Abstract
The wild-type HIV-1 capsid protein (CA) self-assembles in vitro into tubular structures at high ionic strength. We report solid state nuclear magnetic resonance (NMR) and electron microscopy measurements on these tubular CA assemblies, which are believed to contain a triangular lattice of hexameric CA proteins that is similar or identical to the lattice of capsids in intact HIV-1. Mass-per-length values of CA assemblies determined by dark-field transmission electron microscopy indicate a variety of structures, ranging from single-wall tubes to multiwall tubes that approximate solid rods. Two-dimensional (2D) solid state 13C—13C and 15N—13C NMR spectra of uniformly 15N,13C-labeled CA assemblies are highly congested, as expected for a 25.6 kDa protein in which nearly the entire amino acid sequence is immobilized. Solid state NMR spectra of partially labeled CA assemblies, expressed in 1,3-13C2-glycerol medium, are better resolved, allowing the identification of individual signals with line widths below 1 ppm. Comparison of crosspeak patterns in the experimental 2D spectra with simulated patterns based on solution NMR chemical shifts of the individual N-terminal (NTD) and C-terminal (CTD) domains indicates that NTD and CTD retain their individual structures upon self-assembly of full-length CA into tubes. 2D 1H-13C NMR spectra of CA assemblies recorded under solution NMR conditions show relatively few signals, primarily from segments that link the α-helices of NTD and CTD and from the N- and C-terminal ends. Taken together, the data support the idea that CA assemblies contain a highly ordered 2D protein lattice in which the NTD and CTD structures are retained and largely immobilized.
Keywords: solid state NMR, virus structure, protein self-assembly, electron microscopy
Introduction
The genomic material of the HIV-1 virus is contained within a conical capsid shell constructed from ∼1500 copies of the capsid protein (CA).1 The capsid is believed to be formed by a triangular lattice of CA hexamers, with 12 CA pentamers unevenly distributed at the two ends of the cone to produce a closed shell.2 Although much progress has been made toward understanding the structure, stability, and assembly of the HIV-1 capsid, through electron microscopy,3–10 X-ray crystallography,11–20 solution nuclear magnetic resonance (NMR),13,21–27 and various biophysical techniques,28–40 substantial questions remain regarding the details of intermolecular interactions within and between CA hexamers, the structural variations that permit CA to form both hexamers and pentamers, the driving force for curvature and closure of the CA lattice, and the mechanism and pathway for CA self-assembly. In principle, modern solid state NMR techniques, which can provide site-specific molecular-level structural information about noncrystalline protein assemblies,41–49 can contribute to the resolution of these questions. To our knowledge, data reported below represent the first solid state NMR data for retroviral capsid assemblies.
In solution, CA contains globular N-terminal (NTD, residues 1–145) and C-terminal (CTD, residues 150–231) domains that are connected by a short linker (residues 146–149). High-resolution structures of NTD and CTD have been determined by solution NMR and X-ray crystallography.15–24,27,50 In these structures, NTD contains seven α-helical segments, plus a β-hairpin in the first 12 residues. CTD contains four α-helical segments, plus a single-turn 310 helix in the first 4 residues. In solution, NTD exists as a monomer under typical experimental conditions, whereas CTD exists as a dimer.16,18,51 Monomeric CTD can be generated by mutation of certain residues in the dimer interface.21,23
Recombinant wild-type CA self-assembles spontaneously into tubular structures at high ionic strength in vitro,2,3,32,37,40 or in the presence of crowding agents.29 Tubular CA assemblies are believed to be constructed from the same underlying hexamer lattice as the capsids of intact HIV-1, as supported by electron microscopy.3 The R18L mutant of CA has been shown to form highly ordered, planar two-dimensional hexamer lattices that produce high-quality electron diffraction data.4 By fitting the NTD and CTD structures to the density map obtained from the electron diffraction data, Ganser-Pornillos et al. generated a model for the CA hexamer lattice in which the locations and orientations of the two domains and their 11 α-helices were specified.4 More recently, Pornillos et al. have reported X-ray crystal structures of engineered CA hexamers, created by introduction of cysteine substitions (A14C and E45C) and disulfide crosslinking or by fusion to the hexamer-forming CcmK4 protein.11 Mutations were also introduced on the CTD dimerization surface to prevent noncrystalline aggregation. These crystal structures and the electron diffraction model are in good agreement, showing that helices 1, 2, and 3 of NTD form the hexamer core, that contacts between hexamers involve helix 9 of CTD, and that NTD-CTD contacts are primarily intermolecular, involving helices 3, 4, and 7 of NTD and helices 8 and 11 of CTD within a hexamer.
In this article, we describe electron microscopy and solid state NMR measurements on tubular assemblies of full-length, wild-type HIV-1 CA. Transmission electron microscope (TEM) images of negatively stained CA assemblies are in good agreement with earlier results.11 Dark-field images obtained in a tilted-beam (TB-TEM) mode allow mass densities of individual assemblies to be quantified,52 revealing that CA assemblies range from single-walled tubes to nearly solid, multiwalled rods. Two-dimensional (2D) NMR spectra allow assessment of structural order and molecular motions within the CA assemblies. Spectra obtained with solid state NMR methods show that the majority of sites in both NTD and CTD are immobilized in the tubular CA assemblies, and support the idea that the two domains retain their individual structures in full-length CA assemblies. A relatively small number of sites have significant mobility and contribute to spectra obtained under conditions appropriate for solution NMR measurements. These sites can be assigned to segments that link the α-helices of NTD and CTD and to the extreme N- and C-terminal segments.
Results
Characterization of CA assembly morphology and mass-per-length
After adjustment to high ionic strength conditions, wild-type CA spontaneously self-assembles into tubular structures of various diameters as previously reported.2,3,32,37,40 In our hands, self-assembly begins within 5 min at 120 μM protein concentration, 1.0M NaCl, pH 8.0, and room temperature. Tubes exceeding 1 μm in length are observed after 24 h. Figure 1(A) shows a TEM image of negatively stained CA assemblies, with diameters ranging from 25 to 100 nm. Additional images are shown in the Supporting Information (Fig. S1). These assemblies are generally believed to be constructed by wrapping of a 2D triangular lattice of CA hexamers into tubes with various diameters and helicities,3 in analogy to the formation of carbon nanotubes from graphene sheets.53
Figure 1.
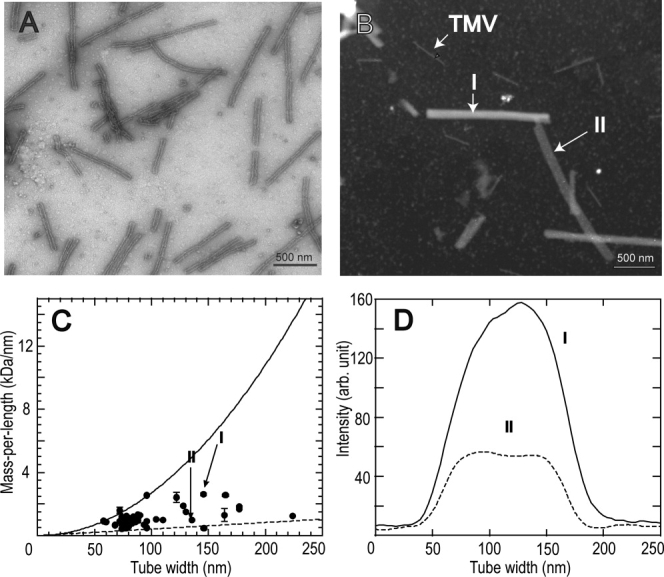
Electron microscopy of tubular HIV-1 CA assemblies. (A) Conventional bright-field image of a negatively stained sample. (B) Dark-field image of an unstained sample with coadsorbed TMV rods, obtained in tilted-beam mode. CA assembly I has a relatively high mass density and is presumably a multiwalled tube. CA assembly II has a lower mass density and is presumably a single-walled tube. (C) Mass-per-length values of individual CA assemblies, plotted against their widths in dark-field images. MPL values were calibrated by TMV rod intensities. Error bars are the root-mean-squared deviation along the length of a single CA assembly. Data points corresponding to multiwalled and single-walled tubes in panel B are indicated. Solid and dashed lines are calculated dependences of MPL on tube width for solid protein rods and single-walled tubes, respectively. (D) Cross sectional intensity profiles of the two CA assemblies in panel B, supporting our interpretation that HIV-1 CA forms both multiwalled and single-walled tubes.
We have recently demonstrated that mass-per-length (MPL) values of protein filaments can be determined from quantitative analyses of the intensities in dark-field images of unstained samples obtained in the tilted-beam mode of a conventional TEM.52 In this dark-field TB-TEM mode, the integrated image intensity of an object is proportional to its mass density and thickness.54 By using co-adsorbed tobacco mosaic virus (TMV) as an internal calibration standard with known MPL, equal to 131 kDa/nm,55 the MPL of a protein assembly can be determined on a single particle basis, providing long-range structural constraints. Figure 1(B) shows a TB-TEM image of unstained CA assemblies with co-adsorbed TMV. Additional TB-TEM images are shown in the Supporting Information (Fig. S1).
