Abstract
Transposable elements are segments of DNA with the unique ability to move about in the genome. This inherent feature can be exploited to harness these elements as gene vectors for diverse genome manipulations. Transposon-based genetic strategies have been established in vertebrate species over the last decade, and current progress in this field indicates that transposable elements will serve as indispensable tools in the genetic toolkit of vertebrate models. In particular, transposons can be applied as vectors for somatic and germline transgenesis, and as insertional mutagens in both loss-of-function and gain-of-function forward mutagenesis screens. The major advantage of using transposons as genetic tools is that they facilitate analysis of gene function in an easy, controlled and scalable manner. Transposon-based technologies are beginning to be exploited to link sequence information to gene functions in vertebrate models. In this article, we provide an overview of transposon-based methods used in vertebrate model organisms, and highlight the most important considerations concerning genetic applications of the transposon systems.
Keywords: Transposon, transgenesis, insertional mutagenesis, chromosome engineering
Introduction
Structural and functional components of the transposon and the transposition process
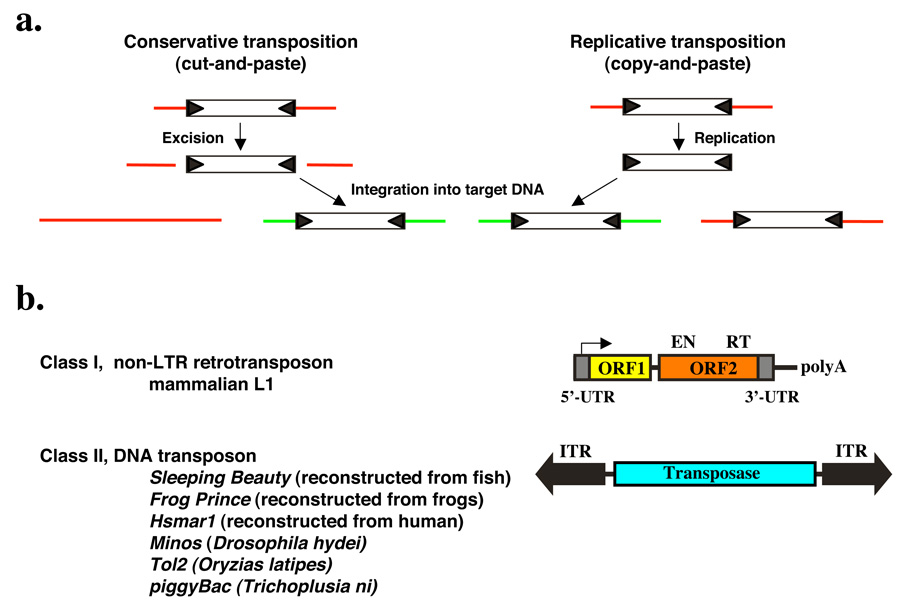
Transposable elements (TEs) are mobile, repetitive, genetic elements that are major components of genomes. Two classes of transposon are distinguished based on their respective transposition mechanisms. The mobility of Class I elements or retrotransposons is achieved through an RNA intermediate mediating a “copy-and-paste” mechanism, whereas Class II or DNA transposons use a DNA-mediated, “cut-and-paste” mode of transposition (Fig. 1a). TE-derived sequences make up about 45% of the human genome, of which retrotransposons form the major type of TEs, whereas DNA transposons contribute to 3% of the genome1. The most abundant transposons in mammals are non-LTR retrotransposons represented by the long interspersed nuclear elements (LINEs) and the short interspersed nuclear elements (SINEs). The major LINEs in humans and rodents (LINE1 or L1) are 6 kbp long and contain two ORFs (Fig. 1b). These encode a nucleic acid binding protein and an enzyme with endonuclease (EN) and reverse transcriptase (RT) activity, respectively2,3. EN generates a single-stranded nick in the target DNA, and RT uses the nicked DNA to prime reverse transcription from the 3'-end of the L1 RNA4,5.
Figure 1. Mechanism of transposition and general organization of Class I and Class II transposable elements.
(a) Schematic representation of the two major mechanisms of transposition. During conservative transposition, the element is excised from the donor DNA (red line), and integrates into a new target DNA (green line). Ligation of the broken ends of the DNA reconstitutes the donor locus. Replicative transposition involves amplification of the element by copying through transcription followed by reverse transcription. The newly made copy gets inserted elsewhere in the genome, but the donor element does not move. (b) Structures and organization of the main types of transposable elements. Class I non-LTR retrotransposon. The element consists of a 5’ untranslated region that has promoter activity (arrow) that drives transcription of the element-encoded genes. ORF1 encodes a nucleic acid binding protein. ORF2 encodes an endonuclease (EN) and a reverse transcriptase (RT). The element has a polyA tail. Class II DNA transposon. The central transposase gene is flanked by terminal inverted repeats (IRs, shown as black arrows). The IRs contain the binding sites for the transposase and sequences that are required for transposase-mediated cleavage.
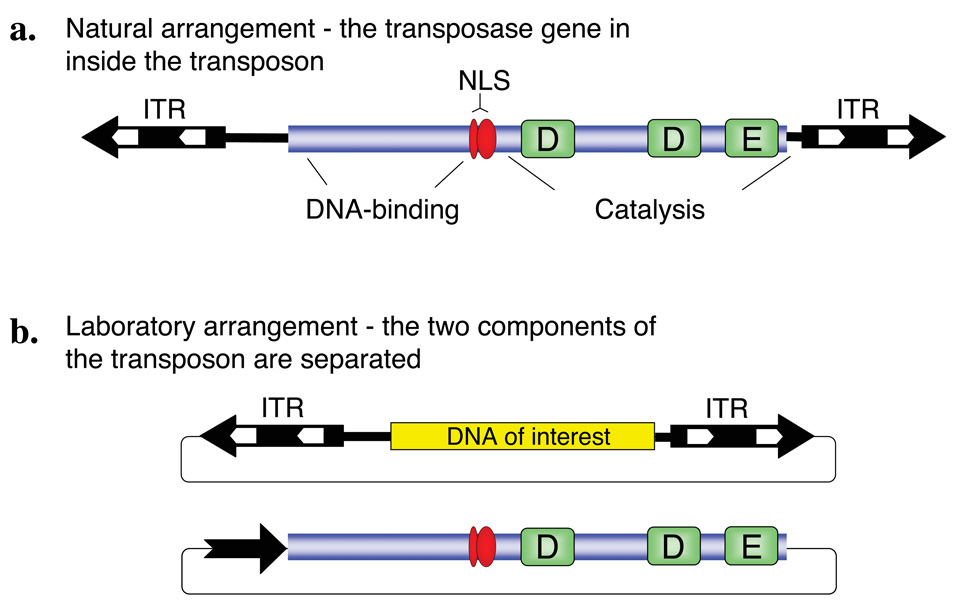
Class II TEs that move in the host genome via a “cut-and-paste” mechanism are simply organized; they encode a transposase protein in their simple genome flanked by inverted terminal repeats (ITRs) that carry transposase binding sites necessary for transposition (Fig. 1b and Fig. 2a). Transposition results in excision of the element from the DNA and subsequent integration into a new sequence environment. The transpositional process can easily be controlled by separating the transposase source from the transposable DNA harboring the ITRs, thereby creating a non-autonomous TE. In such a two-component system, the transposon can only move by trans-supplementing the transposase protein (Fig. 2). Most transposon-based experimental strategies that rely on mobilizing Class II elements utilize this two-component, binary approach, in which practically any sequence of interest can be positioned between the ITR elements according to experimental needs. Although one such two-component system was recently described for a mammalian retrotransposon6, most mammalian retrotransposon mutagenesis systems consist of single-component, autonomous elements.
