Abstract
Unlike most proteins, which are in equilibrium with partially and globally unfolded conformations, kinetically stable proteins (KSPs) are trapped in their native conformations and are often resistant to harsh environment. Based on a previous correlation between kinetic stability (KS) and a protein's resistance to sodium dodecyl sulfate (SDS), we show here a simple method to identify KSPs by SDS-capillary electrophoresis (CE). Control non-KSPs were fully denatured by SDS and formed protein:SDS complexes that exhibited similar mobility in CE. In contrast, KSPs bound fewer SDS molecules, and showed a very different migration time and peak pattern in CE, thereby providing some insight about the structural heterogeneity of SDS:protein complexes and the relative KS of the various proteins.
Keywords: kinetically stable proteins, stability, sodium dodecyl sulfate, hyper-stable proteins, SDS-resistance
Introduction
The thermodynamic equilibrium stability of a protein is determined by the free energy difference between the native and the unfolded states. In contrast, kinetic stability (KS) is determined by the activation energy for unfolding, which is the free energy difference between the native state and the transition state for unfolding. Therefore, the KS of a protein is proportional to (and often represented by) its unfolding rate, and those proteins that unfold slowly due to a high-energy barrier are defined as kinetically stable proteins (KSPs). As KSPs are mostly trapped in their native conformation, they are able to remain active for longer periods of time than would be expected for typical proteins, even under harsh environments (e.g., high temperature, extreme pH, and the presence of proteases).1,2 Another advantage of KS is that it protects the structure of proteins that are prone to misfolding and aggregation,2 a phenomena that is closely linked to various etiologically distinct diseases, such as Parkinson's disease, amyotrophic lateral sclerosis, and Huntington's disease. Transthyretin (TTR) is a well-known example of a disease-related protein that is protected against aggregation by its KS. TTR is a tetrameric protein with a high KS that is compromised by missense mutations, leading to the formation of a monomeric species that is the precursor of aggregates pathologically linked to the disease familial amyloid polyneuropathy.3
Although KS appears to play essential roles in biology and human health, the chemical and physical basis of KS remains poorly understood, probably because of the small number of KSPs known and the lack of simple methods to identify and characterize this property. The most common method to identify KSPs involves measuring the rate of protein unfolding at various concentrations of denaturant and then extrapolating to 0M denaturant. Manning and Colón observed a correlation between KS and the resistance of proteins to denaturation by sodium dodecyl sulfate (SDS), and this led to a simple assay for probing the KS of proteins.4 Recently, Xia et al. extended this method to a diagonal 2D (D2D) SDS-PAGE assay to identify KSPs in complex mixtures of proteins, such as cell lysates.5
Here, we show a simple method to qualitatively identify KSPs by capillary zone electrophoresis (CE). In contrast to SDS-PAGE and capillary gel electrophoresis, in which the mobility of proteins depends on both their charge-to-mass ratio and their size, in CE the separation of proteins is determined only by their charge-to-mass ratio. This is particularly convenient for identifying KSPs because all non-KSP should have similar elusion times regardless of their native size or oligomeric structure, whereas it is expected that KSPs will bind less SDS and thereby exhibit very different mobility. CE is also a powerful tool for protein separation because of its high efficiency, fast analysis time and low requirement for sample consumption.6 Furthermore, CE is useful for probing the interaction between biomolecules7 and the interaction between proteins and SDS. Thus, we applied CE to study four SDS-denatured non-KSPs as a control experiment, and the migration times for each protein were similar, consistent with the similar charge-to-mass ratio of SDS-denatured proteins. However, the four KSPs tested had a significantly different migration time, consistent with the diminished binding of SDS due to their KS. Furthermore, the shape and broadness of the peaks revealed some insight about the SDS:protein complexes and the relative KS of the various proteins.
Results and Discussion
Because of the known correlation between SDS-resistance and KS,5,6 the SDS-PAGE boil–unboil assay was carried out to confirm the KS or lack thereof of the eight control proteins chosen for this study. As expected, the non-KSPs α-chymotrypsin (CHT), glucose dehydrogenase (GD), concanavalin A (ConA), and myoglobin (MYO) were denatured by SDS even when the samples were not heated, resulting in identical migration on the gel as the respective samples that were boiled [Fig. 1(A)]. As proteins that are denatured by SDS bind one molecule of SDS per two amino acids,8 the similar migration of the boiled and unboiled non-KSPs indicate similar extent of denaturation and SDS binding in both cases. In contrast, the KSPs glucose oxidase (GO), streptavidin (SVD), superoxide dismutase (SOD), and subtilisin carlsberg (SCA) were resistant to SDS when the samples were not boiled. The decreased SDS binding led to slower migration on the gel [Fig. 1(B)]. Thus, analysis of these proteins by CE, which is based on the same electrophoretic principle as SDS-PAGE, might yield similar results and establish CE as a simple method to probe the KS of proteins.