CA assemblies with various diameters are clearly visible in the TB-TEM images, and they exhibit obvious variations in brightness. For example, the assembly I in Figure 1(B) exhibits a much higher intensity than the assembly II. The cross sectional intensity distributions of these two assemblies are plotted in Figure 1(D). Interestingly, the intensity for assembly II is almost constant across its diameter, whereas that of assembly I can be fitted with a sinusoidal function. Apparently, assembly II is a hollow tube formed by a single CA layer, flattened upon drying in the vacuum of the TEM sample chamber (or after adsorption to the carbon film of the TEM sample grid), whereas I is a multiwalled tube. When projected onto the plane of the TEM grid, a flattened, single-walled tube would have a constant mass-per-area across its diameter, whereas a multiwalled, rod-like tube would have a sinusoidally varying mass-per-area across its diameter.
Figure 1(C) plots MPL values of individual assemblies against their widths in the TB-TEM images. The MPL of assembly II is close to the calculated value for a single-walled tube, assumed to be constructed from a triangular lattice of CA hexamers with lattice spacings taken from the electron diffraction results of Ganser-Pornillos et al.4 and assumed to be flattened on the TEM grid. The MPL of assembly I lies between the calculated limits of a single-walled tube and a solid cylinder [dashed and solid lines in Fig. 1(D), respectively]. MPL values of all observed assemblies lie between these two limits, indicating that tubular CA assemblies range from single-walled tubes to nearly solid protein cylinders (after drying).
Solid state NMR of uniformly labeled and partially labeled CA assemblies
Figure 2 shows two-dimensional (2D) 13C—13C and 15N—13C NMR correlation spectra of uniformly 15N,13C-labeled CA assemblies, recorded under conditions where only immobilized residues (i.e., rigid protein segments within the tubular assemblies) contribute to the NMR signals. These conditions include Hartmann-Hahn 1H—13C, 1H—15N, and/or 15N—13C cross-polarization driven by nuclear magnetic dipole–dipole couplings,56 high-power 1H decoupling during t1 and t2 periods, magic-angle spinning (MAS), and 13C—13C dipolar recoupling by radio-frequency (rf) pulses synchronized with MAS.57,58 Strong signals are observed, indicating that most of CA is immobilized. Because of the relatively high molecular weight (25.6 kDa), crosspeak signals seriously overlap in both spectra, preventing any specific interpretation of most signals. A small number of resolved crosspeaks attributable to single residues are seen in the 2D 13C—13C spectrum [Fig. 2(A)]. The crosspeak at 10.0 ppm/29.7 ppm can be assigned as the Cδ1—Cγ1 crosspeak of an isoleucine residue. For this crosspeak, the linewidth (full width at half maximum) is 0.7 ppm, consistent with a high degree of molecular conformational order. The sharpest features in the 2D 15N—13C spectrum have 15N linewidths of ∼2.5 ppm, but these features do not necessarily arise from single residues. Rf pulse sequences for 2D 13C—13C and 15N—13C spectroscopy are shown in Supporting Information (Fig. S2).
Figure 2.
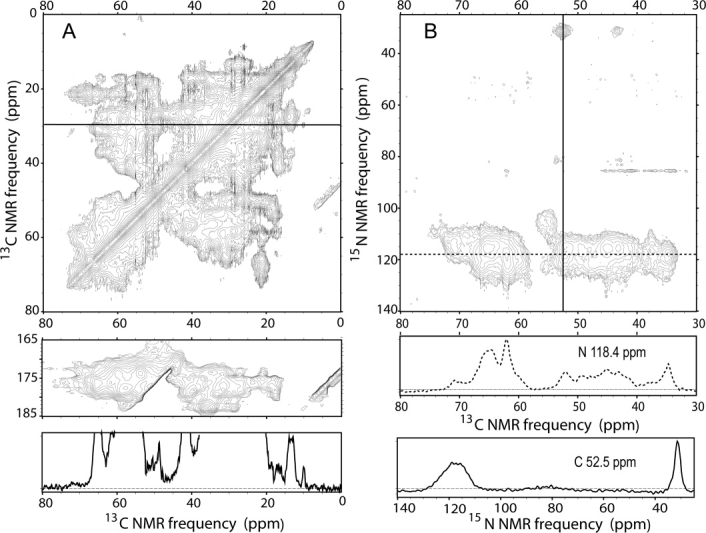
2D 13C—13C (A) and 15N—13C (B) solid state NMR correlation spectra for uniformly 15N,13C-labeled tubular CA assemblies. Spectra were acquired as described in the text. For the 2D 13C-13C spectrum, Gaussian line broadening of 30 Hz (0.20 ppm) and 50 Hz (0.33 ppm) was applied to the ω2 and ω1 dimension, respectively. The 1D slice below the 2D spectrum passes through the Cγ1/Cδ cross peak of a single isoleucine at 29.73 ppm. For the 2D 15N—13C spectrum, no line broadening was applied. Vertical and horizontal 1D slices in solid and dashed lines are extracted at indicated 13C and 15N frequencies.
To reduce spectral congestion, the CA protein was prepared with partial 13C labeling by expression on a medium that contained 1,3-13C2-glycerol as the sole carbon source. Labeling with 1,3-13C2-glycerol has been shown previously to facilitate resonance assignments and extraction of structural constraints in solid state NMR studies of various protein systems.59 As shown in Figure 3(A), the 2D 13C—13C spectrum of the partially 13C-labeled sample shows somewhat better resolution than the corresponding spectrum of the uniformly labeled sample [Fig. 2(A)]. 13C NMR linewidths of roughly 0.5 ppm are observed for several isolated crosspeaks, including isoleucine Cα/Cγ2 and Cγ1/Cδ crosspeaks, threonine Cα/Cγ crosspeaks, and asparagine Cα/Cβ crosspeaks (assignments based on typical chemical shift ranges). Linewidths of roughly 1.0 ppm are observed for alanine Cα/Cβ crosspeaks, isoleucine Cα/Cγ1 crosspeaks, proline Cγ/Cδ crosspeaks, aspartate Cα/Cβ crosspeaks, and other asparagine Cα/Cβ crosspeaks.
Figure 3.
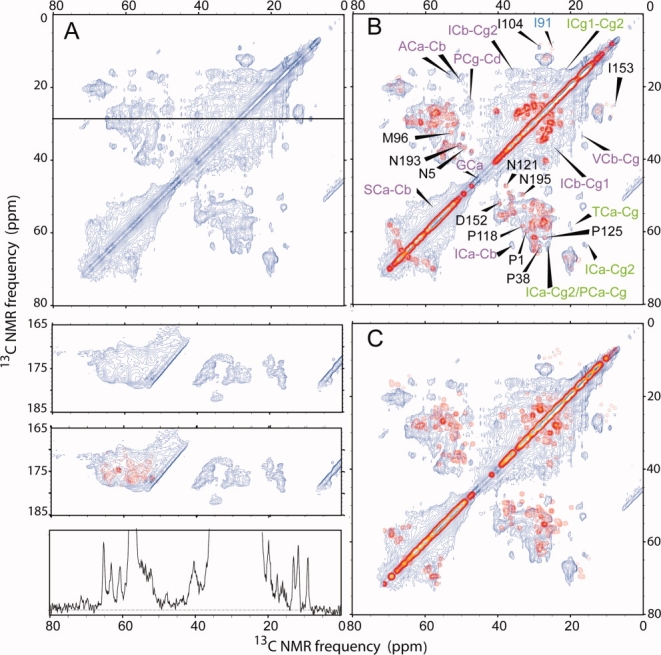
2D 13C—13C solid state NMR correlation spectrum of 1,3-13C2-glycerol-labeled tubular CA assemblies. (A) Experimental spectrum, acquired as described in the text. Gaussian line broadening of 30 Hz (0.20 ppm) and 50 Hz (0.33 ppm) was applied to the horizontal and vertical dimension, respectively. The 1D slice below the 2D spectrum passes through the Cγ1/Cδ cross peak of a single isoleucine at 29.73 ppm, as in Fig. 2. Spectral resolution is improved significantly by partial 13C-labeling. (B) Simulated 2D spectrum in red, generated from chemical shifts observed in solution NMR spectra of NTD and CTD, overlaid on the experimental spectrum. The simulated spectrum assumes an ideal 1,3-13C2-glycerol labeling pattern and includes only one-bond crosspeaks. Additional crosspeaks in the experimental spectrum due to two-bond polarization transfers and nonideal labeling are indicated by green and purple labels, respectively. Well-resolved crosspeaks that can be assigned by comparison with solution NMR chemical shifts are indicated by black labels. The only simulated crosspeak that does not appear in the experimental spectrum is indicated in blue. (C) Typical example of a simulated spectrum generated with randomly chosen chemical shift values for the same residue in the Biological Magnetic Resonance Data Bank, overlaid on the experimental spectrum. [Color figure can be viewed in the online issue, which is available at http://www.interscience.wiley.com.]