Figure 2. Class II DNA transposon system.
(a) Structure of the transposon. The central transposase gene (blue box) is flanked by terminal inverted repeats (IR, black arrows) that contain binding sites for the transposase (white arrows). In case of Sleeping Beauty, the transposase consists of an N-terminal DNA-binding domain, a nuclear localization signal (NLS) and a catalytic domain characterized by the DDE signature. (b) Gene transfer vector system based on a Class II DNA transposon. The transposase coding region can be replaced by a gene of interest (yellow box) within the transposable element. In a typical two-component, binary gene transfer vector system, the transposon can be mobilized if a transposase source is provided in cells; for example, the transposase can be expressed from a separate plasmid vector.
General considerations for the application of transposons as DNA delivery tools
Transposons have been successfully used in invertebrate animal models, including C. elegans 7,8 and Drosophila 9–11 for transgenesis and insertional mutagenesis, but until the reactivation of the Sleeping Beauty (SB) transposon system in 199712,there was no indication of DNA-based transposons in vertebrates sufficiently active for these purposes. Later on, other elements have been shown to catalyze efficient transposition in vertebrate model organisms. For example, the insect TEs piggyBac and Minos have proven to be useful in germline mutagenesis of vertebrates13,14. Moreover, the reconstructed amphibian element Frog Prince 15, the reconstructed human Hsmar1 element16, and the Tol2 element isolated from the medaka fish17 have been found to be active in vertebrates. Finally, a synthetic L1 retrotransposon named ORFeus 18 was shown to undergo efficient transposition in both somatic and germline tissues in transgenic mice19.
The basic criteria for the applicability of a Class II TE in any given model organism are 1) a sufficient level of transpositional activity in the given species, and 2) a lack of endogenous copies in the targeted genome or other strategies that avoid mobilization of resident copies. As host specificity barriers cannot easily be challenged, transposon-based genetic technologies in all of the major model organisms were dependent on the discovery of TEs that are active in the species of interest. The use of TEs originated from distantly related species guarantee fulfillment of the second criterion, if they still show acceptable activity in the desired host. Other, more practical considerations for the design of a particular include cargo capacity of the TE, integration site preference and transposition to linked chromosomal sites (“local hopping”)20.
Capacity for cargo
For mutagenesis purposes, small transposable element vectors can be designed that retain two basic functions: the cis-requirements for transposition and a mutagenic feature designed to disrupt normal gene function. However, size does matter for transgenesis purposes, including the generation of germline-modified laboratory stocks of model species and species of biotechnological interest as well as human gene therapy applications. In these instances, transgene constructs including coding regions of genes with all the necessary transcriptional regulatory elements can exceed several kilobase pairs in size. Tolerance for cargo size varies greatly between TEs. Members of the Tc1/mariner family, including SB, are inhibited by increasing size21. A particular modification of the SB transposon in this respect was the generation of a “sandwich” transposon vector that has two complete SB elements flanking a transgene to be mobilized22. The sandwich SB vector enhanced transposition of large (>10 kb) transgene constructs, and therefore probably represents the vector of choice for transgene constructs that would otherwise transpose poorly due to their size. The piggyBac 13 and Tol2 23,24 transposons appear to be more tolerant to increasing size of cargo, allowing complex transgene designs to be incorporated within the transposon without sacrificing transposition efficiency.
Integration site preference
Integration site preference can greatly influence the utility of transposon vectors for different applications. For example, human gene therapy protocols would require application of transposon vectors showing the least preference to target genes, for obvious safety considerations. On the contrary, mutagenesis screens can capitalize on elements that exhibit a tendency to land in genes. The insertion pattern of most transposons is nonrandom, showing numerous “hotspots” and “cold regions” on a genome-wide scale. Common hotspots represent the main limitation to full genome coverage with individual TE-based vectors. Thus, in this respect, the utility of transposons for mutagenesis is greatly enhanced by the availability of multiple, alternative vector systems with distinct preferences for insertion.
The above considerations will be of paramount significance for insertional mutagenesis screens in vertebrate species. For example, on the level of primary DNA sequence, the Tol2 element does not appear to exhibit a pronounced preference for any sequence for insertion. Similar, L1 retrotransposons seem to have very little if any target preference beyond a preference for a very degenerate, AT-rich sequence25. In contrast, the piggyBac transposon targets the sequence TTAA, whereas all Tc1/mariner TEs, including SB, Frog Prince, Minos and Hsmar1, target their integration into TA dinucleotides. In the case of SB, this preference was studied in detail, and palindromic AT repeats were found to be preferred sites for integration26. However, computational analyses revealed that target selection is determined primarily on the level of DNA structure, not by specific base-pair interactions. It was shown that preferred target sites have a bendable structure and increased distance between the central base-pairs26,27. In the context of chromatin, SB has no preference for transcription units28, and most hits that occur in genes are localized in introns, since this is the largest components, on average, of transcription units. In contrast, piggyBac shows a greater propensity to integrate into transcription units29 with preference to insert around transcription start sites30. Taken together, the preferences of particular elements to integrate into expressed genes versus noncoding DNA, and preferences for integration sites within genes are expected to be substantially different.
