Abstract
The self-assembling MexA-MexB-OprM efflux pump system, encoded by the mexO operon, contributes to facile resistance of Pseudomonas aeruginosa by actively extruding multiple antimicrobials. MexR negatively regulates the mexO operon, comprising two adjacent MexR binding sites, and is as such highly targeted by mutations that confer multidrug resistance (MDR). To understand how MDR mutations impair MexR function, we studied MexR-wt as well as a selected set of MDR single mutants distant from the proposed DNA-binding helix. Although DNA affinity and MexA-MexB-OprM repression were both drastically impaired in the selected MexR-MDR mutants, MexR-wt bound its two binding sites in the mexO with high affinity as a dimer. In the MexR-MDR mutants, secondary structure content and oligomerization properties were very similar to MexR-wt despite their lack of DNA binding. Despite this, the MexR-MDR mutants showed highly varying stabilities compared with MexR-wt, suggesting disturbed critical interdomain contacts, because mutations in the DNA-binding domains affected the stability of the dimer region and vice versa. Furthermore, significant ANS binding to MexR-wt in both free and DNA-bound states, together with increased ANS binding in all studied mutants, suggest that a hydrophobic cavity in the dimer region already shown to be involved in regulatory binding is enlarged by MDR mutations. Taken together, we propose that the biophysical MexR properties that are targeted by MDR mutations—stability, domain interactions, and internal hydrophobic surfaces—are also critical for the regulation of MexR DNA binding.
Keywords: DNA-binding protein, stability, efflux gene regulator, multidrug resistance, MarR family, Biacore, analytical ultracentrifugation, circular dichroism, fluorescence, real-time PCR
Introduction
Acquired multidrug resistance (MDR) in pathogenic microbials is a world-wide threat to human health, and investigating the molecular mechanisms involved is, therefore, of utmost importance.1 The widely adaptable winged-helix protein family2 includes the MarR family of proteins, which is essential for bacterial survival in hostile environments. MarR-like proteins are involved in transcriptional control of virulence factor production, bacterial response to antibiotic and oxidative stresses, and catabolism of environmental aromatic compounds.3 Several MarR–like proteins regulate the expression of efflux pumps, which are multiprotein self-assembly complexes actively extruding chemical compounds with high toxicity to the host organism.4,5 The evolutionary pressure in hostile environments is selective for MDR, because incapacitating the repressor protein leads to continuously high production of the efflux proteins and thus increased survival for the bacteria. Thus, analyzing the structure–function relationships of these proteins is fundamental to overcoming innate and acquired MDR in future drug development.1,6–8
Several structures of members in the MarR family have shed light on their structural properties and possible mechanisms for DNA-recognition.9-14 MexR from the pathogen Pseudomonas aeruginosa belongs to this fold family, where two winged-helix-turn-helix (wHTH) domains are linked by a dimer core formed by four interacting helices N- and C-terminal to the wHTH domains [Fig. 1(A)].10 Different orientations of the DNA-binding domains relative to the dimer core in the crystal structure suggest an “open“ and a “closed“ form where only the latter can be modeled onto DNA.10 Recent structures of the OhrR protein from Bacillus subtilis and Xanthomonas campestris suggest that specific oxidation of a reactive cysteine in the dimer interface propagates structural changes resulting in altered relative orientations of wHTH domains.12,15 Yet another variation on this theme is provided by the structure of HucR from Deinococcus radiodurans, where wHTH domains appear prepositioned for DNA binding.14
Figure 1.
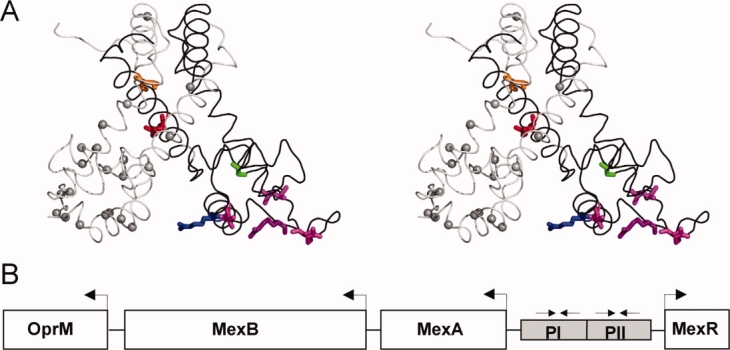
Structural representation of MexR MDR mutation sites and overview of mexAB-oprM operon. A: MexR crystal structure 1LNW, represented by the chain A (black) + B (gray) dimer, which is likely to correspond to the unbound state,10 in stereo view. Mutation sites studied in this work are highlighted in chain A by displaying native sidechains in orange (L13M), red (R21W), green (G58E), blue (R70W), and magenta (T69I, R83H, R91H, L95F). Identified mutation sites in MexR leading to deficient DNA binding and/or multidrug resistance23–25,27,30,35 are indicated with Cα spheres in chain B. The figure was drawn in PyMOL.58 B: Overview of the multidrug efflux operon mexAB-oprM and mexR gene in Pseudomonas aeruginosa. The suggested MexR binding sites PI and PII in mexO are indicated.30
The cellular and molecular events that result in efflux pump regulation in the MarR family are not completely understood. It has been generally considered that DNA binding within the MarR family is regulated by the binding of small organic molecules such as antibiotics and biocides.4,5 Direct molecular observation as to how such regulation would be accomplished has only been obtained for the MarR protein. Although the first MarR structure indicated that salicylic acid binding sites in the wHTH domains would obstruct DNA binding,9 more recent work suggest that the regulatory relevant binding site active at low concentrations of salicylic acid is located in the interface between DNA-binding and dimer region, and hinders the formation of a MarR-marO complex by inducing a large positional shift of the DNA-binding lobe.16 DNA-binding of both MarR and MexR appear to be additionally regulated by specific binding proteins, the binding of which is targetable by multidrug-resistant mutations in vitro.17–19 Recent results propose that MexR, in analogy with OhrR, is a redox-directed regulator, sensing peroxide stress by forming a structurally restricting disulphide across the dimer interface.20 Thus, the entire scene for MarR-family regulation of gene expression is rapidly evolving.
From a clinical perspective, the MexR protein is of particular interest. The P. aeruginosa multidrug efflux system MexA-MexB-OprM is a major contributor to the facile resistance of this organism to multiple antimicrobials.8 MexR regulates the expression of this operon by binding two adjacent DNA boxes, PI and PII, in the MexA-MexR intergenic region21 [Fig. 1(B)]. The emergence of ß-lactam resistance associated with MexAB-OprM in vivo has been shown to be directly linked to mutations affecting its regulator gene mexR.22,23 A range of MexR mutations with highly elevated MIC values to common antibiotics have been identified both from clinical isolates,23–26 by selection for resistant strains24,27 and by combining random mutagenesis and resistance selection.28 These mutations appear scattered over the entire MexR structural scaffold [Fig. 1(A)], as was also observed for the homologous MarR protein.29
MDR mutations in MexR lead to constitutive overexpression of efflux pump proteins,24,25,30 presumably due to deficient DNA binding.20,28 However, because MDR mutants of MexR and related proteins have been so far poorly characterized on a molecular level, the biophysical reason for their disfunction is largely unknown. Two major proposals have been put forward: Weak or abnormal association of the MexR dimer,31 or misfolding of the MexR DNA-binding domain leading to a distorted structure.28 No direct molecular evidence supporting or corroborating either of these hypotheses has been presented so far.
The aim of this work has been to extend the understanding of MexR functionality and how this is perturbed by MDR mutations. To this end, we have used biochemical and biophysical methods to analyze both wt MexR and a well-established set of mexR mutations conferring MDR.20,27,28 Our results show that MDR mutations are distant from the DNA binding surface target stability and internal binding cavities, rather than structure and/or dimerization. Together with our results on DNA binding, our data provides a new angle for interpreting the effects of MDR mutations in MexR, and in the entire MarR family.