Detailed chemical shift assignments are not possible from the 2D spectrum in Figure 3(A) alone, of course. However, a qualitative comparison with the chemical shifts of NTD and CTD determined by solution NMR23,27 can be performed. In Figure 3(B), a simulated 2D 13C—13C spectrum is overlaid with the experimental spectrum, assuming crosspeaks only between the solution NMR chemical shifts of directly bonded 13C sites and assuming the ideal residue-specific labeling patterns expected when 1,3-13C2-glycerol is the sole carbon source.59 Crosspeaks in the simulated 2D spectrum are assumed to have Gaussian shapes with 0.5 ppm linewidths. The resemblance between simulated and experimental crosspeak patterns is quite good in the regions of the 2D spectrum where signals overlap strongly. Site-specific assignments can be made for several crosspeaks [black lettering in Fig. 3(B)], based on agreement with solution NMR assignments. For these assigned crosspeaks, chemical shifts in the solid state NMR spectrum agree with the solution NMR chemical shifts to within a root-mean-squared difference of 0.57 ppm. These results demonstrate that the secondary and tertiary structures of CTD and NTD are largely retained in the full-length CA assemblies.
Certain signals in the experimental 2D spectrum are absent from the simulated spectra. These fall into two categories. Crosspeaks indicated by green lettering in Figure 3(B) are two-bond crosspeaks, due to two-bond 13C polarization transfers in the experiments that are not considered in the simulations. Crosspeaks indicated by magenta lettering in Figure 3(B) are attributable to signals from sites that should not be 13C-labeled, according to the ideal 1,3-13C2-glycerol labeling pattern.59 Comparison of the intensities of these crosspeaks in Figure 3 with the corresponding intensities in Figure 2 indicates that 13C enrichment of these sites is roughly 10–20%. Deviations from the ideal 1,3-13C2-glycerol labeling pattern may be due to the long period (12 h) of CA expression (see Materials and Methods), which we used to ensure a high yield of CA, or to contributions from unanticipated metabolic pathways during protein expression.
To test the sensitivity of the 2D 13C—13C crosspeak patterns to variations in protein structure, we also constructed simulated spectra in which chemical shift values for each amino acid in CA were randomly chosen from entries in the Biological Magnetic Resonance Data Bank (BMRB) for the same amino acid. Agreement with the experimental 2D 13C—13C spectrum was always substantially worse. One example is shown in Figure 3(C), where both the isolated crosspeaks and the regions of overlapping crosspeaks are obviously different in the simulated and experimental spectra. As a further test, Figure 4 shows simulated 2D spectra in which progressive deviations from ±0.25 ppm to ±1.5 ppm were added to the solution NMR chemical shifts of NTD and CTD. The deviations from the chemical shifts of the two domains were chosen randomly within the specified range as input for the spectral simulation. The Cα/Cβ and Cβ/Cγ crosspeaks of threonines and the Cγ1/Cδ crosspeaks of isoleucines are most sensitive to the chemical shift variations. The particular pattern of threonine Cβ/Cγ crosspeaks observed in the experimental spectrum is washed out even with ±0.25 ppm deviations.
Figure 4.
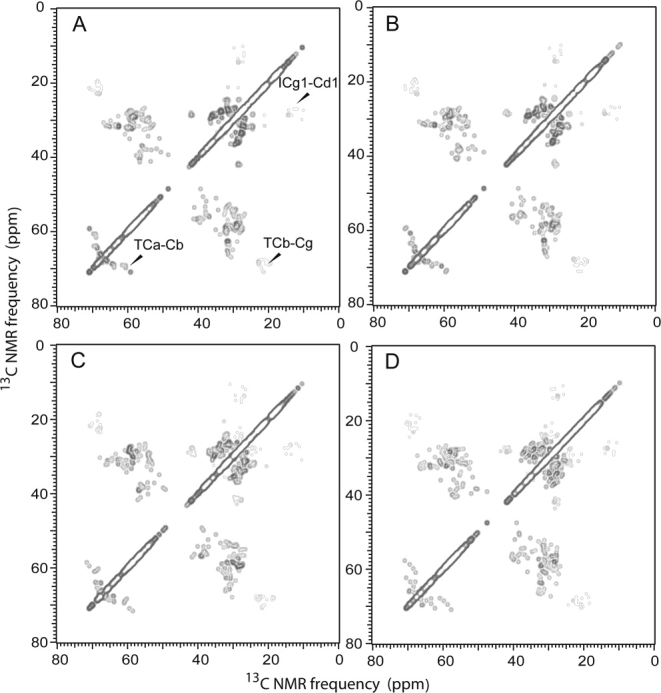
Simulated 2D 13C—13C solid state NMR spectra as in Fig. 3B, but with progressive deviations of chemical shifts from solution NMR values. Deviations for each carbon site were introduced as random values within ±0.25, ±0.50, ±1.00, and ±1.50 ppm ranges for panels A, B, C, and D, respectively.
Spectral simplification by double-quantum filtering
Partial labeling with 1,3-13C2-glycerol improves spectral resolution, as shown in Figure 3, but still does not permit direct chemical shift assignment. For certain residues (i.e., glycine, alanine, serine, cysteine, leucine, valine, and nonaromatic sites of phenylalanine and tyrosine), the 1,3-13C2-glycerol labeling pattern results in 13C nuclei that are separated by at least two bonds from all other 13C nuclei.59 Signals from such nuclei can be suppressed by double-quantum (DQ) filtering, using experimental conditions that excite DQ coherences only between directly bonded 13C pairs, thereby selecting signals that arise only from such 13C pairs. This approach should simplify the solid state NMR spectra considerably.
Figure 5 shows a 2D 13C—13C spectrum of partially labeled CA assemblies, obtained with a pulse sequence that prepares DQ coherences within directly bonded 13C pairs for evolution in the t1 period, converts the DQ coherences to single-spin 13C polarization, allows the 13C polarization to transfer to nearby 13C spins (∼5 Å maximum distance) during a spin diffusion period, and then excites single-quantum (SQ) signals during the t2 period. The spectrum therefore correlates the DQ frequencies of each directly bonded 13C pair (equal to the sum of their two SQ NMR frequencies) with the SQ frequencies within the pair and with SQ frequencies of nearby 13C nuclei. The rf pulse sequence is shown in Supporting Information (Fig. S2).
Figure 5.

2D DQ/SQ solid state 13C NMR spectrum of 1,3-13C2-glycerol-labeled tubular CA assemblies, acquired as described in the text. Gaussian line broadening of 75 Hz (0.50 ppm) and 90 Hz (0.60 ppm) was applied to the ω2 and ω1 dimension, respectively. Crosspeak assignments were determined by comparison with solution NMR chemical shifts for NTD and CTD. Additional assignments are shown in Supporting Information, Figure S3. Intraresidue crosspeaks are labeled in red (within 0.5 ppm of solution NMR shifts) and blue (within 1.0 ppm of solution NMR shifts). Sequential interresidue crosspeaks are labeled in green. 1D slices extracted at the indicated DQ 13C NMR frequencies are shown below the 2D spectrum, with DQ-correlated pairs of peaks marked with red dashed lines. [Color figure can be viewed in the online issue, which is available at http://www.interscience.wiley.com.]
By comparing DQ/SQ crosspeaks in Figure 5 with solution NMR chemical shifts for NTD and CTD,23,27 111 crosspeaks were found to have unique assignments (see Table S1 of Supporting Information). Deviations of SQ 13C NMR frequencies from the solution NMR chemical shift values were less than 0.5 ppm for 80 of these crosspeaks and less than 1.0 ppm for 16 crosspeaks. An additional 15 crosspeaks had deviations greater 1.0 ppm in the SQ dimension, but were assigned to specific sites either because they were uniquely correlated (through spin diffusion) with an assigned pair of DQ/SQ crosspeaks or because they were uniquely correlated (through DQ excitation) with a crosspeak with a smaller deviation and a unique assignment. For clarity, only a subset of these assignments is shown in Figure 5. Full assignments are shown in Supporting Information (Fig. S3). Assignments with green lettering in Figure 5 are sequential assignments, in which the DQ frequency of a 13C pair in one residue is correlated through spin diffusion with the SQ frequency of a 13C site in an adjacent residue. Such crosspeaks result from the spin diffusion period between the t1 and t2 periods in the rf pulse sequence. Signals from 42 different residues were assigned, including 27 residues in NTD and 15 residues in CTD. Most of these are in α-helical segments or within one residue of the end of an α-helix, but signals assigned to residues 5, 13, 90, 91, 95, 96, 108, 152, and 158 are also observed.
All assignments in Figure 5 can be considered tentative, in that they are based on comparisons with reported solution NMR chemical shifts. The high molecular weight of CA precludes direct sequential assignment by 2D and 3D solid state NMR techniques that have been applied to smaller proteins,43,59–63 for reasons of both resolution and sensitivity. Nonetheless, the good agreement between observed and expected crosspeak positions for a large number of crosspeaks indicates that the NTD and CTD structures studied by solution NMR are largely preserved in the CA assemblies.
Observation of mobile residues within CA assemblies
To investigate the possible presence of mobile residues or segments, we performed additional NMR measurements using experimental conditions under which NMR signals from immobile residues are suppressed. These conditions include 1H—13C polarization transfers driven by scalar (rather than dipole–dipole) couplings and low-power 1H decoupling during acquisition of 13C NMR signals, as commonly used in solution NMR studies. Two types of 2D 1H—13C measurements were performed with 13C detection, namely 2D INEPT64,65 and a variant of 2D INEPT with an additional period for 1H—1H polarization transfers through nuclear Overhauser effects after the t1 period and before scalar-driven 1H—13C polarization transfer (2D NOE-INEPT). Pulse sequences are shown in the Supporting Information (Fig. S2). Figure 6 shows the overlaid 2D INEPT (blue) and 2D NOE-INEPT (red) spectra. Compared with the 2D spectrum in Figure 3(A) (obtained under typical solid state NMR conditions), relatively few signals are observed in Figure 6. This indicates that relatively few carbon sites in the tubular CA assemblies have high mobility.