Local hopping
“Local hopping” describes a phenomenon of chromosomal transposition in which transposons have a preference to land into cis-linked sites in the vicinity of the donor locus. Local hopping seems to be a shared feature of eukaryotic cut-and-paste transposons. However, the actual extent and the chromosomal window of local hopping can drastically vary among different TE types, and between different species or even different donor loci when using the same TE. In germline mutagenesis screens in mice using SB, 30–80% of the transposons has been observed to reinsert locally on either side of the transposon donor locus31–33. Even though the piggyBac transposon exhibits local hopping, it has a more random integration pattern than the SB element29. Local hopping can play a significant role in mutagenesis using chromosomally resident transposons. In practical terms, local hopping limits the chromosomal regions accessible to a transposon jumping out of a given chromosomal site. To circumvent this limitation, establishing numerous “launch pads” to initiate transposition out of different loci can be a viable strategy to increase coverage of gene mutations. On the other hand, local hopping can be useful for saturation mutagenesis within limited chromosomal regions for studies of QTLs or gene arrays34. The situation with local hopping is fundamentally different with retrotransposons, due to their life-cycle: in contrast to Class II elements that remain in the nucleus after excision, the genomic RNA molecule produced by retrotransposons needs to be transported to the cytoplasm for production of the protein factors that are required for transposition. Thus, there is no propensity of retrotransposons for local hopping.
Transgenesis
Classical methods to express foreign genes in vertebrates rely on microinjection of nucleic acids into oocytes or fertilized eggs. Two main drawbacks of these approaches are the low rates of genomic integration, and that the injected DNA generally integrates as a concatemer. Both drawbacks can be circumvented utilizing transposition-mediated gene delivery, as it can increase the efficiency of chromosomal integration and facilitates single-copy insertion events. Single units of expression cassettes are presumably less prone to transgene silencing than the concatemeric insertions created by classical methods. In case of transgenesis, a single-copy insertion away from endogenous genes is clearly desirable. The insertional spectrum of Tc1/mariner elements satisfies this need the best, as these elements integrate randomly at the genome level, and do not show pronounced bias for integration into genes. Another particular problem concerning transgenesis is that founders that develop from the injected oocytes or eggs are predominantly mosaic for the transgene, because integration generally occurs relatively late during embryonic development. Therefore, in order to potentiate successful transmission of the transgene through the germline to the next generation, it is necessary to shift the window of integration events as early as possible. The injection of in vitro synthesized mRNA as a transposase source can further enhance the efficiency of this technique due to the more rapid availability of the transposase, resulting in reduced transgene mosaicism in the embryo, and therefore elevated germline transmission rates. This can be facilitated by co-injection of engineered transposons with transposase mRNA. This method has been employed to generate transgenic zebrafish and Xenopus with Tol2 17,35 and SB36,37, transgenic chicken with Tol2 38 and transgenic mice with SB39–41 and piggyBac 13.
Transposon-mediated forward genetic approaches
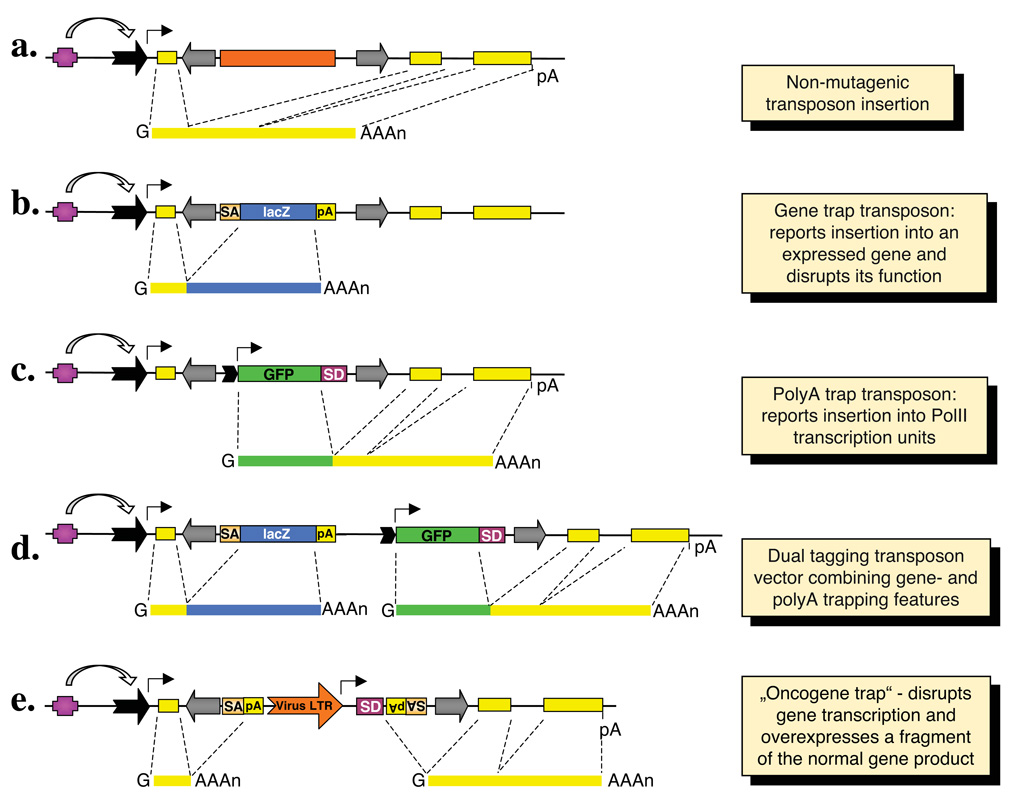
Due to distinct target site preferences, the number of target loci that can be mutagenized by transposon vectors could dramatically increase by combining different transposon systems in genome-wide screens. However, even if a transposon inserts into a gene, it may not have a mutagenic effect. For example, the conventional wisdom dictates that intronic insertions are likely spliced out together with the rest of the intron (Fig. 3a) without having an effect on the expression of a normal gene product. Various technologies have been established to enhance the mutagenicity as well as reporting capabilities of insertional vectors by “trapping” transcription units. Gene trapping is based on the activation of a promoterless reporter gene that is dependent on splicing between the exons of the trapped gene and a splice acceptor site carried by the transposon (Fig. 3b). Thus, gene trap vectors both report the insertion of the transposon into an expressed gene, and have a mutagenic effect by truncating the transcript through imposed splicing. More sophisticated vectors that contain a polyA trap cassette that reports insertion into a Pol II transcription unit have also been developed (Fig. 3c). Because polyA trap cassettes have their own promoters, they can report the insertion into genes irrespective of their expression status in a given cell type. Because polyA trap vectors are not designed to express the downstream exons of the targeted gene at the protein level, polyA trap insertions are unlikely to cause dominant mutagenic effects by themselves. Dual tagging systems that combine both gene trap- and polyA trap elements (Fig. 3d) have been used both in mouse42 and in Drosophila 43. Another possibility to manipulate a trapped transcription unit already proven to be useful is the targeted over- and/or misexpression system (Fig. 3e). By this method, one can overexpress the full-length or a truncated protein product (depending on the position of transposon insertion) of the targeted gene, thereby producing dominant phenotypes by overdosing the affected gene product. This feature has been exploited in genetic screens aiming at the identification of candidate proto-oncogenes in experimental animals44,45 (see below).
