onuchic JN, Deniz AA. Direct single-molecule observation of a protein living in two opposed native structures. Proc Natl Acad Sci USA. 2009;106:10153–10158. doi: 10.1073/pnas.0904461106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Predki PF, Nayak LM, Gottlieb MB, Regan L. Dissecting RNA-protein interactions: RNA-RNA recognition by Rop. Cell. 1995;80:41–50. doi: 10.1016/0092-8674(95)90449-2. [DOI] [PubMed] [Google Scholar]
- 32.Pflugrath JW. The finer things in X-ray diffraction data collection. Acta Crystallogr D Biol Crystallogr. 1999;55:1718–1725. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- 33.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perrakis A, Harkiolaki M, Wilson KS, Lamzin VS. ARP/wARP and molecular replacement. Acta Crystallogr D Biol Crystallogr. 2001;57:1445–1450. doi: 10.1107/s0907444901014007. [DOI] [PubMed] [Google Scholar]
- 35.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 36.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 37.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D Biol Crystallogr. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 38.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, III, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]