Figure 6.
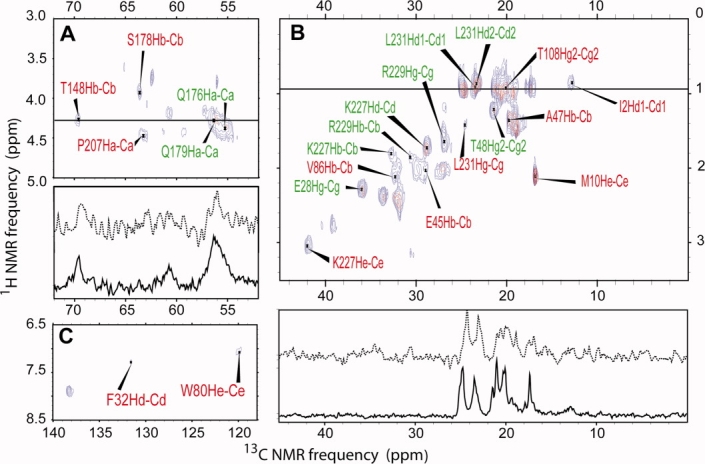
2D 1H—13C INEPT (blue contours) and NOE-INEPT (red contours) spectra of 1,3-13C2-glycerol-labeled tubular CA assemblies, acquired under conditions that select NMR signals from highly mobile sites as described in the text. Lorentzian line broadening of 10 Hz was applied in both dimensions. Crosspeak assignments were determined by comparison with solution NMR chemical shifts for NTD and CTD. Red and green assignments are explained in the text. Panels A, B, and C are different regions of the same 2D spectra. 1D slices from INEPT (solid line) and NOE-INEPT (dashed line) spectra are shown below panels A and B, and were taken at positions indicated by horizontal lines in the 2D spectra. [Color figure can be viewed in the online issue, which is available at http://www.interscience.wiley.com.]
Assignments in Figure 6 were made by comparison of observed crosspeak positions with solution 1H and 13C NMR chemical shifts.23,27 Using the criteria that (a) the 13C and 1H chemical shifts in Figure 6 must agree to within 0.5 ppm and 0.25 ppm, respectively, with the solution NMR values, (b) the assignments must correspond to residues that are not in α-helical segments (unless they are sidechain signals), and (c) the assigned signals in the 2D INEPT spectrum must not also be assigned in the 2D DQ/SQ spectrum, we found unique assignments for 13 2D INEPT crosspeaks. With the additional criterion that (d) the 13C and 1H chemical shifts must agree to within 1.0 ppm and 0.2 ppm, respectively, with random coil values,66 we found unique assignments for an additional 10 crosspeaks (see Table S2 of Supporting Information). This last criterion follows from our reasoning that highly mobile sites within CA assemblies should not have persistent conformational preferences and hence should exhibit chemical shifts close to the random coil values.
2D NOE-INEPT measurements were performed with the goal of observing interresidue crosspeaks that could be used for direct sequential assignment of the mobile residues. No such crosspeaks were observed, and signal intensities were generally lower than in the 2D INEPT measurements. Correlations with the water 1H signal at 4.6 ppm suggest that the aliphatic sidechains that contribute to the 2D NOE-INEPT spectrum are also exposed to solvent.
Figure 7 shows the locations of residues that are assigned in either the 2D DQ/SQ spectrum (blue residues) or the 2D INEPT spectrum (red or green residues, corresponding to signals assigned with criteria a–c or a–d, respectively). The N-terminal β-hairpin segment contains both immobile (residues 4 and 5) and mobile (residues 2 and 10) sites. This is not a paradoxical result, because the mobile sites are in sidechains (Cδ of Ile2 and Cɛ of Met10). Similarly, mobile sites in helix 1 (Cγ of Glu28), helix 3 (Cγ of Thr48), and helix 4 (Cɛ3 of Trp80) are in sidechains. Sites that are assigned in the 2D DQ/SQ spectrum occur in nearly every secondary structure element of CA.
Figure 7.
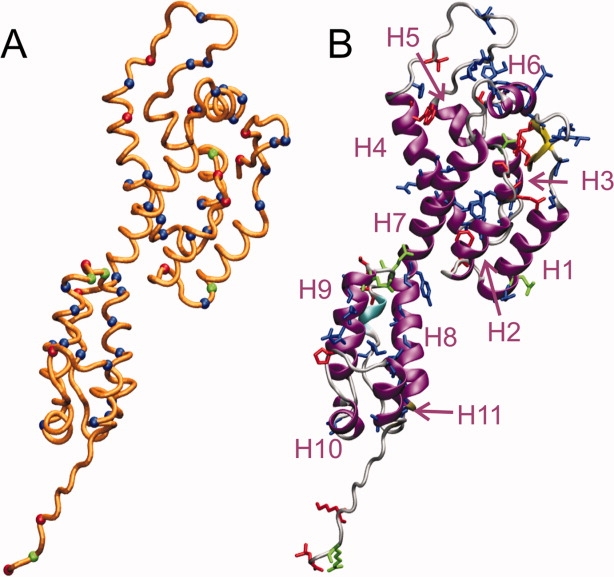
Positions of assigned NMR signals on HIV-1 CA, from spectra of tubular capsid assemblies in Figures 5 and 6. (A) Cα trace of HIV-1 CA, with Cα positions of residues with assigned signals in Figure 5 (i.e., immobile sites) shown in blue and Cα positions of residues with assigned signals in Figure 6 (i.e., highly mobile sites) shown in red or green. Red and green colors indicate different assignment criteria, as explained in the text and shown in Figure 6. (B) Ribbon representation of HIV-1 CA, with residues that have assigned signals shown in blue, red, or green as in panel A. α-helical segments H1–H11 are indicated. [Color figure can be viewed in the online issue, which is available at http://www.interscience.wiley.com.]
For Figure 7, atomic coordinates for full-length, wild-type HIV-1 CA were adapted from Protein Data Bank file 3GV2 by restoration of wild-type residues at positions 184 and 185, addition of residues 220–231 with MOLMOL software,67 prediction of coordinates for missing atoms with VMD software,68 and energy minimization within the TINKER/FFE program (available at http://dasher.wustl.edu/tinker/).
Optical birefringence of CA assemblies
The long tubular nature of CA assemblies suggests that, at high concentrations, these assemblies might form a liquid crystalline phase in which the CA tubes are locally aligned with one another. Such alignment may be useful in future solid state NMR studies that utilize techniques applicable to aligned samples, for example, as demonstrated by solid state NMR studies of filamentous bacteriophages.44 To test this possibility, pelleted CA assemblies were applied to a glass microscope slide, covered with a cover slip, and examined with an optical microscope between crossed polarizers. Figure 8 shows that domains of concentrated CA assemblies with diameters of several hundred microns are strongly birefringent, presumably due to alignment of CA tubes within these domains. It remains to be seen whether alignment of an entire NMR sample can be achieved, mechanically or by application of magnetic or electric fields.
Figure 8.

Optical birefringence of tubular CA assemblies. Assemblies were concentrated to roughly 500 mg/mL by centrifugation, then deposited on a glass microscope slide, covered with a glass cover slip, and viewed between crossed polarizers with an Olympus BX50 microscope equipped with a CCD camera. Images were recorded at 0° (A,B) and 45° (C,D) sample orientations relative to the polarizers. Camera exposure time was adjusted for uniform intensity (A,C) or kept constant (B,D). Pronounced differences in intensity distributions in panels B and D indicated the formation of birefringent domains of CA assemblies, presumably reflecting local alignment of tube axes.
Discussion
The main conclusions to be drawn from the solid state NMR data described above are that NTD and CTD of HIV-1 CA retain their individual structures in tubular CA assemblies, and that nearly the entire CA protein is immobilized in these assemblies. These conclusions follow from the close agreement of solid state 13C NMR chemical shifts in the assemblies with NMR chemical shifts for the individual domains in solution, from the observation that tentative solid state NMR chemical shift assignments based on the solution NMR data cover most of NTD and CTD, and that relatively few NMR signals are observed from the tubular CA assemblies when solution NMR conditions are used. These conclusions are consistent with the results of recent studies of planar CA assemblies by electron diffraction,4 recent crystallographic studies of CA hexamers,11 and earlier electron microscopy of CA tubes.3 The solid state NMR chemical shifts are sensitive to local structural details and local structural distortions. Thus, the solid state NMR data are complementary to electron diffraction and electron microscopy data, which have been interpreted with the assumption that NTD and CTD structures are undistorted.3,4 Solid state NMR measurements were performed on noncrystalline, self-assembled tubes of wild-type CA, and are therefore complementary to crystallographic studies that involve mutagenesis, crosslinking, or fusion protein constructs and that examine CA in a three-dimensional crystalline state.11
The relatively sharp 13C NMR lines in solid state NMR spectra (0.5–1.0 ppm linewidths) indicate that our CA assemblies are nearly structurally homogeneous at the molecular level, despite the variations in tube diameters and MPL discussed eariler. Similar linewidths are observed in solid state NMR spectra of certain amyloid fibrils under similar measurement conditions,69–72 for example, although sharper lines are observed in spectra of microcrystalline proteins.60,63 Significantly broader 13C NMR lines (>3 ppm) are typically observed in solid state NMR spectra of highly disordered proteins and peptides.48,73,74 It is worth noting that a tubular assembly cannot be perfectly structurally homogeneous, because curvature necessarily breaks the perfect translational and rotational symmetry of an ideal planar triangular lattice of CA hexamers. Curvature generally produces three sets of structurally inequivalent CA molecules (oriented differently relative to the long axis of the tube). This symmetry-breaking and variations in tube diameter probably contribute to the observed NMR linewidths. If solid state NMR measurements were performed on planar CA assemblies, as in the electron diffraction studies,4 sharper lines might be observed.