Figure 1.
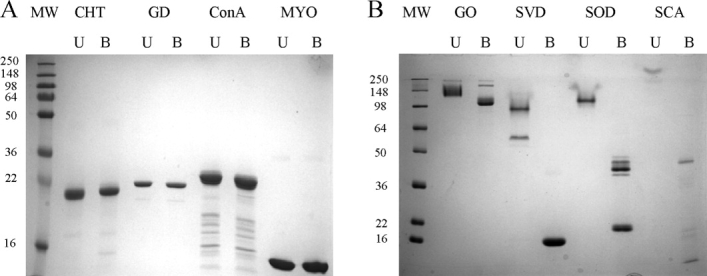
SDS-PAGE of (A) non-KSP and (B) KSP samples that were unboiled (U) or boiled (B) in 1% SDS. The non-KSPs α-chymotrypsin (CHT), glucose dehydrogenase (GD), concanavalin A (ConA), and myoglobin (MYO) showed no migration difference before and after boiling. The unboiled KSPs glucose oxidase (GO), streptavidin (SVD), superoxide dismutase (SOD), and subtilisin carlsberg (SCA) migrated less than their boiled counterparts. None of the samples contained reducing agent to avoid disrupting the native disulfide bonds of GO and SOD, as this might compromise their KS. Therefore, boiled GO and SOD migrated as a dimer due to non-native intermolecular disulfide bonds.
Figure 2 shows the CE data for the four non-KSPs and four KSPs analyzed by SDS-PAGE (Fig. 1). Consistent with the similar charge-to-mass ratios of the boiled and unboiled samples of non-KSPs, their migration times ranged from 13.4–14.2 min under the instrumental settings of this study [Fig. 2(A–D)]. Since KSPs bind few SDS molecules, it was anticipated that the CE migration time would be faster for the KSPs. Indeed, when incubated in SDS for 10 min at room temperature, each KSP exhibited a major peak(s) with a migration time of 6–7 min [Fig. 2(E–H)]. As expected, when the KSP samples were boiled in SDS buffer for 10 min before CE analysis, there was complete disappearance of the faster peaks and an increase in peaks with a migration time (14–15 min) similar to that of the non-KSPs. Thus, CE can separate proteins that are resistant and susceptible to SDS-induced denaturation with excellent resolution.
Figure 2.
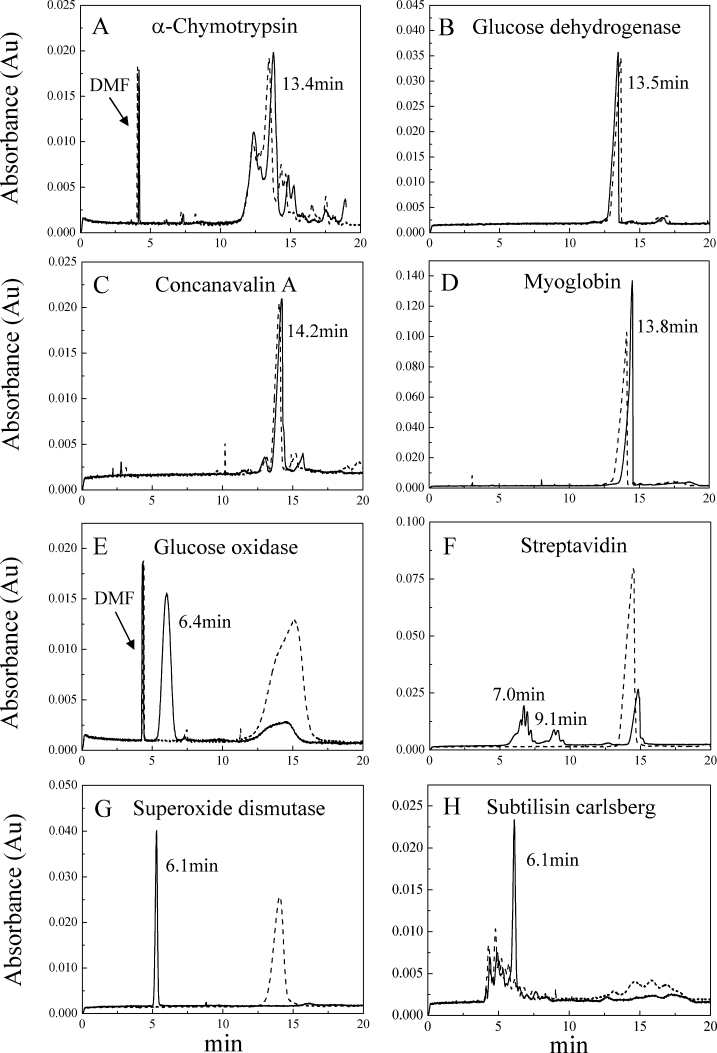
Electropherograms showing the migration of unboiled and boiled samples of non-KSPs (A–D) and KSPs (E–H). Black solid lines and dash lines represent the data of samples incubated in SDS that were not boiled or boiled, respectively. Samples were incubated in 20 mM sodium phosphate buffer (pH 7.4) containing 1% (w/v) SDS for 10 min. The electropherograms of unboiled and boiled non-KSPs showed little difference, but unboiled KSPs had significantly faster migration.