Results
MexR-MDR mutants confer derepression of the MexB-Opr operon and target critical structural regions in the MarR family
To assay the efficiency of repression by MexR-MDR mutants relative to the WT protein, a real-time PCR experiment was performed where the extent of MexR and MexB mRNA production from the MexAB-OprM operon was assayed. Because MexR acts as a repressor to this operon, increased production of MexR as well as MexB indicates reduced MexR affinity to the PI and/or PII sites. The results of real-time PCR of cDNA preparations showed that the mutants produced 5–91 times more mRNA for MexR and MexB than the wild type strain (Table I). Thus, all MDR mutants analyzed show deficient repression, but to highly varying degrees. The ratio between the amount of mRNAs produced for MexR and MexB suggest that although two mutations suggest a slightly higher tendency to disturb MexR repression (G58E, L95F), overall, the repression of both genes are similarly affected by the mutations.
Table I.
Effects of mexR Mutations on the Production of mexR and mexB mRNA
| P. aeruginosa Strain | MexR Mutation | mexR mRNA | mexB mRNA | mexR/mexB mRNA Ratio | Sequence-Related Critical Positions in Other MarR Proteins |
|---|---|---|---|---|---|
| PAO1 | – | 1 | 1 | 1 | |
| K4 | L13M | 10 | 12 | 0.8 | C15 OhrR12 |
| T7 | R21W | 63 | 48 | 1.3 | His51 HucR14,32 |
| T3 | G58E | 81 | 34 | 2.4 | |
| T6 | T69I | 8 | 6 | 1.3 | T72A MarR,3,29 S104A HucR32,33 |
| T10 | R70W | 77 | 91 | 0.9 | R73C MarR3,29 |
| K2 | R83H | 38 | 19 | 2 | R118A Huc32,33 |
| K3 | R91H | 5 | 6 | 0.9 | R94C MarR3,29 |
| T4 | L95F | 47 | 26 | 1.8 |
The amount of mRNA for MexR and MexB in the mutated strains as judged from CT data is presented relative to PAO1 (wild type strain). Because MexR-wt acts as a repressor of mexR and mexB, increased mRNA production for these genes implies that the repressive function of MexR has been impaired by the mutation. The experiment was done once for each strain; thus, the error in the relative expression is estimated to be ±20%. Within this error limit, significant derepression is observed for all mutants.
The identified MexR-MDR mutations map onto widely dispersed parts of the MexR structure [Fig. 1(A)]. Mutations R83H, R91H, and L95F are located in or directly adjacent to the important “wing” of the DNA binding motif, and their reducing effect on repression is most likely caused by altering critical DNA-binding determinants. The molecular causes for impaired DNA binding by mutations at MexR positions L13, R21, G58, T69, and R70 are less evident. Previous sequence analysis of the MarR family have aligned residue L13 both with a conserved hydrophobic core in the dimer region9,14 and with C15 in OhrR, where oxidation leads to altered subdomain orientations and consequent derepression.12 A majority of MarR proteins have a conserved Arg, Lys, Asn, or Gln corresponding to MexR residue 21 or 22 in the dimer core,3,12 thus suggesting a conserved functional role at this position. Mutations surrounding MexR-G58 have been observed to be detrimental for DNA binding.28,31 The conserved R70 was found in a critical hydrogen bond across the dimer interface in the MarR crystal structure,9 however, this hydrogen bond was absent in the MexR crystal structure.10 The effect of T69I on repression is minor in comparison to R70W (Table I). To focus our study on critical internal interactions in MexR, we decided to investigate the biophysical properties of MexR-MDR mutants MexR-L13M, MexR-R21W, MexR-G58E, MexR-R70W, and the MexR-wt protein in the absence and presence of DNA.
MexR-wt binds to both binding sites in the MexR operator with high affinity, whereas mutant MexR proteins show no significant DNA binding
To estimate DNA binding affinities for MexR-wt and -MDR mutants, Biacore measurements were used. In agreement with rt-PCR assays, none of MexR-L13M, MexR-R21W, MexR-G58E, or MexR-R70W bound DNA with any significance compared with the background reference surface at protein concentrations in the low micromolar range. In contrast, MexR-wt bound DNA at high affinity to the previously recognized PI and PII sites in mexO21 [Fig. 1(B)]. Simple 1:1 Langmuir binding models could not fit the binding event. For both PI and PII, a “two-state reaction conformational change” model34 was required to fit the sensorgrams, indicating a conformational transition and/or induced oligomerization on binding. Both for PI and PII, the best fit for PI—MexR-wt binding was obtained for a dimer model, whereas tetramer and monomer models showed less significance (Fig. 2, Table II). Consequently, it appears that MexR-wt binds the PI and PII sites as a dimer, with slightly higher affinity to the PI site.
Figure 2.
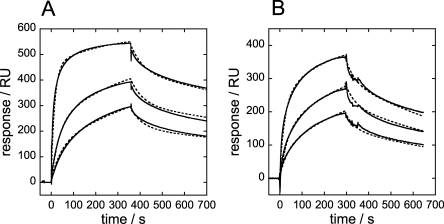
SPR sensorgrams for MexR-wt binding to mexO. Sensorgrams (solid lines) and fitted curves (broken lines) for MexR-wt binding to immobilised (A) PI-DNA using 4, 1, and 0.5 μM analyte, (B) PII-DNA using 4, 2, and 1 μM analyte. A two-state reaction conformational change model was used for MexR binding to PI and PII.
Table II.
SPR Derived Binding Constants for MexR-wt Binding to mexO
| DNA Box | Oligomeric State | Ka | X2/R(max) |
|---|---|---|---|
| PIa | Dimer | 1.2·107 | 0.08 |
| PIIa | Dimer | 2.7·106 | 0.05 |
Predicted association constants (Ka, M) of MexR-wt, PI-, PII-, and PI-PII-DNA, calculated from Biacore measurements. Fits to experimental data are derived using a “two-state reaction conformational change” model where independent fits were made assuming different oligomeric states. The corresponding fits are shown in Figure 2. The goodness of the fit was judged by X2/R(max), as described in Experimental Procedures. Less accurate fits showed showed X2/R(max) of 0.1 (PI, monomer, tetramer), 0.15 (PII, monomer), and 0.18 (PII, tetramer) and significant visual discrepancies in the fit.
MexR oligomerization in the apo state
To address oligomerization of MexR in the absence of DNA, analytical ultracentrifugation (AUC) experiments were performed. Single-species analysis of the resulting data show corresponding MWs of 50 kDa, 92 kDa, and 75 kDa for MexR-wt, MexR-R70W, and MexR-R21W, respectively. MexR-L13M and MexR-G58E lack W and were less soluble than MexR-wt, which precluded AUC measurements. Because the expected mass of a MexR dimer, as observed in the crystal structure10 is only 34 kDa, equilibria including multiple dimers had to be taken into account in the analysis. Models for dimer–monomer, monomer–tetramer, dimer–tetramer, tetramer–hexamer, and dimer–hexamer equilibria were attempted, but proper fits were not obtained until a dimer–octamer equilibrium was considered (Fig. 3). Although higher order oligomerization cannot be excluded, evaluation of the AUC data showed no indication of disrupted dimer in the absence of DNA, or of additional oligomers with lower complexity than octamer.
Figure 3.
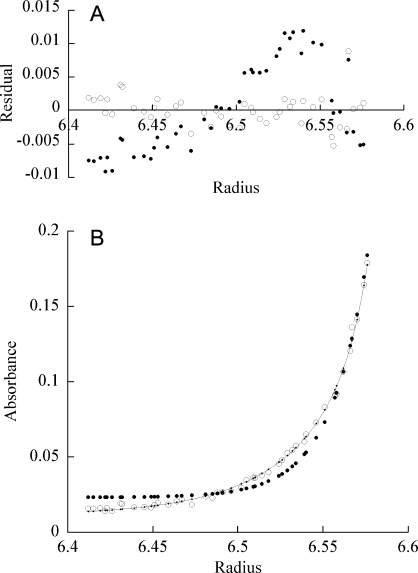
Oligomerization of free MexR as judged by AUC. Representative plots of sedimentation equilibrium experiments of MexR R21W at 20°C and 30,000 rpm. A: Residuals for dimer-octamer equilibrium (empty circles) and dimer-tetramer equilibrium (filled circles). B: R21W experimental values (empty circles), calculated fit of a dimer-octamer equilibrium (solid line) and calculated data points for a fitted equilibrium between dimer and tetramer (filled circles).