Figure 3. Summary of the basic gene trapping strategies.
On top, a hypothetical transcription unit is depicted with an upstream regulatory element (purple box), a promoter (black arrow), three exons (yellow boxes) and a polyadenylation signal (pA). Major classes of transposon-based trapping constructs and spliced transcripts are shown below. Transposon inverted repeats are indicated by gray arrows. (a) An intronic transposon insertion is typically not mutagenic, because the transposon is spliced out from the primary RNA transcript together with the targeted intron sequences. (b) Gene trapping cassettes contain a splice acceptor (SA) followed by a reporter gene and a pA. The reporter is only expressed when transcription starts from the promoter of an endogenous transcription unit. Thus, the expression of the reporter follows the expression pattern of the trapped gene. (c) Polyadenylation [poly(A)] traps contain a promoter followed by a reporter gene and a splice donor (SD) site, but they lack a pA signal. Therefore, reporter gene expression depends on splicing to downstream exon/s of a Pol II transcription unit containing a pA. (d) The “dual tagging” vectors are based on both gene- and poly(A) trapping of a targeted transcription unit. (e) The oncogene trap contains SA signals followed by pA signals in both orientations to disrupt transcription, as well as a strong, viral enhancer/promoter that drives transcription towards the outside of an inserted transposon, and thereby overexpresses a gene product.
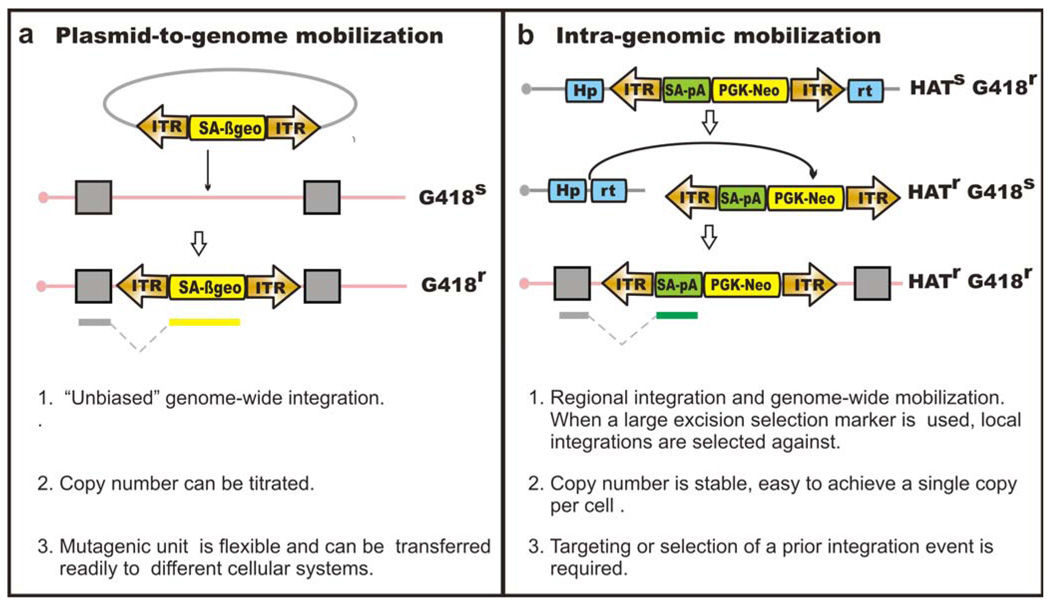
Another important consideration when fine-tuning the parameters of a mutagenesis screen is copy number of the insertional mutagen. Specifically, in any screen, the insertion site of the mutagen (the transposon) must be determined to correlate a phenotype with a genotype; for this purpose, a single mutation per cell is advantageous. In other applications, multiple copies of the mutagen are integral for the screen design. For example, in one application, multi-copy integration of the piggyBac transposon was used in combination with Cre/LoxP to generate deletions, duplications and translocations in vivo 46. The copy number of the mutagen for a specific application may affect the methods used. In cell culture systems ex vivo, transposon delivery can be achieved by transfection of plasmid DNA harboring the transposons or, alternatively, by mobilizing a single transposon, which has been placed in the genome by gene targeting or a prior transposition event. Transfection-based, “plasmid-to-genome” delivery (Fig. 4a) gives relatively unbiased genome-wide integrations; however, careful titration of the amount of the donor and transposase plasmid is required to provide the appropriate copy number per cell of the transposon. Intra-genomic, “genome-to-genome” mobilization (Fig. 4b) can be selected based on excision and re-integration33, enabling efficient genome-wide mutagenesis and tight control over the copy number.
Figure 4. Transposon delivery methods in ES cells.
Gene trap based loss-of-function mutagenesis is shown here as an example. (a) plasmid-to-genome mobilization. Cells with mutagenic transposon insertions can be selected in G418. (b) Intra-genomic mobilization. Upon transposase expression, the transposon will be excised from the donor site and re-integrate at a different genomic location. Enrichment of such cells can be achieved by selecting for transposition excision and reintegration. Using Hprt as an excision selection marker, cells with the transposon excised from the donor site will be Hprt-proficient and therefore HAT-resistant.