The faster migration of the SDS-resistant proteins is consistent with the conditions of the CE experiment. We used a fused silica capillary to separate the proteins. At our experimental pH of 7.4, the silanol groups of the interior wall of the capillary are ionized and negatively charged. Therefore, the positively charged cations of the buffer solution will interact with the silanoate groups and form a mobile cation layer. When normal polarity is applied with the anode (+) at the sample inlet and the cathode (−) at the sample outlet, the mobile cation layer is pulled in the direction of the negatively charged cathode. Because these cations are solvated, the bulk buffer solution migrates with the mobile layer, producing the electro-osmotic flow (EOF). Both non-KSPs and KSPs move towards the cathode due to the EOF. However, the protein:SDS complexes of the denatured proteins are highly negatively charged and experience more repulsion from the cathode (outlet), resulting in slower migration than the relatively SDS-free unboiled samples of KSPs.
An interesting feature of the data in Figure 2 is that in addition to the main peak(s) observed at 6–7 min for the unboiled KSP samples, GO and SVD exhibited one or more smaller peaks in the 14–15 min region. This suggests that the samples might have contained some fraction of denatured or misfolded protein, or alternatively, it may be related to the kinetics of SDS-induced denaturation. Since SDS appears to denature proteins irreversibly by interacting with them during their transient periods of partial and global unfolding, proteins that have lower KS and are marginally resistant to SDS will be eventually denatured by SDS at longer incubation times. In our case, we chose 10 min as a convenient incubation time and did not see significant changes in the data compared to hours of incubation. However, by varying the incubation times with SDS, it might be possible to measure the kinetics of SDS-induced denaturation for some proteins, and thereby obtain quantitative information about their KS. SOD exhibits a single sharp peak at about 6 min [Fig. 2(G)] consistent with a homogenous native state conformation of high KS. Therefore, when comparing the data in Figure 2 it is reasonable to suggest that SOD could have a higher KS than the other KSPs. Time-depend studies might allow a quantitative comparison of the KS of these proteins.
Another interesting observation is the presence of broader and/or multiple peaks present in the unboiled samples of the KSPs, SVD, and SCA. This peak heterogeneity may be a useful feature for identifying the presence of different population of species under native conditions. Unboiled SVD exhibited two broad faster peaks (7 and 9 min) that suggest the presence of multiple SVD species with different sensitivity to SDS. The existence of more than one type of protein:SDS species was also found by Gudiksen K. L. et al.,9 who observed a mixture of protein:SDS complexes with similar migration times that resulted in broadening of the peak. They suggested that the various peaks result from the different number of SDS molecules binding to various protein conformations. Because of the high sensitivity of CE to species with different charge-to-mass ratio, this method could be particularly useful for probing the conformational heterogeneity of the native state of KSPs. Thus, the CE method described could be convenient for probing the conformational heterogeneity of proteins that might exist in more than one conformation that differ in their sensitivity to SDS.
In conclusion, these results demonstrate that CE is a quick method to identify KSP. Regardless of their native size or oligomeric structure, all non-KSPs will have similar elusion times (e.g., 13–14 min in our study) in CE, whereas KSPs will have much different (6–9 min in our study) mobility because they bind less SDS. This provides a fast, convenient, and sample-efficient way to identify KSPs. Another advantage of using CE to identify KSPs is that the mobility, the number of peaks, and the sharpness of the peaks may reveal valuable and unique information about the conformational heterogeneity of KSPs and the extent of protein KS.