Previous measurements of nuclear spin relaxation parameters by solution NMR have provided information about molecular motions within CTD and NTD.22,26 Published solution NMR measurements involve backbone amide signals and are, therefore, confined to backbone motions, whereas our solid state NMR measurements involve both backbone and sidechain signals. Spin relaxation parameters in solution are also primarily sensitive to motions on the nanosecond or sub-nanosecond time scales (limited by the overall molecular tumbling time), whereas our solid state NMR measurements are sensitive to motions on time scales up to roughly 100 μs (i.e., the inverse of nuclear magnetic dipole–dipole coupling strengths). In addition, solution NMR measurements were performed on the isolated CTD and NTD in unassembled states, whereas solid state NMR measurements were performed on full-length CA in the assembled state. Therefore, exact correspondence between solution NMR data and solid state NMR results is not expected. Nonetheless, certain points of agreement can be noted. In the case of NTD, measurements by Campos-Olivas and Summers26 show below-average 1H—15N nuclear Overhauser effects and 15N T2 relaxation rates (i.e., enhanced internal backbone motion) in residues 86–97 and 141–151, and to a lesser extent in residues 1–10. These results may be consistent with our assignment of signals in 2D INEPT spectra to Cδ of Ile2, Cɛ of Met10, Cβ of Val86, and Cβ of Thr148. In the case of CTD, measurements by Alcaraz et al.22 show below-average 1H—15N nuclear Overhauser effects and 15N T2 relaxation rates at residues 153, 155, 159, 170, 191, 204–208, and 225. These results may be consistent with our assignment of signals in 2D INEPT spectra to Cα of Pro207 and to sidechain sites of Lys227, Arg229, and Leu231.
Finally, solid state NMR studies of CA assemblies may be extended in at least several directions: (1) Formation of closed capsid shells is believed to require both pentamers and hexamers of CA, although pentamers have not been directly observed in structural studies to date. It has been suggested that structural differences between CA hexamers and CA pentamers may be localized to intermolecular NTD-NTD and NTD-CTD interfaces, with minimal differences within the NTD and CTD structures.11 In principle, solid state NMR measurements on closed CA assemblies that contain a relatively large fraction of pentamers, such as relatively small spherical assemblies that have been reported for the R18L mutant of CA,4 could provide direct information regarding this issue; (2) although the recent crystal structures of hexameric CA provide high-resolution structural information about NTD-NTD interactions that stabilize the hexamers, high-resolution information about CTD–CTD interactions that are believed to drive the assembly of hexamers into two-dimensional sheets is not available because these interactions were deliberately disrupted by W184A and M185A substitutions.11 The resolution of the structural model of planar CA assemblies derived from electron diffraction data is not sufficient to reveal sidechain conformations and sidechain–sidechain interactions.4 Therefore, although crystal structures of dimeric CTD have been described,16,18 it has not been established unequivocally that the details of intermolecular interactions involving helix 9 in CTD observed in these crystal structures are retained in noncrystalline assemblies of full-length CA; (3) numerous questions remain regarding the self-assembly pathways for CA, both in vitro and within HIV-1. For example, does the formation of closed CA assemblies (conical or spherical) or long tubular assemblies proceed in vitro by the addition of CA dimers to a growing lattice of hexamers, by aggregation of small lattice fragments into larger lattices, or by some other mechanism? At what point does curvature develop? Do nonclosed structures form first, then anneal to closed structures? Solid state NMR measurements on freeze-trapped intermediates in the early stages of self-assembly may provide answers to these and other questions.
Materials and Methods
Protein expression and purification
Plasmids containing the gene for wild type HIV-1 CA were obtained from the laboratories of Profs. W. I. Sundquist and M.F. Summers. After amplification, the plasmids were transformed into Escherichia coli BL21(DE3) cells for expression. Competent cells were added into 50 mL 2× YT microbial medium with 100 μg/mL ampicillin, 34 μg/mL chloramphenicol, and 75 μg/mL streptomycin and grown at 37°C overnight to OD600 ∼5. Minimal medium (1.5 L) were prepared by modifying the method described earlier75–77: 50 mL 30× salt solution (1M Na2HPO4·7H2O, 1M KH2PO4, and 100 mM Na2SO4), 300 μL 5000× trace metal solution (100 mM FeCl3, 1M CaCl2, 500 mM MnCl2, 500 mM ZnSO4, 400 mM CoCl2, 400 mM CuCl2, 400 mM NiCl2, 400 mM Na2MoO4, 400 mM Na2SeO3, 400 mM H3BO3, 100 mM FeCl3, and 400 mM EDTA), 3 g glucose and 2.25 g NH4Cl, 1.5 mL 2M MgCl2, 1.5 mL 50 mg/mL thiamine, 20 μL 10 g/100 mL yeast extract. Three liters of this medium with unlabeled d-glucose, NH4Cl, and antibiotics were inoculated with the overnight culture to OD600 = 0.05. The cells were grown to OD600 ∼2 at 37°C with 250 RPM shaking and spun down by 17,700g centrifugation for 20 min. Cell pellets were immediately transported into warm fresh medium with labeled 13C6-d-glucose, 15NH4Cl, and antibiotics. Protein expression was initiated by addition of 1 mM IPTG after 20 min of growth in the fresh medium. The cells were harvested by centrifugation at 17,700g for 20 min after 3 h when OD600 reached ∼4. The pellets were stored at −80°C or purified immediately.
CA was purified with a procedure slightly modified from a published protocol.2 The harvested cells were resuspended in 350 mL lysis solution (17.5 mL 0.5M Tris, pH 8.0, 3.5 m. 5M NaCl, 0.175 mL β-mercaptoethanol (βME), 0.35 mL 0.1M protease inhibitor, and 7 mL 100 mg/mL lysozyme) on ice with stirring for 1 h. The solution was then sonicated (Branson Sonifier model 250) at duty cycle 40, amplitude 4 for 16 min as stirring on ice. The cell debris was spun down by centrifugation at 17,700g for 30 min. The protein was precipitated from the supernatant by addition of (NH4)2SO4 to a final concentration of 25% with stirring on ice for 45 min. The precipitate was collected by centrifugation at 17,700g for 1 h. Pellets were redissolved in 40 mL Sepharose buffer A (25 mM KMOPS at pH 6.9 with 5 mM βME). The protein solution was then dialyzed against 4 L Sepharose buffer A overnight, filtered, and loaded onto a SP Sepharose column. After washing with 2 columns of Sepharose buffer A, the protein was eluted at around 40 mL by running a linear gradient of Sepharose buffer B (25 mM KMOPS at pH 6.9 with 1M NaCl, and 5 mM βME) over 300 mL. The eluted protein solution was concentrated to 10 mL and loaded onto a Superdex 75 size exclusion column. CA was eluted at around 60 mL by washing the column with S75 buffer (10 mM Tris, pH = 8.0, with 50 mM NaCl and 5 mM βME).
The partially 13C-labeled CA sample was prepared in the same manner, but with 13C6-d-glucose replaced by 1,3-13C2-glycerol. The protein expression period in glycerol medium after inducing was extended to about 12 h to improve yield. The protein yield for glucose-labeled CA was about 27 mg/L, and for glycerol-labeled CA was about 30 mg/L.
CA protein assembly
Purified CA was concentrated to about 5 mg/mL by centrifugal filtration. Assembly was initiated by addition of NaCl to a final concentration of 1M at room temperature. After confirmation of tube formation by TEM images, the CA assemblies were concentrated by 436,000g centrifugation as gel-like dense pellets and packed into 3.2 mm diameter thin-walled MAS rotors for solid state NMR experiments. Each solid state NMR sample contained ∼0.6 μmol of CA.
Electron microscopy
To prepare the negatively stained samples, 5 μL of the CA assembly solution was adsorbed to glow-discharged carbon films on electron microscope grids (300 mesh copper with lacey carbon support) for 2 min, blotted with filter paper, rinsed twice with 5 μL of deionized water, blotted, stained for 30 s with 5 μL of 3% uranyl acetate, blotted again, and dried in air. Images were recorded with an FEI Morgagni TEM operating at 80 keV.