MexR secondary structure content is highly similar in mutants, wt, and wt-DNA complexes
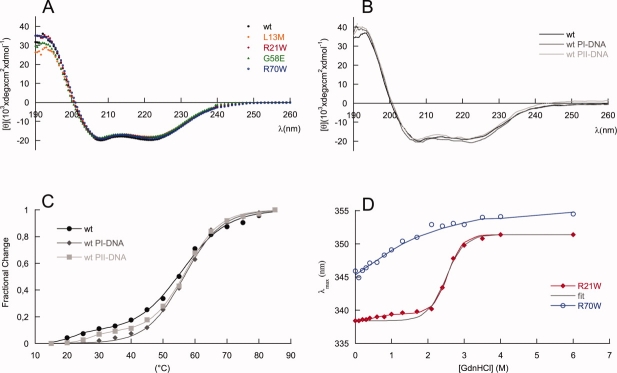
The secondary structure content of MexR-wt and the four targeted mutants was analyzed by circular dichroism spectroscopy [Fig. 4(A)]. The secondary structure content of MexR-wt in solution is very close to that in the crystal structure (Table III), considering that loose structures that are characterized as “disordered” in the crystal structure are often classified as “turns” in the solution CD analysis. All MexR mutants L13M, R21W, G58E, and R70W have slightly lower ellipticities compared with MexR-wt [Fig. 4(A)]. However, the corresponding lowering of the total amount of secondary structure is less than 5% for all mutants (Table III). Conformational changes in secondary structure for these MDR mutants are thus minor, and they do not by themselves explain the dramatic reduction in repressor function. In complex with DNA, a slight increase in helicity could be noted for MexR-wt when binding to PI, whereas changes on binding to PII were minor [Fig. 4(B), Table III].
Figure 4.
MexR structural effects of mutations and DNA binding. Far UV CD spectra of (A) MexR-wt (filled circle, black), MexR-L13M (empty rhomb, orange), MexR-R21W (filled rhomb, red), MexR-G58E (filled triangle, green), MexR-R70W (empty circle, blue) and (B) MexR-wt (solid black line) in complex with DNA binding boxes PI (solid line, dark grey) and PII (solid line, light grey) show that the structural effects of the studied MDR mutations are negligible. C: Thermal denaturation measurements at 222 nm for MexR-wt (filled circle, black), MexR-wt with PI-DNA (filled rhomb, dark grey), and MexR-wt with PII-DNA (filled square, light grey) indicate a less stable DNA-binding domain that is stabilized on DNA binding. The corresponding fits for two- and three-state denaturation (Table 4) are shown as solid lines. D: Gdn-HCl induced unfolding of MexR R21W (filled rombs, red) and R70W (empty circles, blue) indicate that the most stable MexR region is located in the dimer interface, while the DNA-binding region is unstable. Two-state (black) and three-state (red) fits (Table 4) to R21W data are shown. The R70W data were not sufficient for a full thermodynamic fit, thus the blue line only represents a graphic best fit.
Table III.
Secondary Structure Evaluations from Circular Dichroism Data
| MexR Variants and Complexes | Helix (%) | Sheet (%) | Turns (%) | Disordered (%) |
|---|---|---|---|---|
| wt | 61 | 5 | 12 | 21 |
| wt + PI | 63 | 6 | 10 | 21 |
| wt + PII | 61 | 8 | 10 | 21 |
| L13M | 58 | 7 | 14 | 21 |
| R21W | 59 | 7 | 12 | 22 |
| G58E | 57 | 6 | 13 | 24 |
| R70W | 61 | 6 | 12 | 21 |
| Crystal structure (1LNW) | 60 | 6 | 6 | 28 |
Evaluations of solution structure content are based on CD spectra given in Figure 4(A,B). Structure content in the crystal structure was averaged from the eight chains in the unit cell.10 Average standard deviations for the fractional structure content derived from CD spectra was: ±2.4% for helix, ±1.0% for beta structure, ±1.5% for turns, and ±1.4% for disordered regions.
DNA binding stabilizes a less stable state and increases the cooperativity of the major unfolding transition
The stability of MexR-wt and its DNA-bound complexes was analyzed by thermal unfolding followed by CD at 222 nm, where helical unfolding is best detected. The thermal unfolding of MexR-wt shows two transitions: A major unfolding transition with a Tm of 55.6°C, unfolding ∼80% of the native structure, and a minor unfolding transition at lower Tm (below 30°C), which is accompanied by a much smaller change in ellipticity [Fig. 4(C)]. A three-state denaturation with two transitions is sufficient to fit MexR-wt unfolding (Table IV, Methods). The minor transition is highly dependent on the presence or absence of DNA. Indeed, denaturation of the MexR-wt–PI complex lacks a minor transition, and fits with high cooperativity to a two-state denaturation, suggesting that the minor transition corresponds to a less stable DNA-binding region which is stabilized in the MexR-DNA complex. In the MexR-wt - PII complex, the minor transition is still visible but shifted to higher Tm compared with in Mex-wt alone. The major transition in both the MexR-PI and MexR-PII complex is only slightly increased (Table III) but significantly steeper compared with apo-MexR, suggesting that the MexR core region, although distant from the immediate DNA-binding region, is both slightly more stable and significantly more cooperatively folded in complex with DNA. This is consistent with the requirement of a conformational transition model in the analysis of Biacore data for MexR-wt DNA binding (Fig. 2, Table II). The unfolding of the MexR-PI complex as one single cooperatively folded unit is in agreement with the higher degree of increased folding in the MexR-PI complex (Table III) and its slightly higher DNA affinity compared with the MexR-PII complex (Table II).
Table IV.
Thermal and Chemical Denaturation Midpoints
| MexR Variants and Complexes | 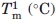 |
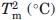 |
Tm (°C) | 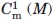 |
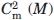 |
|---|---|---|---|---|---|
| wt | <20a | 55.6 | |||
| wt PI | 56.6 | ||||
| wt PII | 26.8 | 57.1 | |||
| L13M | 47.8 | ||||
| R21W | 33.6 | 70.9 | 0.6 | 2.6 | |
| G58E | 32.6 | 62.6 | |||
| R70W | 25.2 | 56.4 | <1a |
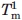 and
and 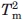 refers to the thermal melting points of the first and second transition in a three-state denaturation, whereas Tm refers to the melting temperature in a two-state transition.
refers to the thermal melting points of the first and second transition in a three-state denaturation, whereas Tm refers to the melting temperature in a two-state transition. 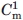 and
and 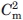 refer to the corresponding chemical midpoint of denaturation.35 The corresponding fits are shown in Figure 4(C,D).
refer to the corresponding chemical midpoint of denaturation.35 The corresponding fits are shown in Figure 4(C,D).
The Tm of the first transitions in MexR-wt, MexR-wt-PII DNA, and MexR-R70W are less accurate because the entire transition was not complete within the temperature range of the experiment; these Tm's are, therefore, to be interpreted only qualitatively. Each experiment was performed thrice with average standard deviations of 0.5°C for 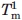 , 0.3°C for
, 0.3°C for 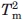 , and 0.8°C for Tm. The chemical denaturation experiment was performed once, with estimated errors of ±0.2M in
, and 0.8°C for Tm. The chemical denaturation experiment was performed once, with estimated errors of ±0.2M in 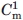 and ±0.1M in
and ±0.1M in 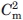 .
.