Mutagenesis ex vivo: mutagenesis in stem cells
Ex vivo mutagenesis complements in vivo screening for the purposes of gene function discovery at a cellular level. Pluripotent mouse embryonic stem (ES) cells are attractive models for in vitro mutagenesis, as they possess several features not available with other cell types, including their amenability to sophisticated genetic manipulation of their genome, contribution to mouse germ lines, the ability to differentiate into many cell types and the stability of their diploid genome. Gene targeting technology has been very successful in generating altered alleles of specific genes, allowing individual gene function to be dissected at cellular and whole-animal levels. However, this gene-by-gene based approach does not facilitate gene discoveries related to a particular pathway of interest on a genome-wide scale. International consortia are involved in the generation of ES cells with a mutation in each and every mouse gene by a combination of gene trapping and gene targeting47. Once complete, this mutant ES cell resource will be very powerful in addressing genome functions in both hypothesis-driven and non-hypothesis driven manners. The parallel generation of thousands of mutations is an important alternative technology, which allows phenotype-based screens to be conducted for non-hypothesis based discovery of gene functions. Although chemical-based mutagenesis can achieve such a goal, the identification of causal mutations is difficult48. Genome-wide insertional mutagenesis provides a powerful and high-throughput means to ascribe functions to genes associated with particular biological pathways.
Recessive genetic screens in ES cells
Insertional mutagenesis in mammalian cells is challenged by the diploid genome. Both copies of a gene are nearly always required to be inactivated to evoke a phenotypic change. The probability of generating bi-allelic mutations of a single locus by two independent “hits” is extremely low. However, single allele mutations can undergo loss-of-heterozygosity (LOH), and in some cases a high dose of G418 can be used to select homozygote mutants if a neomycin resistant cassette is included in the insertional mutagen49. However, the rate of such an event occurring is approximately 10−5 per cell per generation50. Additionally, genome context affects the expression of the neomycin resistance gene; therefore, the optimal dose for homozygote selection is locus-specific, which makes the selection of genome-wide homozygote mutants from a pool of heterozygote mutants impractical.
An alternative system has been developed in ES cells that combines a Blm-deficient genetic background with insertional mutagenesis. Blm-deficiency promotes the conversion of cells that bear mutations generated by the insertional mutagen as single alleles into bi-allelic mutations (Fig. 5a). Blm-deficient ES cells display a high rate of homologous recombination between homologous chromosomes, and the conversion rate from a heterozygous to a homozygous state (LOH rate) is elevated more than tenfold compared to wild-type cells50,51. A typical recessive genetic screen using Blm-deficient ES cells (Fig. 5b) includes four parts: 1. genome-wide mutagenesis and selection of mutants with the insertional mutagen; 2. mutant pool propagation to provide sufficient generations for homozygote conversion; 3. phenotype-driven screening of the biological pathway of interest to isolate candidate mutants; 4. candidate validation in terms of mutant locus identification, homozygosity status, phenotype rescue and functional relevance to the biology of interest. Since the homozygote mutants are rare compared to their heterozygote counterparts, strong selection is required to isolate the relevant mutants from a background of cells with irrelevant mutations. Recessive screens using such a system have been successfully conducted51,52. Two screens used retroviruses as the insertional mutagen and this revealed non-random genome coverage of the mutagenesis, partly due to the biased integration patterns of the retrovirus53. As discussed above, DNA transposons such as piggyBac have been shown to have a more random genome-wide distribution, clearly distinctive from the retroviruses that have insertional “hotspots” across the genome. As a proof of principle, piggyBac mutagenesis (conducted by plasmid transfection) was used to construct a mutant pool in Blm-deficient ES cells and this was screened to identify components involved in the DNA mismatch repair (MMR) pathway54. Four known components of the MMR pathway genes were recovered by piggyBac based mutagenesis, whereas in the previous retrovirus-based screen, only one known component and a novel gene were isolated.
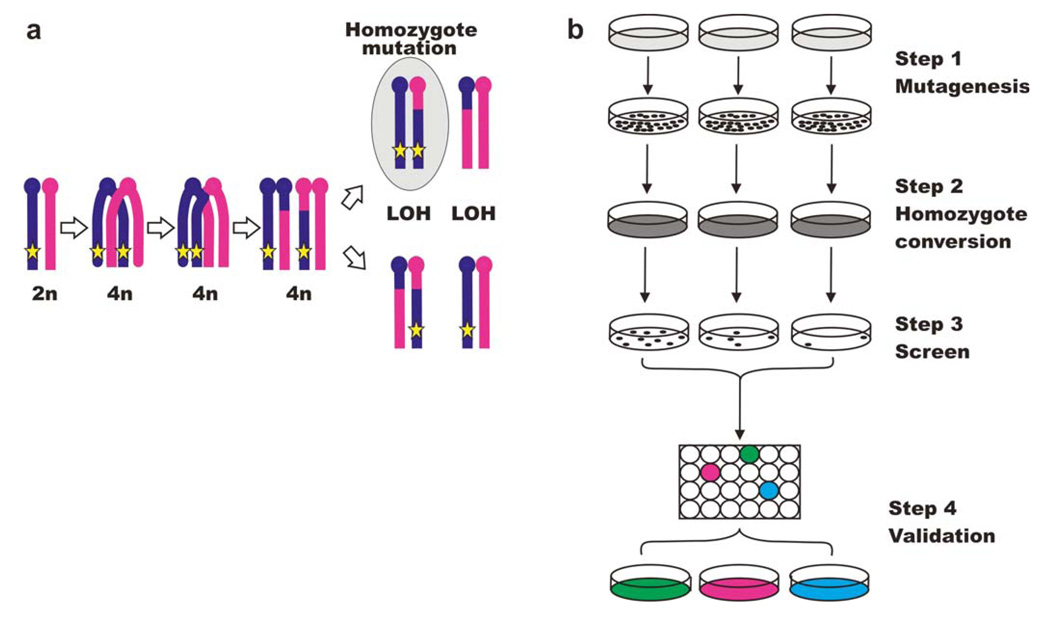
Figure 5. Recessive genetic screens in Blm-deficient ES cells.
(a) Non-sister mitotic homologous recombination gives rise to homozygote mutants. The star represents the integration site of an intertional mutagen. The genotype highlighted in grey contains a bi-allelic mutation. (b) Flow chart for recessive genetic screens using a Blm-deficient ES cells system.