For MPL measurements, unstained samples were prepared by co-adsorbing 5 μL of the CA assembly solution with 1 μL of TMV solution (0.23 mg/mL) for 5 min, followed by blotting and drying in air. Carbon films for MPL measurements were ∼6 nm thick, estimated from the volume of evaporated carbon and the geometry of our carbon evaporation chamber, and were glow-discharged immediately before sample preparation. Dark field images were acquired and analyzed as previously described.52
Nuclear magnetic resonance
Solid state NMR experiments on both uniformly 15N,13C-labeled and 1,3-13C2-glycerol-labeled CA assemblies were performed with a Varian InfinityPlus spectrometer operating at 150.7 MHz 13C NMR frequency and a Varian triple-channel balun MAS NMR probe with 3.2 mm rotors. Sample heating by rf irradiation was significant, due to the 1M NaCl concentration in CA assembly samples, requiring that cooling gas at −80°C be supplied to the variable-temperature stack of the NMR probe to maintain sample temperatures in the 30–40°C range. The temperature of the sample was higher than that of the cooling gas both because the cooling gas warmed by ∼50°C while passing through the variable-temperature stack and because the rf irradiation heated the sample directly during measurements. Sample temperatures were determined from measurements of the 1H NMR frequency of water in the sample under the experimentally relevant rf power loads.78
For 2D 13C—13C spectra in Figures 2 and 3, two-pulse phase-modulated (TPPM) 1H decoupling79 was used in both the t1 period (128 points, 31.2 μs increment) and the t2 period (1024 points, 15 μs dwell for the uniformly 15N,13C-labeled sample; 1600 points, 15 μs dwell for the 1,3-13C2-glycerol-labeled sample). Continuous wave decoupling was used for the rest of the pulse sequence. The decoupling field strength was 96–98 kHz. Mixing periods were 1.846 ms, with finite-pulse radio-frequency-driven recoupling (fpRFDR)58 using 18 μs 13C π pulses. MAS frequencies were 13.00 kHz. Total experiment times were ∼6 days with a 2.0 s recycle delay for the uniformly 15N,13C-labeled sample and 5 days with a 2.7 recycle delay for the 1,3-13C2-glycerol-labeled sample.
For the 2D 15N—13C spectrum in Figure 2, 100 t1 points were acquired with a 100 μs increment. In the mixing period, 15N spin polarization was transferred to directly bonded 13Cα sites with a 3.8 ms cross-polarization step, followed by a 2.667 ms fpRFDR period for 13C—13C polarization transfers before the t2 period (512 points, 15 μs dwell). Proton decoupling conditions were the same as in 2D 13C—13C spectra. The MAS frequency was 12.00 kHz. The total experiment time was ∼7 days, with a 3.0 s recycle delay.
For the 2D DQ/SQ spectrum in Figure 5, four rotor periods of SPC5 recoupling57 were used to create DQ coherences. After the t1 period (180 points 20.8 μs increment), DQ coherences were converted to single-spin polarization by another SPC5 period, and a 200 ms radio-frequency-assisted spin diffusion (RAD) period was employed to permit intraresidue and interresidue 13C—13C polarization transfers before the t2 period (512 points, 15 μs dwell).80,81 TPPM decoupling fields of 152 kHz and 98 kHz were used during SPC5 and t2 periods, respectively, and continuous wave decoupling of 98 kHz was used for the rest of the sequence. The MAS frequency was 12.00 kHz. The total experiment time was ∼5 days, with a 2.0 s recycle delay.
2D 1H—13C INEPT and NOE-INEPT experiments in Figure 6 were performed with a Varian InfinityPlus spectrometer operating at 100.4 MHz 13C NMR frequency and a Varian T3 MAS NMR probe with 3.2 mm rotors. Following t1 evolution on the 1H channel (150 points, 66.7 μs increment), an optional 500 ms period for longitudinal 1H—1H spin polarization exchange was followed by polarization transfer to 13C spins by a refocused INEPT period (3.4 ms, determined empirically to maximize 13C NMR signals). During the t2 period (2048 points, 35 μs dwell), WALTZ-16 decoupling82 was applied with an 8 kHz 1H rf field. MAS at 5.00 kHz was used, primarily to reduce line broadening due to magnetic field inhomogeneity. The total experiment time was ∼9 days, with a 1.0 s recycle delay.
All NMR data were processed with NMRpipe,83 and plotted with Sparky software (available at http://www.cgl.ucsf.edu/home/sparky/).
Acknowledgments
We thank Prof. W. I. Sundquist and Prof. M. F. Summers for providing plasmids for HIV-1 CA protein expression. We also thank Prof. Sundquist and Prof. N. R. Krishna for providing solution NMR chemical shifts of NTD and CTD, respectively. We thank Dr. R. B. Wickner and Dr. Sorin Luca for help with transformation and protein expression.
References
- 1.Ganser-Pornillos BK, Yeager M, Sundquist WI. The structural biology of HIV assembly. Curr Opin Struct Biol. 2008;18:203–217. doi: 10.1016/j.sbi.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ganser-Pornillos BK, von Schwedler UK, Stray KM, Aiken C, Sundquist WI. Assembly properties of the human immunodeficiency virus type 1 CA protein. J Virol. 2004;78:2545–2552. doi: 10.1128/JVI.78.5.2545-2552.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li S, Hill CP, Sundquist WI, Finch JT. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature. 2000;407:409–413. doi: 10.1038/35030177. [DOI] [PubMed] [Google Scholar]
- 4.Ganser-Pornillos BK, Cheng A, Yeager M. Structure of full-length HIV-1 CA: a model for the mature capsid lattice. Cell. 2007;131:70–79. doi: 10.1016/j.cell.2007.08.018. [DOI] [PubMed] [Google Scholar]
- 5.Briggs JAG, Wilk T, Welker R, Krausslich HG, Fuller SD. Structural organization of authentic, mature HIV-1 virions and cores. EMBO J. 2003;22:1707–1715. doi: 10.1093/emboj/cdg143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benjamin J, Ganser-Pornillos BK, Tivol WF, Sundquist WI, Jensen GJ. Three-dimensional structure of HIV-1 virus-like particles by electron cryotomography. J Mol Biol. 2005;346:577–588. doi: 10.1016/j.jmb.2004.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ganser BK, Li S, Klishko VY, Finch JT, Sundquist WI. Assembly and analysis of conical models for the HIV-1 core. Science. 1999;283:80–83. doi: 10.1126/science.283.5398.80. [DOI] [PubMed] [Google Scholar]
- 8.Briggs JAG, Riches JD, Glass B, Bartonova V, Zanetti G, Krausslich HG. Structure and assembly of immature HIV. Proc Natl Acad Sci USA. 2009;106:11090–11095. doi: 10.1073/pnas.0903535106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wright ER, Schooler JB, Ding HJ, Kieffer C, Fillmore C, Sundquist WI, Jensen GJ. Electron cryotomography of immature HIV-1 virions reveals the structure of the CA and SP1 gag shells. EMBO J. 2007;26:2218–2226. doi: 10.1038/sj.emboj.7601664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grattinger M, Hohenberg H, Thomas D, Wilk T, Muller B, Krausslich HG. In vitro assembly properties of wild-type and cyclophilin-binding defective human immunodeficiency virus capsid proteins in the presence and absence of cyclophilin A. Virology. 1999;257:247–260. doi: 10.1006/viro.1999.9668. [DOI] [PubMed] [Google Scholar]
- 11.Pornillos O, Ganser-Pornillos BK, Kelly BN, Hua YZ, Whitby FG, Stout CD, Sundquist WI, Hill CP, Yeager M. X-ray structures of the hexameric building block of the HIV capsid. Cell. 2009;137:1282–1292. doi: 10.1016/j.cell.2009.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartonova V, Igonet S, Sticht J, Glass B, Habermann A, Vaney MC, Sehr P, Lewis J, Rey FA, Krausslich HG. Residues in the HIV-1 capsid assembly inhibitor binding site are essential for maintaining the assembly-competent quaternary structure of the capsid protein. J Biol Chem. 2008;283:32024–32033. doi: 10.1074/jbc.M804230200. [DOI] [PubMed] [Google Scholar]
- 13.Kelly BN, Kyere S, Kinde I, Tang C, Howard BR, Robinson H, Sundquist WI, Summers MF, Hill CP. Structure of the antiviral assembly inhibitor CAP-1 complex with the HIV-1 CA protein. J Mol Biol. 2007;373:355–366. doi: 10.1016/j.jmb.2007.07.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelly BN, Howard BR, Wang H, Robinson H, Sundquist WI, Hill CP. Implications for viral capsid assembly from crystal structures of HIV-1 gag(1-278) and CA(133-278) Biochemistry. 2006;45:11257–11266. doi: 10.1021/bi060927x. [DOI] [PubMed] [Google Scholar]