MexR-wt dimer- and DNA-binding regions have different intrinsic stability
To further dissect the molecular origin of the two melting transitions, we analyzed the fluorescence of the two MexR mutants R21W and R70W in a GdnHCl-induced unfolding experiment [Fig. 4(D), Table IV]. Because MexR-wt lacks tryptophan, the single tryptophans in R21W and R70W should probe the local stability in the dimer-forming and DNA-binding region respectively [Fig. 1(A)] providing the overall structure is maintained as suggested by CD experiments. Native MexR-R70W Trp fluorescence emits at 345 nm suggesting an initially partially exposed tryptophan. On adding denaturant, the emission is slightly shifted in a low cooperative transition which is complete at ∼3M GuHCl, but where the midpoint cannot be estimated. In contrast, MexR-R21W Trp fluorescence emission in the absence of denaturant is highly shifted to lower wavelengths indicating a compactly folded dimerization region in the native state. The unfolding of the dimer region, as reflected in the GuHCl-induced unfolding of MexR-R21W, shows two transitions: the first unfolding transition at low Cm induces only a small wavelength shift, whereas the second transition at higher Cm shifts tryptophan emission from 338 to 352 nm in a highly cooperative manner, indicating a complete unfolding transition [Fig. 4(D)]. The completeness of the two transitions enabled a three-state unfolding analysis, yielding ΔG(H2O) of 1.5 and 7.3 kcal/mol for the respective transitions.35 The lack of tryptophans in MexR-wt, MexR-L13M, and MexR-G58E, and the incomplete transition for R70W, prevented full thermodynamic analysis for all mutants.
The high cooperativity, high ΔG(H2O) and high Cm for the major transition observed in R21W fits well with the major, higher Tm transition in the thermal melting analysis and the unfolding of a highly packed dimer region, whereas the low-cooperativity unfolding transition of MexR-R70W at lower Cm and ΔG(H2O) agrees with the lower Tm of the first transition in the thermal melting experiments, and thus reflects the unfolding of a less stable region within the DNA-binding domain. The minor transition for MexR-R21W could well-correspond to unfolding within the DNA-binding region of MexR-R21W being sensed by the dimer region. However, reversely, once the DNA-binding region has been unfolded, the fluorescence in R70W is not affected by unfolding of the dimer region.
MDR single point mutations in MexR significantly affect protein stability in both DNA-binding and dimer regions
Considering the small effects on stabilization on DNA binding, and the minor changes in secondary structure in the MDR mutants, the large variation in stability for the MDR mutants was unexpected [Fig. 5(A), Table IV]. As in MexR-wt (Fig. 4), all proteins show a major unfolding transition corresponding to the unfolding of the dimer region, but melting transition temperatures (Tm) range from 47°C to 71°C [Table IV, Fig. 5(A)]. These changes in Tm compared with MexR-wt are considerable because the MDR mutants studied here all have single-amino acid substitutions. Based on the major melting transition related to the dimer region, MexR-R21W and MexR-G58E are both more stable than MexR-wt, MexR-L13M is less stable, and MexR-R70W maintains similar stability as MexR-wt. Furthermore, all proteins except MexR-L13M also showed a minor unfolding transition at lower Tm corresponding to unfolding within the DNA-binding domain. Although the unfolding of MexR-L13M was fitted to a two-state denaturation, all other unfolding curves were fitted to a three-state denaturation with two unfolding transitions (Table IV).
Figure 5.
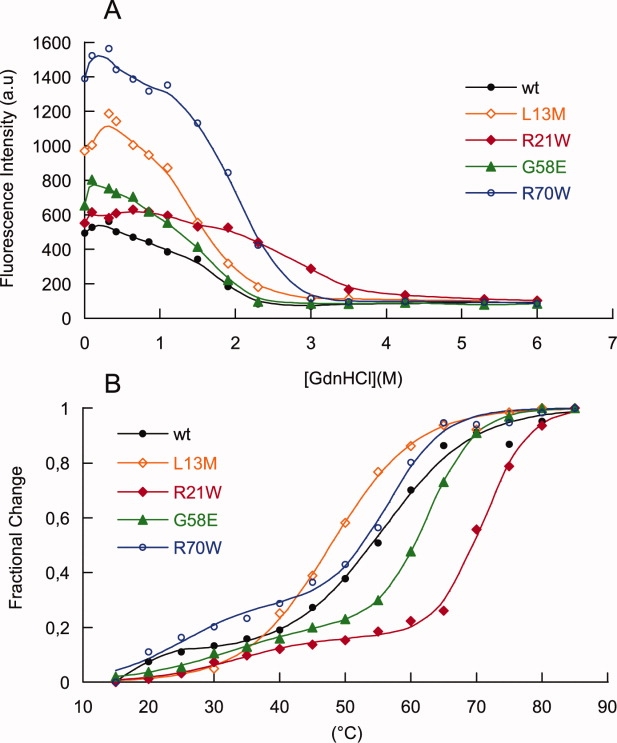
Stability of MexR and mutants as detected by thermal stability and ANS binding measurements. A: ANS binding at 480 nm of MexR wt and mutants labeled as above, analyzed from fluorescence emission as a function of GuHCl concentration. The fluorescence of native MexR-wt (0 M GuHCl) incubated with ANS in the presence of PI- or PII-DNA was slightly higher than without DNA (+7.5 and +7.9%, respectively, data not shown in figure). B: Thermal stability measurements recording the CD ellipticity at 222 nm of MexR-wt (filled circle, black), MexR-L13M (empty rhomb, orange), MexR-R21W (filled rhomb, red), MexR-G58E (filled triangle, green), and MexR-R70W (empty circle, blue).
Notably, mutations affecting stability within the dimer- or DNA-binding region also affect the unfolding transition of the neighboring domain in the same direction (Table IV). Although the partial superposition of the two unfolding transitions and the incompleteness of the low-Tm transition for all mutants except R21W within the accessible temperature range hindered reliable evaluation of the thermodynamic parameters beyond the analysis of Tm values, it is obvious that the G58E mutation in the wHTH does not only significantly increase the stability of the DNA-binding region but also that of the dimer region. Similar but weaker effect is observed for R70W. The R20W mutation in the dimer region significantly increases the stability in both domains, whereas L13M that drastically reduces dimer stability also appears to shift the transition in the DNA-binding domain to a temperature window outside of the measurable experimental range. Thus, stability of the DNA-binding region and the dimer region appear interdependent.
MDR mutations increase hydrophobic surface accessibility in the dimer region
To further investigate the properties of the native, partially folded and unfolded states of MexR, ANS binding was analyzed as a function of denaturant both for the MexR-MDR mutants and for MexR-wt, and for the native state in the absence and presence of DNA. Both native and partially denatured states of MexR-wt and MexR-MDR mutants clearly bind ANS, as judged from the high fluorescence at 480 nm [Fig. 5(A)]. Furthermore, all MDR mutants show increased ANS binding compared with MexR-wt, and ANS binding is retained until the MexR dimer is completely unfolded (3M GuHCl and above).
Significant ANS binding to a natively folded structure can be explained either by the presence of exposed hydrophobic surfaces in the folded state or by molten-globule properties of the native state. It is well-known that DNA-binding domains with molten-globule properties fold to a compact structure on DNA binding with reduced ANS fluorescence.36,37 However, for MexR-wt, we observed that ANS binding to the MexR-PI and MexR-PII complexes is even slightly higher than in the free state (+7.5 % and +7.9%, respectively), thus giving no evidence for molten-globule-like DNA-binding domains. Molten-globule properties in the dimerization region are unlikely, because both the cooperativity and the Tm of the dimerization domain is high for wt and mutants. Also, mutational effects on dimerization were already shown to be minimal. Therefore, our data does not give evidence that the increased ANS binding to MDR mutants is due to increased structural disorder in the mutated proteins compared with wt.
Although MexR does not show any hydrophobic surfaces of sufficient size for efficient ANS binding,10 an internal hydrophobic cavity in the dimer interface was recently found to bind the MexR repressor protein ArmR, which inserts its ligand-binding arm into the cavity through a channel-like opening between the DNA-binding domain and the dimer region.19 This cavity is also accessible through an opening between the two DNA-binding domains, and it has been suggested as a ligand-binding surface for regulatory molecules.10 Two of the mutations studied in this work, R21W and L13M, line this cavity, and G58E and R70W are located at the two respective entrances. Notably, all these mutants show a higher effective ANS binding until the dimer region unfolds, although the structure content of the mutants remain the same as MexR-wt. Taken together, our results suggests that the MexR-MDR mutant proteins studied here open up access to, or extend the surface of, the regulatory ligand-binding internal cavity of MexR; testing this hypothesis requires further structural analysis.