In addition to genome coverage, other factors are important for the recovery of homozygote mutants in the Blm-deficient ES cell system. The number of cells derived from each independent insertion needs to be large enough to obtain homozygote mutants, given that mitotic recombination rates will vary along the chromosomes with lower rates towards the centromeric regions. It is also useful to sector the pools into sub-pools, as this will limit the “jack-pot” effect of a single mutant that converts to homozygosity early during pool expansion.
Mutagenesis in vivo
Insertional mutagenesis using engineered transposable elements can be one of the most productive and versatile approaches toward disrupting and manipulating genes on a genome-wide scale. By far the most productive approach for this is to set up a two-component experimental system, in which transposition is controlled by trans-supplementation of the transposase (Fig. 2). This enables the generation of transgenic stocks, each containing a separate component of the binary transposon system in its genome: one component, encoding the transposase, is carried by the “jumpstarter” strain, which, upon inter-crossing, efficiently mobilizes the second component, a non-autonomous transposon in the genome of the “mutator” strain (Fig. 6). This experimental setup is especially useful for directing transposition events to particular tissues or organs by tissue-specific promoters driving transposase expression. Importantly, once integrated, transposase-deficient nonautonomous transposons are stable in the absence of the transposase.
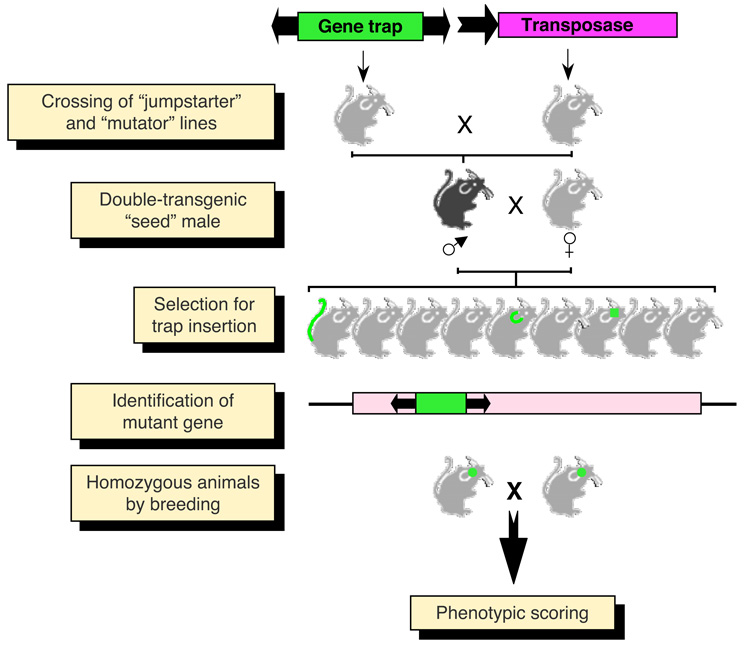
Figure 6. In vivo germline mutagenesis of the mouse with transposable elements.
Breeding of “jumpstarter” and “mutator” stocks induces transposition in the germline of double-transgenic “seed” males. The transposition events that take place in germ cells are segregated in the offspring. Animals with transposition events need to be bred to homozygosity in order to visualize the phenotypic effects of recessive mutations. Mutant genes can easily be cloned by different PCR methods making use of the inserted transposon as a unique sequence tag.
All the Class II transposon vectors used in vertebrate insertional mutagenesis to date are versions of gene trapping insertional mutagenic constructs (Fig. 3), equipped with elevated mutagenicity and reporting properties20. For the detection of gene trap insertions in vivo, fluorescent reporters such as GFP have been widely used. In the mutator stock, the GFP gene trap must be in an inactive state. These two stocks are crossed to bring the two components of the transposon system together, and transposition of the gene trap transposons is expected to occur in the sperm cells of F1 double-transgenic males (referred to as “seed” mice, Fig. 6). Such males will be bred to wild type females to segregate the different insertion events in their sperm cells in separate F2 animals (Fig. 6). Transposon insertion sites can easily be established by PCR protocols from genomic DNA isolated from GFP-positive pups, which can be further characterized by RT-PCR of composite transcripts made up by sequences of the insertional vector and the endogenous gene. In addition, similar to the GAL4/UAS system in Drosophila, a conditional, tetracycline-regulated system has been shown to be applicable to TE-mediated insertional mutagenesis55.
SB has been successfully used for forward genetics approaches in mice. In these experiments, chromosomally resident transposon vectors were mobilized in double-transgenic animals that were either ubiquitously expressing the transposase32,56–58 or were expressing the transposase in the male germline using the protamine 1 (Prm1) promoter31. Recently, SB-based insertional mutagenesis was also established in the rat by using essentially the same experimental approach59. In the mouse system, segregating the transposition events by mating the seed males to wild-type females revealed that up to 90% of the progeny can carry transposon insertions57, and a single sperm of a seed male can contain, on average, two insertion events56. The germline of such a founder was estimated to harbor approximately 10,000 different mutations58. Importantly, transposition of gene trap transposons identified mouse genes with ubiquitous and tissue-specific expression patterns, and mutant/lethal phenotypes were easily obtained by generating homozygous animals32,58,60,61. Other studies34 showed that local saturation mutagenesis of a genomic region is a realistic goal using the SB transposon system with a chromosomally resident transposon donor site. The Minos transposon has also been shown to mobilize in mice by transposase expression in the oocytes using ZP314 and in the lymphocytes using CD2 promoters62. PiggyBac has also been used in coinjection experiments in mice13. The activity of the Tol2 element has already been demonstrated in mouse ES cells63 and in vivo in the mouse liver23. In zebrafish, SB and Tol2 have been shown to be useful for insertional mutagenesis in coinjection experiments64–68. As discussed above, the availability of a battery of vector systems based on diverse TEs will undoubtedly increase genome coverage in mutagenesis screens.
Basic design of recessive, loss-of-function screens in the mouse germline using the L1 retrotransposon
The basic design of setting up mutagenesis using the L1 retrotransposon in mice is similar to the scheme shown in Fig. 6, except that only a single transgenic stock based on an autonomous retrotransposon construct needs to be established; this carries a transcriptionally (and hence transpositionally) active L1 element. One system that has been described in detail19 utilized an ORFeus transgene driven by a constitutive promoter (CAG), and marked by a retrotransposition indicator cassette, in which a GFP marker is disrupted by an intron (Fig. 7). An insertion resulting from a retrotransposition event lacks the intron (and thus has the potential to express GFP) because the primary transcript pruduced by the retrotransposon will undergo splicing (Fig. 7). Germline insertion frequency was estimated to be about 30%, and the genomic distribution of de novo retrotransposon insertions revealed ~28 % of the events occurring in RefSeq genes, and a uniform ditribution of intragenic insertions along the targeted genes.