- 15.Ternois F, Sticht J, Duquerroy S, Krausslich HG, Rey FA. The HIV-1 capsid protein C-terminal domain in complex with a virus assembly inhibitor. Nat Struct Mol Biol. 2005;12:678–682. doi: 10.1038/nsmb967. [DOI] [PubMed] [Google Scholar]
- 16.Worthylake DK, Wang H, Yoo SH, Sundquist WI, Hill CP. Structures of the HIV-1 capsid protein dimerization domain at 2.6 angstrom resolution. Acta Crystallogr Sect D. 1999;55:85–92. doi: 10.1107/S0907444998007689. [DOI] [PubMed] [Google Scholar]
- 17.Berthet-Colominas C, Monaco S, Novelli A, Sibai G, Mallet F, Cusack S. Head-to-tail dimers and interdomain flexibility revealed by the crystal structure of HIV-1 capsid protein (p24) complexed with a monoclonal antibody Fab. EMBO J. 1999;18:1124–1136. doi: 10.1093/emboj/18.5.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gamble TR, Yoo SH, Vajdos FF, von Schwedler UK, Worthylake DK, Wang H, McCutcheon JP, Sundquist WI, Hill CP. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science. 1997;278:849–853. doi: 10.1126/science.278.5339.849. [DOI] [PubMed] [Google Scholar]
- 19.Momany C, Kovari LC, Prongay AJ, Keller W, Gitti RK, Lee BM, Gorbalenya AE, Tong L, McClure J, Ehrlich LS, Summers MF, Carter C, Rossmann MG. Crystal structure of dimeric HIV-1 capsid protein. Nat Struct Biol. 1996;3:763–770. doi: 10.1038/nsb0996-763. [DOI] [PubMed] [Google Scholar]
- 20.Gamble TR, Vajdos FF, Yoo SH, Worthylake DK, Houseweart M, Sundquist WI, Hill CP. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell. 1996;87:1285–1294. doi: 10.1016/s0092-8674(00)81823-1. [DOI] [PubMed] [Google Scholar]
- 21.Alcaraz LA, del Alamo M, Barrera FN, Mateu MG, Neira JL. Flexibility in HIV-1 assembly subunits: solution structure of the monomeric C-terminal domain of the capsid protein. Biophys J. 2007;93:1264–1276. doi: 10.1529/biophysj.106.101089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alcaraz LA, del Alamo M, Mateu MG, Neira JL. Structural mobility of the monomeric C-terminal domain of the HIV-1 capsid protein. FEBS J. 2008;275:3299–3311. doi: 10.1111/j.1742-4658.2008.06478.x. [DOI] [PubMed] [Google Scholar]
- 23.Wong HC, Shin R, Krishna NR. Solution structure of a double mutant of the carboxy-terminal dimerization domain of the HIV-1 capsid proteint. Biochemistry. 2008;47:2289–2297. doi: 10.1021/bi7022128. [DOI] [PubMed] [Google Scholar]
- 24.Tang C, Ndassa Y, Summers MF. Structure of the N-terminal 283-residue fragment of the immature HIV-1 gag polyprotein. Nat Struct Biol. 2002;9:537–543. doi: 10.1038/nsb806. [DOI] [PubMed] [Google Scholar]
- 25.Bosco DA, Eisenmesser EZ, Pochapsky S, Sundquist WI, Kern D. Catalysis of cis/trans isomerization in native HIV-1 capsid by human cyclophilin A. Proc Natl Acad Sci USA. 2002;99:5247–5252. doi: 10.1073/pnas.082100499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Campos-Olivas R, Summers MF. Backbone dynamics of the N-terminal domain of the HIV-1 capsid protein and comparison with the G94D mutant conferring cyclosporin resistance/dependence. Biochemistry. 1999;38:10262–10271. doi: 10.1021/bi990991x. [DOI] [PubMed] [Google Scholar]
- 27.Gitti RK, Lee BM, Walker J, Summers MF, Yoo S, Sundquist WI. Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science. 1996;273:231–235. doi: 10.1126/science.273.5272.231. [DOI] [PubMed] [Google Scholar]
- 28.Lidon-Moya MC, Barrera FN, Bueno M, Perez-Jimenez R, Sancho J, Mateu MG, Neira JL. An extensive thermodynamic characterization of the dimerization domain of the HIV-1 capsid protein. Prot Sci. 2005;14:2387–2404. doi: 10.1110/ps.041324305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.del Alamo M, Rivas G, Mateu MG. Effect of macromolecular crowding agents on human immunodeficiency virus type 1 capsid protein assembly in vitro. J Virol. 2005;79:14271–14281. doi: 10.1128/JVI.79.22.14271-14281.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.del Alamo M, Mateu MG. Electrostatic repulsion, compensatory mutations, and long-range non-additive effects at the dimerization interface of the HIV capsid protein. J Mol Biol. 2005;345:893–906. doi: 10.1016/j.jmb.2004.10.086. [DOI] [PubMed] [Google Scholar]
- 31.Kuznetsov YG, Victoria JG, Low A, Robinson WE, Fan H, McPherson A. Atomic force microscopy imaging of retroviruses: human immunodeficiency virus and murine leukemia. Scanning. 2004;26:209–216. doi: 10.1002/sca.4950260409. [DOI] [PubMed] [Google Scholar]
- 32.Douglas CC, Thomas D, Lanman J, Prevelige PE. Investigation of N-terminal domain charged residues on the assembly and stability of HIV-1 CA. Biochemistry. 2004;43:10435–10441. doi: 10.1021/bi049359g. [DOI] [PubMed] [Google Scholar]
- 33.del Alamo M, Neira JL, Mateu MG. Thermodynamic dissection of a low affinity protein-protein interface involved in human immunodeficiency virus assembly. J Biol Chem. 2003;278:27923–27929. doi: 10.1074/jbc.M304466200. [DOI] [PubMed] [Google Scholar]
- 34.BonHomme M, Wong S, Carter C, Scarlata S. The pH dependance if HIV-1 capsid assembly and its interaction with cyclophilin A. Biophys Chem. 2003;105:67–77. doi: 10.1016/s0301-4622(03)00063-2. [DOI] [PubMed] [Google Scholar]
- 35.Mateu MG. Conformational stability of dimeric and monomeric forms of the C-terminal domain of human immunodeficiency virus-1 capsid protein. J Mol Biol. 2002;318:519–531. doi: 10.1016/S0022-2836(02)00091-8. [DOI] [PubMed] [Google Scholar]
- 36.Ehrlich LS, Liu TB, Scarlata S, Chu B, Carter CA. HIV-1 capsid protein forms spherical (immature-like) and tubular (mature-like) particles in vitro: structure switching by pH-induced conformational changes. Biophys J. 2001;81:586–594. doi: 10.1016/S0006-3495(01)75725-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gross I, Hohenberg H, Krausslich HG. In vitro assembly properties of purified bacterially expressed capsid proteins of human immunodeficiency virus. Eur J Biochem. 1997;249:592–600. doi: 10.1111/j.1432-1033.1997.t01-1-00592.x. [DOI] [PubMed] [Google Scholar]
- 38.Misselwitz R, Hausdorf G, Welfle K, Hohne WE, Welfle H. Conformation and stability of recombinant HIV-1 capsid protein p24 (rp24) Biochim Biophys Acta. 1995;1250:9–18. doi: 10.1016/0167-4838(95)00041-r. [DOI] [PubMed] [Google Scholar]
- 39.Ehrlich LS, Agresta BE, Gelfand CA, Jentoft J, Carter CA. Spectral-analysis and tryptic susceptibility as probes of HIV-1 capsid protein structure. Virology. 1994;204:515–525. doi: 10.1006/viro.1994.1565. [DOI] [PubMed] [Google Scholar]
- 40.Ehrlich LS, Agresta BE, Carter CA. Assembly of recombinant human immunodeficiency virus type 1 capsid protein in vitro. J Virol. 1992;66:4874–4883. doi: 10.1128/jvi.66.8.4874-4883.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tycko R. Molecular structure of amyloid fibrils: insights from solid state NMR. Q Rev Biophys. 2006;39:1–55. doi: 10.1017/S0033583506004173. [DOI] [PubMed] [Google Scholar]
- 42.Paravastu AK, Leapman RD, Yau WM, Tycko R. Molecular structural basis for polymorphism in Alzheimer's β-amyloid fibrils. Proc Natl Acad Sci USA. 2008;105:18349–18354. doi: 10.1073/pnas.0806270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jehle S, van Rossum B, Stout JR, Noguchi SM, Falber K, Rehbein K, Oschkinat H, Klevit RE, Rajagopal P. Alpha B-crystalla hybrid solid state/solution state NMR investigation reveals structural aspects of the heterogeneous oligomer. J Mol Biol. 2009;385:1481–1497. doi: 10.1016/j.jmb.2008.10.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Opella SJ, Zeri AC, Park SH. Structure, dynamics, and assembly of filamentous bacteriophages by nuclear magnetic resonance spectroscopy. Ann Rev Phys Chem. 2008;59:635–657. doi: 10.1146/annurev.physchem.58.032806.104640. [DOI] [PubMed] [Google Scholar]
- 45.Lorieau JL, Day LA, McDermott AE. Conformational dynamics of an intact virus: order parameters for the coat protein of Pf1 bacteriophage. Proc Natl Acad Sci USA. 2008;105:10366–10371. doi: 10.1073/pnas.0800405105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu TY, Schaefer J. REDOR NMR characterization of DNA packaging in bacteriophage T4. J Mol Biol. 2008;382:1031–1042. doi: 10.1016/j.jmb.2008.07.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jaroniec CP, MacPhee CE, Bajaj VS, McMahon MT, Dobson CM, Griffin RG. High-resolution molecular structure of a peptide in an amyloid fibril determined by magic angle spinning NMR spectroscopy. Proc Natl Acad Sci USA. 2004;101:711–716. doi: 10.1073/pnas.0304849101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sharpe S, Kessler N, Anglister JA, Yau WM, Tycko R. Solid state NMR yields structural constraints on the V3 loop from HIV-1 gp120 bound to the 447-52d antibody Fv fragment. J Am Chem Soc. 2004;126:4979–4990. doi: 10.1021/ja0392162. [DOI] [PubMed] [Google Scholar]