Discussion
The aim of this work has been to investigate whether biophysical analysis of MexR-wt and MexR-MDR mutant proteins could provide further clues as to how MexR mutations confer MDR to P. aeruginosa. Our particular focus has been set on investigating properties of mutations outside of the DNA binding surface, because a critical issue for MarR-type protein functionality appears to be the ease in which DNA binding is compromised by mutations located all over the protein sequence.
We show that in four-key multidrug resistant MexR proteins with mutations outside of the DNA-binding surface, the structure content is nearly identical to MexR-wt and there is no biophysical evidence suggesting misfolding or the presence of monomeric states. These results contrast previous proposals based on indirect observations and mere visualization of mutation sites without further experimental data.28,29,31,38 Analytical ultracentrifugation data revealed that at micromolar concentrations, both wt and mutant MexR proteins are prevalently dimeric. Indeed, the dimeric state of proteins in the MarR family is well-documented both biophysically32 and structurally.9–12,14 All mutants in this study maintain a near-native secondary structure content, which would be hard to conceptualize for a monomeric state, considering the intertwined helices in the dimer region [Fig. 1(A)]. Furthermore, because we show that MexR mRNA is produced to similarly high levels as the efflux pump proteins for a disregulated gene, misfolded and/or monomeric proteins would be a burden to a viable MDR P. aeruginosa strain and should, therefore, not be selected for in MDR development.
MexR-wt binding to the PI- and PII-DNA boxes [Fig. 1(B)] show high-affinity binding to both sites as measured by Biacore. The affinity toward the PI site is slightly higher (Table II), and it seems to better stabilize the structure of the MexR DNA-binding domain [Fig. 4(C)]. However, a recent review of DNA affinities for MarR and its homologues ExpG, PecS, OhrR, and HucR3 indicates that MexR binds a single PI or PII box with ∼50-fold lower affinities compared with closely related MarR family members. Although MexR DNA binding is best described by a dimer in our Biacore experiments, the distance between the middle of the PI and PII boxes as determined by footprinting studies (31 bp)21 corresponds well to three complete turns of the DNA helix, suggesting that two MexR dimers bind the adjacent PI and PII palindromes on the same face of the DNA helix. Cooperative DNA binding to two consecutive DNA boxes, mediated by protein dimer–dimer contacts and DNA bending, may thus be required by MexR for full repression, and additional structural and biochemical studies should clarify this further.
The current study reveals that the MexR dimer in the apo state features unstably prefolded DNA-binding wHTH domains and a stably folded dimer region. Indeed, wHTH domains are known to possess widely varying as well as highly context-dependent stabilities.39–41 Complex formation with DNA is required for MexR to form a jointly folded entity with a single, highly cooperative, unfolding transition. In this context, it is interesting that the structural homologue HucR, which is prefolded for DNA-binding, displays a single cooperative melting transition at 51°C already in the absence of DNA;14,42 this Tm is close to what is observed for the dimer region in MexR both in its apo and DNA-bound states.
In this work, we show that MDR mutations distant from the DNA-binding region significantly affect MexR subdomain stabilities without affecting secondary structure content. Thus, the changes in stability cannot be attributed to changes in structure compared with the wt protein, rather, the magnitude of these changes requires considering that the mutations may affect interdomain interactions. We find that interdomain crosstalk between the dimer region and the DNA-binding region is present already in the apo state, because mutations affecting the stability in one region also affect the stability of the other, and unfolding probed by a fluorophore in the dimer region (W21) senses the unfolding of the distant DNA-binding domain. Such interactions convey allosteric interactions between oligomerization entities and DNA-binding domains in the Lac repressor (reviewed in Ref. 43) as well as in TetR,44 and may be central to the functionality of the MexR dimer. The molecular details of these interactions remain to be resolved.
If the MDR mutations affect interdomain interactions, this may alter the relative orientations of the DNA-binding domain versus the dimer region. Indeed, the specific binding of well-defined ligands observed for MarR, HucR, and OmpR results in distinct reorientation of well-structured DNA-binding domains toward an equally stable dimer region.12,14–16 However, for MexR the molecular properties appear different: First, the crystal structure suggest a conformational ensemble of various relative interdomain orientations in the apo state, where only some are compatible with DNA binding,10 and second, the DNA-binding domain is significantly less stable than the core dimer region as shown here. Stabilizing mutations such as R21W and G58E may limit the conformational range of MexR interdomain orientations to those not compatible with DNA binding, as does the MexR antirepressor protein ArmR,19 or may increase the propensity for conformations allowing for the formation of disulphide bridges, which severely restrict interdomain orientation and was recently suggested as a regulatory mechanism.20 For the less stable MexR-L13M, gene repression is much less affected compared with the other mutants analyzed (Table I), suggesting that this mutant despite its lower stability is still fairly efficient, presumably because it still samples a considerable range of DNA-binding conformations. Although it is still unknown whether the molecularly widely diverse efflux substrates of the MexAB-OprM efflux pump (reviewed in Ref. 17) are also effectors of MexR derepression by direct binding to MexR, such effectors could well act by affecting the distribution of the conformational ensemble rather than by inducing lock-and-key structural effects.45
The increased ANS binding to the dimer region in MDR mutants presents further clues as to possible mechanisms for MexR derepression. Binding of a wide variety of effector molecules suggested to deregulate MexR repression should be facilitated by the presence of an adaptive hydrophobic cavity. Such binding pockets have been described structurally for at least three multidrug binding transcription regulators including QacR, TtgR, and LmrR.26,46,47 To this end, it is interesting that all of the analyzed MDR mutations in MexR show extended ANS binding to a hydrophobic surface in the native protein. The hydrophobic surface is in the dimer region, which remains accessible until the dimer unfolds and dissociates at high concentrations of denaturant. Although no larger hydrophobic patches are present on the surface of MexR, internal hydrophobic grooves in the dimer region have been suggested to bind antirepressing ligands.10,19 Thus, it is reasonable to assume that these grooves, or their access routes, are enlarged in the multidrug-resistant MexR mutant proteins, possibly leading to larger adaptivity and/or accessibility and thereby enhancing deactivation of MexR repressive activity as well as the production of efflux pumps and concomitant antibiotics resistance.
Taken together, our results extend the current view on MexR regulation of the MexAB-OprM operon by showing that correlated domain stabilities and internal hydrophobic grooves are critical intrinsic features of the MexR protein that are sensitive to mutations, and directly relate to DNA binding. Mutations in MexR distant from the DNA binding surface alter the intrinsic stability of both DNA-binding and dimer regions in a correlated way, and extend, or facilitate access to, the internal hydrophobic cavity suggested to participate in derepression mechanisms. Based on these results, further biophysical and structural studies of MexR-MDR mutants hold promise to increase our understanding of gene regulation in P. aeruginosa and how it is disrupted in antibiotics resistance.
Experimental Procedures
Protein cloning, expression, and purification
Nine MexR proteins were chosen for subcloning and overexpression in Escherichia coli: The native mexR gene (441 bp) from the P. aeruginosa strain PAO503 (met-9011) and eight MexR mutants conferring multiresistance in P. aeruginosa isolates: K2 (R83H), K3 (R91H), K4 (L13M), T3 (G58E), T4 (L95F), T6 (T69I), T7 (R21W), and T10 (R70W).27 PCR amplified mexR genes cloned into pGEX-4T-3, an N-terminal thrombin-cleavable GST-tagged expression vector, were expressed and acquired onto glutathione-Sepharose 4B beads by incubation for 2 h at 4°C (Pharmacia Biotech, Uppsala, Sweden). After PBS wash, MexR was cleaved from the bound fusion protein using 40 U of thrombin per liter cell culture. Cleavage was monitored by MALDI analysis. To remove residual GST and thrombin, heparin sepharose (GE healthcare) was used for final purification using a NaCl/10 mM NaPO4 gradient elution buffer at pH 7.4. After purification, all MexR proteins were more than 95% pure as judged by SDS-page and MALDI analysis, and contained no sign of covalent MexR dimers. The purified MexR proteins were stored in 20 mM NaPO4, 150 mM NaCl, 1 mM DTT, at −20°C. Great care was taken to avoid covalent dimer formation during protein preparation and storage, which was all performed under reducing conditions. Furthermore, MALDI was regularly used to check that covalently bonded MexR dimers were not formed in wild type or mutant proteins during the course of experiments.