Materials and Methods
Chemicals and regents
All proteins and disodium phosphate were purchased from Sigma-Aldrich (St. Louis, MO) except for GD and SVD. GD was purchased from Fluka (Milwaukee, WI) and SVD was from Calbiochem (San Diego, CA). Monosodium phosphate was purchased from Mallinckrodt (Paris, KY).
SDS-PAGE assay
All electrophoresis protein samples were ∼0.25 mg/mL with 1% (w/v) SDS (∼35 mM) in 25 mM tris (hydroxymethyl) aminomethane (Tris) buffer (pH 6.8). Protein samples were unheated or boiled for 10 min before loading onto a 12% acrylamide gel. Running buffer contained 25 mM Tris base, 0.2M glycine, and 0.1% SDS. The gels were stained with Coomassie blue.
Capillary electrophoresis
All CE experiments were carried out using a Beckman PACE-MDQ system at 25°C. The fused silica capillary was 55.5 cm in length (45.0 cm to the detector window) with the inner diameter of 75 μm. The running buffer was 20 mM sodium phosphate buffer (pH 7.4) containing 1% (w/v) SDS. Some samples contained 0.65 mM dimethylformamide as a neutral marker. All proteins were dissolved in running buffer immediately before testing to reach the final concentration of 1 mg/mL and were filter with 0.2 μm filters (Nalgene, Rochester, NY). The boiled and unboiled samples were incubated in the same running buffer for 10 min before CE analysis. The separation voltage was 20 KV with normal polarity and samples were injected on the capillary at 1 psi for 5 s. Proteins were detected at 214 nm.
Glossary
Abbreviations:
- CE
capillary electrophoresis
- CHT
α-Chymotrypsin
- ConA
Concanavalin A
- D2D SDS-PAGE
diagonal 2-dimensional SDS-PAGE
- E. coli
Escherichia coli
- GD
Glucose dehydrogenase
- GO
Glucose oxidase
- KS
kinetic stability
- KSPs
kinetically stable proteins
- MYO
Myoglobin
- SDS
sodium dodecyl sulfate
- SVD
Streptavidin
- SOD
Superoxide dismutase
- SCA
Subtilisin Carlsberg
- TTR
transthyretin.
References
- 1.Cunningham EL, Jaswal SS, Sohl JL, Agard DA. Kinetic stability as a mechanism for protease longevity. Proc Natl Acad Sci USA. 1999;96:11008–11014. doi: 10.1073/pnas.96.20.11008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Plaza Del Pino IM, Ibarra-Molero B, Sanchez-Ruiz JM. Lower kinetic limit to protein thermal stability: a proposal regarding protein stability in vivo and its relation with misfolding diseases. Proteins. 2000;40:58–70. doi: 10.1002/(sici)1097-0134(20000701)40:1<58::aid-prot80>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 3.Kelly JW, Colón W, Lai Z, Lashuel HA, Mcculloch J, Mccutchen SL, Miroy GJ, Peterson SA. Transthyretin quaternary and tertiary structural changes facilitate misassembly into amyloid. Adv Protein Chem. 1997;50:161–181. doi: 10.1016/s0065-3233(08)60321-6. [DOI] [PubMed] [Google Scholar]
- 4.Manning M, Colón W. Structural basis of protein kinetic stability: resistance to sodium dodecyl sulfate suggests a central role for rigidity and a bias toward β-sheet structure. Biochemistry. 2004;43:11248–11254. doi: 10.1021/bi0491898. [DOI] [PubMed] [Google Scholar]
- 5.Xia K, Manning M, Hesham H, Lin Q, Bystroff C, Colón W. Identifying the subproteome of kinetically stable proteins via diagonal 2D SDS/PAGE. Proc Natl Acad Sci USA. 2007;104:17329–17334. doi: 10.1073/pnas.0705417104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fonslow BR, Yates JR., III Capillary electrophoresis applied to proteomic analysis. J Sep Sci. 2009;32:1175–1188. doi: 10.1002/jssc.200800592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schou C, Heegaard NH. Recent applications of affinity interactions in capillary electrophoresis. Electrophoresis. 2006;27:44–59. doi: 10.1002/elps.200500516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reynolds JA, Tanford C. The gross conformation of protein-sodium dodecyl sulfate complexes. J Biol Chem. 1970;245:5161–5165. [PubMed] [Google Scholar]
- 9.Gudiksen KL, Gitlin I, Whitesides GM. Differentiation of proteins based on characteristic patterns of association and denaturation in solutions of SDS. Proc Natl Acad Sci USA. 2006;103:7968–7972. doi: 10.1073/pnas.0602816103. [DOI] [PMC free article] [PubMed] [Google Scholar]