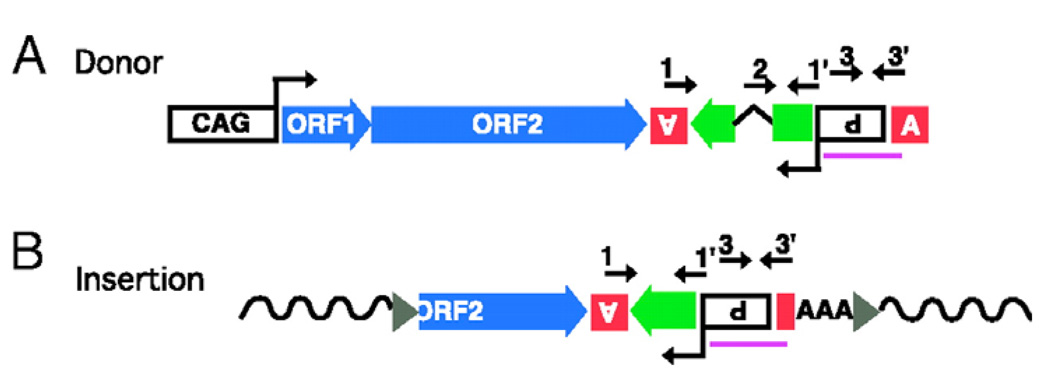
Figure 7. Synthetic L1/ORFeus transgene and progeny retrotransposon insertions.
(a) The transgene construct or donor element consists of the following sequence elements from 5’ to 3’: (i) a composite CMV IE enhancer/modified chicken β-actin promoter, designated “CAG”. (ii) synthetic L1 ORF1, ORF2 and 5’ portion of 3’UTR. (iii) Herpes simplex virus thymidine kinase poly(A) signal (boxed inverted letter A) in antisense orientation to polyadenylate gfp mRNA. (iv) gfp (green block arrow), a modified version of EGFP coding sequence. The gfp ORF is in antisense orientation relative to L1 and interrupted by intron 2 of human β-globin gene, which is in sense orientation relative to L1; gfp serves as a “retrotransposition indicator gene”. (v) Rous sarcoma virus LTR promoter in antisense orientation relative to L1, which drives gfp transcription (boxed inverted P for promoter). (vi) β-globin poly(A) signal (boxed upright letter A). Numbered arrows above the diagram represent locations of genotyping PCR primers. Region used to generate Southern blotting probes is indicated (purple line). (b) Structure of a representative progeny element. A typical progeny insertion is 5’ truncated, intronless, ends in a poly(A) tail (AAA) and is flanked by target site duplications (gray triangles) and target genomic DNA sequences (wavy solid lines). Primers 1 and 1’ (intron flanking primers) amplify a longer product when derived from the donor element (A) than from the progeny insertions (B); product length differs by the length of the intron. Primers 2 and 1’ (primer 2 is the “intron spanning” primer that spans the splice junction give rise to a product only from progeny retrotransposition events. Primers 3 and 3’ are control primers that give rise to products of constant length for donor and progeny elements.
From the perspective of use as mutagenesis tools, L1 retrotransposons have several potential advantages/features. 1) As endogenous elements, they are presumably optimized for their native host species. Indeed, they have been shown to cause a wide variety of insertions in both the mouse and human germ lines, as well as in human and other mammalian tumors. 2) Because of their “copy-and-paste” mechanism of retrotransposition, the donor copy of the element is not touched by any transposase. Rather, the active nucleic acid species that donates the newly inserted material is an RNA molecule. While this places some constrains on the system (e.g., one cannot insert polyadenylation signals in the sense orientation for fear of truncating the element), it has the great advantage that the donor element is rock-stable. 3) Because donor elements can be driven by external “cellular” promoters with essential upstream elements that are not transcribed, it is possible to design them so that they transpose once and only once (Fig. 7). 4) Retrotransposons can be tagged with an intron used as an “indicator” of retrotransposition (Fig. 7). This provides a convenient assay for retrotransposition even in the presence of a donor element copy, or even dozens to hundreds of such copies, using either an “intron flanking” or “intron spanning” PCR approach19. 5) Because both the proteins and genomic RNA of the element derive from the same molecule, and this is utterly dependent on the element’s promoter, it is possible to control the retrotransposition very sensitively, and tissue specifically, by controlling RNA expression. For example, it is possible to interpose a “STOP” signal between a strong promoter and the body of an element and if this is flanked by site-specific recombination sites (loxP), it is possible to activate retrotransposition using tissue specific or (in principle) chemically controlled recombinases such as Cre6.
Other features of L1 elements can pose problems to their implementation as mutagenesis tools. Most significantly, in 90% of progeny transposition events, rearrangements are observed. Typically, the element has a 5’ truncation ranging in length from a few base pairs to >5 kb. Most of these truncations remove more than half of the 6 kb element, resulting in many progeny insertions of <1 kb. A subset of insertions show a 5’ inversion, or 5’ inversion combined with truncation. An elegant model called “twin priming” explains how such structures might be generated at a high frequency69. In addition, the terminal target site duplications (TSDs) as well as the 3’ polyA sequence may vary in length, and the complexities of these structures can create problems in determining sites of new insertion.