RNA purification, synthesis of cDNA, and real-time PCR
P. aeruginosa strain PAO503 (met-9011) and its multiresistant mutant isolates K2, K3, K4, T3, T4, T6, T7, and T10 were harvested from LB in the logarithmic phase by centrifugation at 3000g for 10 min at 4°C. Pellets were resuspended on ice in 10 mM Tris, pH 8.0 to isolate total RNA (High Pure RNA Isolation Kit, Roche Diagnostics), which was aliquoted and stored at −70°C until used in the cDNA preparation reaction. One microgram of total RNA was taken as a template in the RT (reverse transcriptase)-PCR reaction in 20-μL volumes as described in first Strand cDNA Synthesis Kit for RT-PCR, Roche. In the RT-PCR reaction, we used random primers p(dN)6. The cDNA was stored at −20°C until used in real-time PCR in the LightCycler (LC; Roche). A master mix containing 4 mM MgCl2 and 2 μM each gene specific primers (mexR and mexB), was prepared on ice using LightCycler-FastStart DNA Master SYBR Green I kit (Roche), and 18-μL master mix was mixed with 2 μL cDNA in the capillaries placed in the LightCycler. The temperature profile was 95°C for 600 s, 45 cycles at 95°C for 15 s, 62°C for 5 s, and 72°C for 16 s. The temperature transition rate was 20°C/s. Melting curve analysis was done in one cycle with three segments: 95°C for 60 s, 67°C for 60 s, and 95°C for 0 s with a temperature transition rate of 20°C/s for first and second segments and 0.1°C for third segment. Crossing point values (CTs) from the real-time PCR products were used to quantify the amounts of the mRNA produced.
DNA target sequences for biophysical studies
Double-stranded oligonucleotides corresponding to the MexR DNA-binding sites on the mexA-mexR intergenic regions [Fig. 1(B)] 21 were synthesized by solid phase synthesis (Invitrogen) as follows (only the upper strand is shown): PI: 5′-TGT AAA TGT GGT TGA TCC AGT CAA CTA TTT TGC TT-3′; PII: 5′-CTT ATT TTA GTT GAC CTT ATC AAC CTT GTT TCA G-3′. For Biacore studies, the forward DNA strand was labeled with biotin. The two complementary strands were annealed at 90°C with subsequent cooling on the bench.
SPR measurements and ligand immobilization procedures
SPR measurements were conducted at 760 nm and 25°C in a fully automatic Biacore 2000 instrument (Biacore, GE Healthcare, Uppsala, Sweden) equipped with four flow cells. HBS-N (10 mM hepes, 0.15M NaCl, 3 mM EDTA, pH 7.4, from Biacore) was used as running and sample buffer. Streptavidin-coated Biacore SA-chips were conditioned with three sequential 1-min injections of 1M NaCl in 50 mM NaOH at a flow rate of 10 μL/min before ligand immobilization. The 5′-end biotinylated dsDNA ligands were diluted to 4 μg/mL in HBS-N buffer and injected until 200–300 RU was captured in fc2, followed by two 1-min injections of 0.05% (w/v) SDS to remove any unbound DNA. Complementary ssDNA was injected to confirm that the immobilized DNA was double stranded. A biotinylated random dsDNA sequence was immobilized in fc1 and used as a reference. Blocking of un-reacted biotin-binding sites was performed by a 1-min injection of a 0.5 mg/mL biotin solution. Kinetic measurements were conducted at flow rates of 10–20 μL/min and the analyte was injected over the surface for 5 min. The MexR analyte concentrations used was 4, 1, and 0.5 μM over the PI surface and 4, 2, and 1 μM over PII. Two subsequent 1-min injections of 0.05% (w/v) SDS were used to regenerate the surface between the injections of MexR. No signs of mass transport limitations were observed. Sensorgrams were evaluated using the BiaEvaluation software version 4.1 (Biacore, GE Healthcare, Uppsala, Sweden). The quality of the fits was determined by visual inspection of the residual plots and by evaluation of the X2/Rmax ratio, where X2 corresponds to the average squared residual per data point and Rmax is the maximally obtained RU (response units).48 A model was judged appropriate if it could fit to the data with a X2/Rmax ratio below 0.1.
Analytical ultracentrifugation
All ultracentrifugation experiments were performed in a Beckman Coulter Optima XL-I Analytical Ultracentrifuge using an An-50 Ti rotor and six-sector cell. All measurements were recorded at 20°C in 1× PBS buffer, pH 7.2. Concentrations of MexR-wt, MexR-R21W, and MexR-R70W were 30 μM, 10 μM, and 12 μM, respectively. Lower concentrations where not possible to use due to limitations in the sensitivity of the UV-detector below the absorbance of 0.1 at 280 nm. Sedimentation equilibrium experiments were conducted using rotor speeds between 3000–18,000 rpm for MexR-wt, 6000–30,000 for MexR-R21W, and 6000–35,000 for MexR R70W. The background was corrected for each sample well-against well containing 1× PBS buffer only. Calculations and curve fitting were performed using the Microcal Origin Program,49 and Sednterp50 was used for calculations of solvent density and partial specific volume at 22°C.
Circular dichroism experiments
Circular dichroism spectra were recorded using a CD6 spectrodichrograph (Jobin-Yvon Instruments SA, Longjumeau, France). For secondary structure predictions, spectra in the far-UV region (190–240 nm) were recorded at 22°C with 0.5 nm increments in a 0.5 mm quartz cell, and six scans were averaged for each spectrum. Secondary structure estimates of wild type and mutant MexR was made using DICHROWEB33,51 and the algorithms CDSSTR,52,53 CONTIN,54,55 and Selcon3.56 Thermal denaturation spectra were recorded at 210–240 nm with 5°C intervals from 15–85°C using a 0.5 mm quartz cell, 15 μM protein in 10 mM sodium phosphate buffer, pH 7.2. Thermal melting midpoints (Tm) were estimated from fraction of unfolding curves fitting equations for two- and three-state transitions, using TableCurve (Jandel Scientific).
Fluorescence spectroscopy measurements
Fluorescence spectra of tryptophan-containing MexR mutants were recorded on a Hitachi F-4500 Fluorescence spectrophotometer at 20°C in a 1-cm path length quartz cell using a bandwidth of 5 nm for both excitation and emission. All spectra are averages of three scans, with buffer values subtracted. After a coarse screen to find the range of transition, 17 MexR-R21W and 18 MexR-R70W samples were prepared and incubated for 20 h at 25°C with a protein concentration of 3 μM and GdnHCl concentrations ranging from 0 to 6M at regular intervals. Excitation was performed at 295 nm, and emission spectra were recorded from 300 to 400 nm.
To examine ANS binding, MexR-wt and mutants were incubated at 4°C in 20 h with 0–6M GdnHCl. Before reading, the samples were incubated with a 20-fold excess of ANS for 10 min. Spectra of MexR-wt in the absence or presence of DNA were obtained using a protein concentration of 0.5 μM and a fourfold molar excess of ANS in the absence or presence of molar equivalents of PI or PII-DNA. All spectra were recorded on a Horiba Jobin-Yvon Fluoromax-4 instrument in a 1-cm quarts fluorescence cell, with an excitation wavelength of 380 nm, emission wavelengths of 400–600 nm, and bandwidths of 5 nm (GdnHCl experiments) and 2.5 nm (DNA-binding experiments).
Determination of protein concentrations
All UV-absorption measurements were recorded using a Varian Cary 100 Bio UV spectrophotometer at 20°C. Concentration of wt MexR, MexR-L13M, and MexR-G58E were determined by a carefully calibrated Bradford assay. Mutants MexR-R21W and MexR R70W were always combined with UV-absorption measurements at 280 nm with calculated extinction coefficients57 in 6M GdnHCl, 0.02M phosphate buffer, pH 6.5, of 1615M−1 cm−1for MexR-wt, MexR-L13M, MexR-G58E, and 7115M−1 cm−1 for MexR-R21W and MexR R70W.