- 49.Wasmer C, Lange A, Van Melckebeke H, Siemer AB, Riek R, Meier BH. Amyloid fibrils of the HET-s(218-289) prion form a β solenoid with a triangular hydrophobic core. Science. 2008;319:1523–1526. doi: 10.1126/science.1151839. [DOI] [PubMed] [Google Scholar]
- 50.Ndassa YM, Tang C, Shuja S, Summers MF. Structure determination of the HIV-1 matrix-capsid fusion protein. Biophys J. 2001;80:60A–60A. [Google Scholar]
- 51.Ivanov D, Tsodikov OV, Kasanov J, Ellenberger T, Wagner G, Collins T. Domain-swapped dimerization of the HIV-1 capsid C-terminal domain. Proc Natl Acad Sci USA. 2007;104:4353–4358. doi: 10.1073/pnas.0609477104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen B, Thurber KR, Shewmaker F, Wickner RB, Tycko R. Measurement of amyloid fibril mass-per-length by tilted-beam transmission electron microscopy. Proc Natl Acad Sci USA. 2009;106:14339–14344. doi: 10.1073/pnas.0907821106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dresselhaus MS, Dresselhaus G, Saito R. Physics of carbon nanotubes. Carbon. 1995;33:883–891. [Google Scholar]
- 54.Zeitler E, Bahr GF. Contributions to the quantitative interpretation of electron microscope pictures. Exp Cell Res. 1957;12:44–65. doi: 10.1016/0014-4827(57)90293-8. [DOI] [PubMed] [Google Scholar]
- 55.Namba K, Pattanayek R, Stubbs G. Visualization of protein-nucleic acid interactions in a virus: refined structure of intact tobacco mosaic virus at 2.9 Å resolution by x-ray fiber diffraction. J Mol Biol. 1989;208:307–325. doi: 10.1016/0022-2836(89)90391-4. [DOI] [PubMed] [Google Scholar]
- 56.Pines A, Gibby MG, Waugh JS. Proton-enhanced NMR of dilute spins in solids. J Chem Phys. 1973;59:569–590. [Google Scholar]
- 57.Hohwy M, Rienstra CM, Jaroniec CP, Griffin RG. Fivefold symmetric homonuclear dipolar recoupling in rotating solids: application to double quantum spectroscopy. J Chem Phys. 1999;110:7983–7992. [Google Scholar]
- 58.Ishii Y. 13C-13C dipolar recoupling under very fast magic angle spinning in solid state nuclear magnetic resonance: applications to distance measurements, spectral assignments, and high-throughput secondary structure determination. J Chem Phys. 2001;114:8473–8483. [Google Scholar]
- 59.Higman VA, Flinders J, Hiller M, Jehle S, Markovic S, Fiedler S, van Rossum BJ, Oschkinat H. Assigning large proteins in the solid state: a MAS NMR resonance assignment strategy using selectively and extensively 13C-labelled proteins. J Biomol NMR. 2009;44:245–260. doi: 10.1007/s10858-009-9338-7. [DOI] [PubMed] [Google Scholar]
- 60.Castellani F, van Rossum B, Diehl A, Schubert M, Rehbein K, Oschkinat H. Structure of a protein determined by solid state magic-angle-spinning NMR spectroscopy. Nature. 2002;420:98–102. doi: 10.1038/nature01070. [DOI] [PubMed] [Google Scholar]
- 61.Franks WT, Kloepper KD, Wylie BJ, Rienstra CM. Four-dimensional heteronuclear correlation experiments for chemical shift assignment of solid proteins. J Biomol NMR. 2007;39:107–131. doi: 10.1007/s10858-007-9179-1. [DOI] [PubMed] [Google Scholar]
- 62.Siemer AB, Ritter C, Steinmetz MO, Ernst M, Riek R, Meier BH. 13C,15N resonance assignment of parts of the HET-s prion protein in its amyloid form. J Biomol NMR. 2006;34:75–87. doi: 10.1007/s10858-005-5582-7. [DOI] [PubMed] [Google Scholar]
- 63.Igumenova TI, Wand AJ, McDermott AE. Assignment of the backbone resonances for microcrystalline ubiquitin. J Am Chem Soc. 2004;126:5323–5331. doi: 10.1021/ja030546w. [DOI] [PubMed] [Google Scholar]
- 64.Morris GA, Freeman R. Enhancement of nuclear magnetic-resonance signals by polarization transfer. J Am Chem Soc. 1979;101:760–762. [Google Scholar]
- 65.Burum DP, Ernst RR. Net polarization transfer via a J-ordered state for signal enhancement of low-sensitivity nuclei. J Magn Reson. 1980;39:163–168. [Google Scholar]
- 66.Wishart DS, Bigam CG, Holm A, Hodges RS, Sykes BD. 1H,13C, and 15N random coil NMR chemical-shifts of the common amino acids. 1. Investigations of nearest-neighbor effects. J Biomol NMR. 1995;5:67–81. doi: 10.1007/BF00227471. [DOI] [PubMed] [Google Scholar]
- 67.Koradi R, Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graphics. 1996;14:51–56. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 68.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 69.Heise H, Celej MS, Becker S, Riede D, Pelah A, Kumar A, Jovin TM, Baldus M. Solid state NMR reveals structural differences between fibrils of wild-type and disease-related A53T mutant α-synuclein. J Mol Biol. 2008;380:444–450. doi: 10.1016/j.jmb.2008.05.026. [DOI] [PubMed] [Google Scholar]
- 70.Luca S, Yau WM, Leapman R, Tycko R. Peptide conformation and supramolecular organization in amylin fibrils: constraints from solid state NMR. Biochemistry. 2007;46:13505–13522. doi: 10.1021/bi701427q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kloepper KD, Zhou DH, Li Y, Winter KA, George JM, Rienstra CM. Temperature-dependent sensitivity enhancement of solid state NMR spectra of α-synuclein fibrils. J Biomol NMR. 2007;39:197–211. doi: 10.1007/s10858-007-9189-z. [DOI] [PubMed] [Google Scholar]
- 72.Petkova AT, Buntkowsky G, Dyda F, Leapman RD, Yau WM, Tycko R. Solid state NMR reveals a pH-dependent antiparallel β-sheet registry in fibrils formed by a β-amyloid peptide. J Mol Biol. 2004;335:247–260. doi: 10.1016/j.jmb.2003.10.044. [DOI] [PubMed] [Google Scholar]
- 73.Hu KN, Havlin RH, Yau WM, Tycko R. Quantitative determination of site-specific conformational distributions in an unfolded protein by solid state nuclear magnetic resonance. J Mol Biol. 2009;392:1055–1073. doi: 10.1016/j.jmb.2009.07.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Havlin RH, Tycko R. Probing site-specific conformational distributions in protein folding with solid state NMR. Proc Natl Acad Sci USA. 2005;102:3284–3289. doi: 10.1073/pnas.0406130102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shewmaker F, Kryndushkin D, Chen B, Tycko R, Wickner RB. Two prion variants of Sup35p have in-register parallel β-sheet structures, independent of hydration. Biochemistry. 2009;48:5074–5082. doi: 10.1021/bi900345q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shewmaker F, Ross ED, Tycko R, Wickner RB. Amyloids of shuffled prion domains that form prions have a parallel in-register β-sheet structure. Biochemistry. 2008;47:4000–4007. doi: 10.1021/bi7024589. [DOI] [PubMed] [Google Scholar]
- 77.Shewmaker F, Wickner RB, Tycko R. Amyloid of the prion domain of Sup35p has an in-register parallel β-sheet structure. Proc Natl Acad Sci USA. 2006;103:19754–19759. doi: 10.1073/pnas.0609638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dvinskikh SV, Castro V, Sandstrom D. Heating caused by radiofrequency irradiation and sample rotation in 13C magic angle spinning NMR studies of lipid membranes. Magn Reson Chem. 2004;42:875–881. doi: 10.1002/mrc.1477. [DOI] [PubMed] [Google Scholar]
- 79.Bennett AE, Rienstra CM, Auger M, Lakshmi KV, Griffin RG. Heteronuclear decoupling in rotating solids. J Chem Phys. 1995;103:6951–6958. [Google Scholar]
- 80.Morcombe CR, Gaponenko V, Byrd RA, Zilm KW. Diluting abundant spins by isotope edited radio frequency field assisted diffusion. J Am Chem Soc. 2004;126:7196–7197. doi: 10.1021/ja047919t. [DOI] [PubMed] [Google Scholar]
- 81.Takegoshi K, Nakamura S, Terao T. 13C-1H dipolar-assisted rotational resonance in magic-angle-spinning NMR. Chem Phys Lett. 2001;344:631–637. [Google Scholar]
- 82.Shaka AJ, Keeler J, Frenkiel T, Freeman R. An improved sequence for broad-band decoupling: WALTZ-16. J Magn Reson. 1983;52:335–338. [Google Scholar]
- 83.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRpipe: a multidimensional spectral processing system based on Unix pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]