Acknowledgments
We thank Prof. Jan-Ingvar Flock for stimulating discussions in the initial stages of this work, and Dr. Kaisong Zhou, Dr. Anna-Katrine Museth, Assoc. Prof. Magdalena Svensson, Prof. Bengt-Harald Jonsson, and Prof. Uno Carlsson for generous experimental advice.
References
- 1.Alekshun MN, Levy SB. Molecular mechanisms of antibacterial multidrug resistance. Cell. 2007;128:1037–1050. doi: 10.1016/j.cell.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 2.Gajiwala KS, Burley SK. Winged helix proteins. Curr Opin Struct Biol. 2000;10:110–116. doi: 10.1016/s0959-440x(99)00057-3. [DOI] [PubMed] [Google Scholar]
- 3.Wilkinson SP, Grove A. Ligand-responsive transcriptional regulation by members of the MarR family of winged helix proteins. Curr Issues Mol Biol. 2006;8:51–62. [PubMed] [Google Scholar]
- 4.Poole K. Efflux-mediated resistance to fluoroquinolones in gram-positive bacteria and the mycobacteria. Antimicrob Agents Chemother. 2000;44:2595–2599. doi: 10.1128/aac.44.10.2595-2599.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poole K. Efflux-mediated antimicrobial resistance. J Antimicrob Chemother. 2005;56:20–51. doi: 10.1093/jac/dki171. [DOI] [PubMed] [Google Scholar]
- 6.Schumacher MA, Brennan RG. Structural mechanisms of multidrug recognition and regulation by bacterial multidrug transcription factors. Mol Microbiol. 2002;45:885–893. doi: 10.1046/j.1365-2958.2002.03039.x. [DOI] [PubMed] [Google Scholar]
- 7.Godsey MH, Heldwein EEZ, Brennan RG. Structural biology of bacterial multidrug resistance gene regulators. J Biol Chem. 2002;277:40169–40172. doi: 10.1074/jbc.R200018200. [DOI] [PubMed] [Google Scholar]
- 8.Schweizer HP. Efflux as a mechanism of resistance to antimicrobials in Pseudomonas aeruginosa and related bacteria: unanswered questions. Genet Mol Res. 2003;2:48–62. [PubMed] [Google Scholar]
- 9.Alekshun MN, Levy SB, Mealy TR, Seaton BA, Head JF. The crystal structure of MarR, a regulator of multiple antibiotic resistance, at 2.3 A resolution. Nat Struct Biol. 2001;8:710–714. doi: 10.1038/90429. [DOI] [PubMed] [Google Scholar]
- 10.Lim D, Poole K, Strynadka NC. Crystal structure of the MexR repressor of the mexRAB-oprM multidrug efflux operon of Pseudomonas aeruginosa. J Biol Chem. 2002;277:29253–29259. doi: 10.1074/jbc.M111381200. [DOI] [PubMed] [Google Scholar]
- 11.Wu RY, Zhang RG, Zagnitko O, Dementieva I, Maltzev N, Watson JD, Laskowski R, Gornicki P, Joachimiak A. Crystal structure of Enterococcus faecalis SlyA-like transcriptional factor. J Biol Chem. 2003;278:20240–20244. doi: 10.1074/jbc.M300292200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hong M, Fuangthong M, Helmann JD, Brennan RG. Structure of an OhrR-ohrA operator complex reveals the DNA binding mechanism of the MarR family. Mol Cell. 2005;20:131–141. doi: 10.1016/j.molcel.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 13.Chen PR, Bae T, Williams WA, Duguid EM, Rice PA, Schneewind O, He C. An oxidation-sensing mechanism is used by the global regulator MgrA in Staphylococcus aureus. Nat Chem Biol. 2006;2:591–595. doi: 10.1038/nchembio820. [DOI] [PubMed] [Google Scholar]
- 14.Bordelon T, Wilkinson SP, Grove A, Newcomer ME. The crystal structure of the transcriptional regulator HucR from Deinococcus radiodurans reveals a repressor preconfigured for DNA binding. J Mol Biol. 2006;360:168–177. doi: 10.1016/j.jmb.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 15.Newberry KJ, Fuangthong M, Panmanee W, Mongkolsuk S, Brennan RG. Structural mechanism of organic hydroperoxide induction of the transcription regulator OhrR. Mol Cell. 2007;28:652–664. doi: 10.1016/j.molcel.2007.09.016. [DOI] [PubMed] [Google Scholar]
- 16.Saridakis V, Shahinas D, Xu X, Christendat D. Structural insight on the mechanism of regulation of the MarR family of proteins: high-resolution crystal structure of a transcriptional repressor from Methanobacterium thermoautotrophicum. J Mol Biol. 2008;377:655–667. doi: 10.1016/j.jmb.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 17.Daigle DM, Cao L, Fraud S, Wilke MS, Pacey A, Klinoski R, Strynadka NC, Dean CR, Poole K. Protein modulator of multidrug efflux gene expression in Pseudomonas aeruginosa. J Bacteriol. 2007;189:5441–5451. doi: 10.1128/JB.00543-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Domain F, Bina XR, Levy SB. Transketolase A, an enzyme in central metabolism, derepresses the marRAB multiple antibiotic resistance operon of Escherichia coli by interaction with MarR. Mol Microbiol. 2007;66:383–394. doi: 10.1111/j.1365-2958.2007.05928.x. [DOI] [PubMed] [Google Scholar]
- 19.Wilke MS, Heller M, Creagh AL, Haynes CA, McIntosh LP, Poole K, Strynadka NC. The crystal structure of MexR from Pseudomonas aeruginosa in complex with its antirepressor ArmR. Proc Natl Acad Sci USA. 2008;105:14832–14837. doi: 10.1073/pnas.0805489105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen H, Hu J, Chen PR, Lan L, Li Z, Hicks LM, Dinner AR, He C. The Pseudomonas aeruginosa multidrug efflux regulator MexR uses an oxidation-sensing mechanism. Proc Natl Acad Sci USA. 2008;105:13586–13591. doi: 10.1073/pnas.0803391105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Evans K, Adewoye L, Poole K. MexR repressor of the mexAB-oprM multidrug efflux operon of Pseudomonas aeruginosa: identification of MexR binding sites in the mexA-mexR intergenic region. J Bacteriol. 2001;183:807–812. doi: 10.1128/JB.183.3.807-812.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ziha-Zarifi I, Llanes C, Kohler T, Pechere JC, Plesiat P. In vivo emergence of multidrug-resistant mutants of Pseudomonas aeruginosa overexpressing the active efflux system MexA-MexB-OprM. Antimicrob Agents Chemother. 1999;43:287–291. doi: 10.1128/aac.43.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jalal S, Wretlind B. Mechanisms of quinolone resistance in clinical strains of Pseudomonas aeruginosa. Microb Drug Resist. 1998;4:257–261. doi: 10.1089/mdr.1998.4.257. [DOI] [PubMed] [Google Scholar]
- 24.Srikumar R, Tsang E, Poole K. Contribution of the MexAB-OprM multidrug efflux system to the beta-lactam resistance of penicillin-binding protein and beta-lactamase-derepressed mutants of Pseudomonas aeruginosa. J Antimicrob Chemother. 1999;44:537–540. doi: 10.1093/jac/44.4.537. [DOI] [PubMed] [Google Scholar]
- 25.Oh H, Stenhoff J, Jalal S, Wretlind B. Role of efflux pumps and mutations in genes for topoisomerases II and IV in fluoroquinolone-resistant Pseudomonas aeruginosa strains. Microb Drug Resist. 2003;9:323–328. doi: 10.1089/107662903322762743. [DOI] [PubMed] [Google Scholar]
- 26.Brooks BE, Piro KM, Brennan RG. Multidrug-binding transcription factor QacR binds the bivalent aromatic diamidines DB75 and DB359 in multiple positions. J Am Chem Soc. 2007;129:8389–8395. doi: 10.1021/ja072576v. [DOI] [PubMed] [Google Scholar]
- 27.Jalal S, Wretlind G, Gotoh N, Wretlind B. Rapid identification of mutations in a multidrug efflux pump in Pseudomonas aeruginosa. APMIS. 1999;107:1109–1116. doi: 10.1111/j.1699-0463.1999.tb01516.x. [DOI] [PubMed] [Google Scholar]
- 28.Saito K, Eda S, Maseda H, Nakae T. Molecular mechanism of MexR-mediated regulation of MexAB-OprM efflux pump expression in Pseudomonas aeruginosa. FEMS Microbiol Lett. 2001;195:23–28. doi: 10.1111/j.1574-6968.2001.tb10492.x. [DOI] [PubMed] [Google Scholar]
- 29.Alekshun MN, Kim YS, Levy SB. Mutational analysis of MarR, the negative regulator of marRAB expression in Escherichia coli, suggests the presence of two regions required for DNA binding. Mol Microbiol. 2000;35:1394–1404. doi: 10.1046/j.1365-2958.2000.01802.x. [DOI] [PubMed] [Google Scholar]
- 30.Dupont P, Hocquet D, Jeannot K, Chavanet P, Plesiat P. Bacteriostatic and bactericidal activities of eight fluoroquinolones against MexAB-OprM-overproducing clinical strains of Pseudomonas aeruginosa. J Antimicrob Chemother. 2005;55:518–522. doi: 10.1093/jac/dki030. [DOI] [PubMed] [Google Scholar]
- 31.Adewoye L, Sutherland A, Srikumar R, Poole K. The mexR repressor of the mexAB-oprM multidrug efflux operon in Pseudomonas aeruginosa: characterization of mutations compromising activity. J Bacteriol. 2002;184:4308–4312. doi: 10.1128/JB.184.15.4308-4312.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilkinson SP, Grove A. Negative cooperativity of uric acid binding to the transcriptional regulator HucR from Deinococcus radiodurans. J Mol Biol. 2005;350:617–630. doi: 10.1016/j.jmb.2005.05.027. [DOI] [PubMed] [Google Scholar]
- 33.Lobley A, Whitmore L, Wallace BA. DICHROWEB: an interactive website for the analysis of protein secondary structure from circular dichroism spectra. Bioinformatics. 2002;18:211–212. doi: 10.1093/bioinformatics/18.1.211. [DOI] [PubMed] [Google Scholar]
- 34.Karlsson R, Falt A. Experimental design for kinetic analysis of protein-protein interactions with surface plasmon resonance biosensors. J Immunol Methods. 1997;200:121–133. doi: 10.1016/s0022-1759(96)00195-0. [DOI] [PubMed] [Google Scholar]
- 35.Santoro MM, Bolen DW. A test of the linear extrapolation of unfolding free energy changes over an extended denaturant concentration range. Biochemistry. 1992;31:4901–4907. doi: 10.1021/bi00135a022. [DOI] [PubMed] [Google Scholar]
- 36.Twigg PD, Parthasarathy G, Guerrero L, Logan TM, Caspar DL. Disordered to ordered folding in the regulation of diphtheria toxin repressor activity. Proc Natl Acad Sci USA. 2001;98:11259–11264. doi: 10.1073/pnas.191354798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Narayanan SS, Pal SK. Nonspecific protein-DNA interactions: complexation of alpha-chymotrypsin with a genomic DNA. Langmuir. 2007;23:6712–6718. doi: 10.1021/la063586x. [DOI] [PubMed] [Google Scholar]
- 38.Suman G, Khan M, Sabitha K, Jamil K. Mutation in mexR-gene leading to drug resistance in corneal keratitis in human. Indian J Exp Biol. 2006;44:929–936. [PubMed] [Google Scholar]
- 39.Hardy JA, Nelson HC. Proline in alpha-helical kink is required for folding kinetics but not for kinked structure, function, or stability of heat shock transcription factor. Protein Sci. 2000;9:2128–2141. doi: 10.1110/ps.9.11.2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yan H, Liao X. Amino acid substitutions in a long flexible sequence influence thermodynamics and internal dynamic properties of winged helix protein genesis and its DNA complex. Biophys J. 2003;85:3248–3254. doi: 10.1016/S0006-3495(03)74742-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen L, Chen LR, Zhou XE, Wang Y, Kahsai MA, Clark AT, Edmondson SP, Liu ZJ, Rose JP, Wang BC, Meehan EJ, Shriver JW. The hyperthermophile protein Sso10a is a dimer of winged helix DNA-binding domains linked by an antiparallel coiled coil rod. J Mol Biol. 2004;341:73–91. doi: 10.1016/j.jmb.2004.05.044. [DOI] [PubMed] [Google Scholar]
- 42.Wilkinson SP, Grove A. HucR, a novel uric acid-responsive member of the MarR family of transcriptional regulators from Deinococcus radiodurans. J Biol Chem. 2004;279:51442–51450. doi: 10.1074/jbc.M405586200. [DOI] [PubMed] [Google Scholar]
- 43.Kalodimos CG, Boelens R, Kaptein R. Toward an integrated model of protein-DNA recognition as inferred from NMR studies on the Lac repressor system. Chem Rev. 2004;104:3567–3586. doi: 10.1021/cr0304065. [DOI] [PubMed] [Google Scholar]
- 44.Reichheld SE, Davidson AR. Two-way interdomain signal transduction in tetracycline repressor. J Mol Biol. 2006;361:382–389. doi: 10.1016/j.jmb.2006.06.035. [DOI] [PubMed] [Google Scholar]
- 45.Wright PE, Dyson HJ. Linking folding and binding. Curr Opin Struct Biol. 2009;19:31–38. doi: 10.1016/j.sbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alguel Y, Meng C, Teran W, Krell T, Ramos JL, Gallegos MT, Zhang X. Crystal structures of multidrug binding protein TtgR in complex with antibiotics and plant antimicrobials. J Mol Biol. 2007;369:829–840. doi: 10.1016/j.jmb.2007.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Madoori PK, Agustiandari H, Driessen AJ, Thunnissen AM. Structure of the transcriptional regulator LmrR and its mechanism of multidrug recognition. EMBO J. 2009;28:156–166. doi: 10.1038/emboj.2008.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Biacore. Biaevaluation Software Handbook. Biacore AB: Uppsala Sweden; 2004. June 2004. [Google Scholar]
- 49.Johnson ML, Correia JJ, Yphantis DA, Halvorson HR. Analysis of data from the analytical ultracentrifuge by nonlinear least-squares techniques. Biophys J. 1981;36:575–588. doi: 10.1016/S0006-3495(81)84753-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hayes D, Laue T, Philo J. Sednterp. Durham, NH: University of New Hampshire; 1995. [Google Scholar]
- 51.Whitmore L, Wallace BA. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 2004;32:W668–W673. doi: 10.1093/nar/gkh371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Compton LA, Johnson WC. Analysis of protein circular dichroism spectra for secondary structure using a simple matrix multiplication. Anal Biochem. 1986;155:155–167. doi: 10.1016/0003-2697(86)90241-1. [DOI] [PubMed] [Google Scholar]
- 53.Sreerama N, Woody RW. Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal Biochem. 2000;287:252–260. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- 54.Provencher SW, Glockner J. Estimation of globular protein secondary structure from circular dichroism. Biochemistry. 1981;20:33–37. doi: 10.1021/bi00504a006. [DOI] [PubMed] [Google Scholar]
- 55.van Stokkum IH, Spoelder HJ, Bloemendal M, van Grondelle R, Groen FC. Estimation of protein secondary structure and error analysis from circular dichroism spectra. Anal Biochem. 1990;191:110–118. doi: 10.1016/0003-2697(90)90396-q. [DOI] [PubMed] [Google Scholar]
- 56.Sreerama N, Woody RW. A self-consistent method for the analysis of protein secondary structure from circular dichroism. Anal Biochem. 1993;209:32–44. doi: 10.1006/abio.1993.1079. [DOI] [PubMed] [Google Scholar]
- 57.Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 58.DeLano WL. 2002. http://www.pymol.orgThe PyMOL Molecular Graphics System DeLano Scientific, Palo Alto, CA, USA.