Basic design of dominant, gain-of-function screens in the soma
As an alternative to the loss-of-function approaches, targeted over- and/or misexpression has been shown to be efficient in somatic tissues of mice using SB. Such screens are especially useful for the generation of experimental cancers in animal models. Forward genetic approaches for cancer gene discovery are attractive, because they allow unbiased, whole-genome screens for cancer genes70. Though this approach is similar to the application of retroviruses in cancer screens, TEs allow the recovery of tumors in tissues previously not amenable to such genomic approaches, including the liver and the brain. The “oncogene trap” SB transposon (Fig. 3e) previously used for somatic mutagenesis contains splice acceptors in both orientations followed by a polyadenylation signals as well as sequences from the murine stem cell virus (MSCV) LTR that contains enhancer/promoter elements44,45. Accordingly, this mutagenic transposon can induce loss-of-function mutations in tumor suppressor genes as well as promote gain-of-function over-expression of proto-oncogenes near the genomic insertion sites. Thus, by design, such screens can capitalize on TEs with an intronic preference of intragenic insertion, such as SB. As tumor formation is hypothesized to require multiple hits in cancer genes in the same cell71, it is thought that having more transposons to mobilize would allow the generation of multiple insertions and a cumulative mutagenic effect. Thus, insertional mutagenesis with SB in somatic tissues of mice has been approached with mutator lines harboring transposon donor loci containing many copies (in the range between 25 and 358) of the transposon vector in the form of concatemeric arrays44,45. These mutator lines are crossed with stocks that express the SB transposase to generate double-transgenic animals (Fig. 8) similar to the seed founders described above, except that the experimental tumors will develop in somatic tissues of the double-transgenic animals as a result of dominant mutations, so that there is no need in such a screen for a further round of crossing. In the published studies, somatic mobilization of the oncogene trap transposons accelerated tumor formation (mostly sarcomas) in a p19Arf-deficient cancer-predisposed genetic background44, as well as the formation of leukemia and medulloblastoma in wild-type animals45. The next step in the procedure is to isolate the transposon insertions from tumor samples (Fig. 8) by using high-throughput PCR methods72, and determine which one(s) are causative with respect to tumor formation. This is not a trivial task, as such tumors may contain hundreds of transposon insertions at different chromosomal locations. In order to determine which insertions tagged cancer genes, common insertion site (CIS) analysis is performed. CIS analysis identifies repeated occurrence of insertions in particular genes in tumors thad had been collected from different animals derived from the same mutator lines or, to have even higher statistical power, from different mutators. Candidate oncogenes are validated by transgenic models (in tissue culture or in vivo, Fig. 8), for which transposons can be applied as powerful gene vectors73. In order to devise customized screens for cancer development, a current approach is pointing towards establishing mouse lines conditionally expressing the transposase74. One approach is to express the transposase from tissue-specific promoters. The second is to generate a Cre recombinase-inducible transposase allele, and take advantage of the many existing Cre strains to induce mutagenesis in specific tissues in mice74.
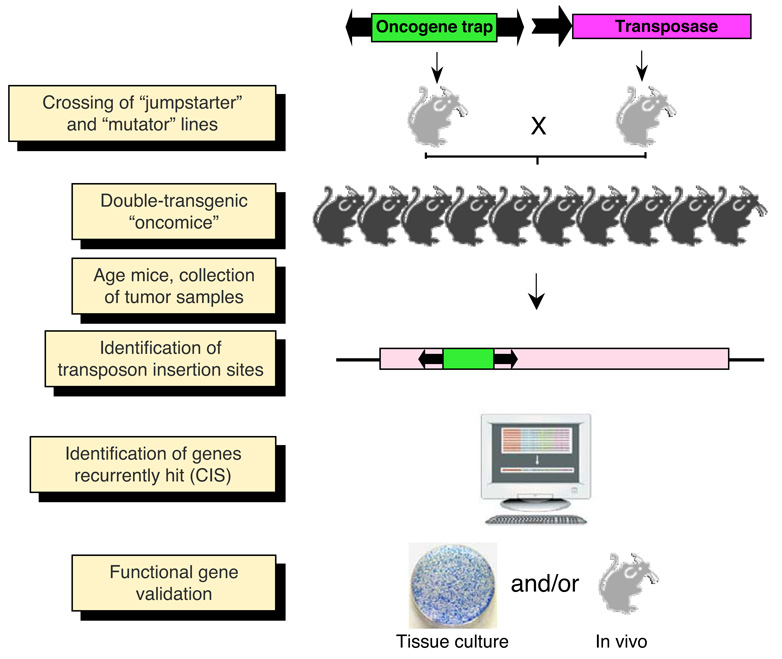
Figure 8. Somatic mutagenesis in the mouse with transposable elements.
Breeding of “jumpstarter” and “mutator” stocks induces transposition in the soma of double-transgenic animals (“oncomice”). In case of tissue-specific screens, a third genotype containing a tissue-specific Cre allele has to be crossed in. The crosses can be made either in wild-type or in specific cancer-predisposed genetic backgrounds. Transposition in somatic cells leads to random insertional mutations, and animals are aged for tumor development. Transposon insertions are cloned from genomic DNA isolated from tumor samples, and are subsequently mapped and annotated with respect to mutagenized genes. Those genes repeatedly mutated in multiple, independent tumors are designated as common insertion sites or CIS. These candidate cancer genes are functionally validated.
Future projections
The characteristics of the transposon toolkit are being unveiled, enabling an informed choice of the specific transposon most suitable for each experimental design. One obvious immediate application of transposon-based technologies will be to provide an alternate source of mutations to complement current efforts in Europe and North-America to mutagenize all mouse genes. Further, recent advances in reprogramming of various differentiated cell types back to induced pluripotent stem-cell like cells (iPS) opens up the field of stem cell research beyond the mouse75. Transposon-based mutagenesis technologies developed in mouse ES cells are directly applicable to human iPS cells, which should facilitate the identification of genetic determinants involved in physio/pathological pathways in human cells isolated from patients with specific genetic diseases76. A relatively unexplored, but highly relevant area is animal transgenesis beyond the laboratory model species. The transposon technologies currently in hand are immediately testable in large animal species of agricultural and biotechnological importance, including cattle, sheep and pig. Finally, it is now becoming amenable to create libraries of gene knockouts and to thereby establish new animal models of human disease for therapeutic and pharmaceutical intervention in species in which ES cell/homologous recombination-based knockout technology has not been established. For example, QTLs implicated in cardiovascular diseases could be dissected in the future using transposon-mediated insertional mutagenesis in the rat system, the preferred model for cardiovascular biology77. Thus, transposon-based technologies have enormous potential to develop powerful genomic tools for vertebrate models with the vision of creating a bridge between physiology and genetics.
Supplementary Material
Table 1.
| Transposon name |
Transposon family |
Tolerated cargo size |
Target site | Chromatic integration pattern |
Systems tested |
|---|---|---|---|---|---|
|
Sleeping Beauty Reconstructed from Fish |
Tc1/mariner | Increased cargo size exponentially decreases the efficiency of transposition in cultured cells |
TA | No preference for genes. Gene hits dominantly in introns |
Cultured vertebrate cell lines, mouse, rat, zebrafish, medaka fish, Xenopus |
|
Frog Prince Reconstructed from Rana pipiens (Northern leopard frog) |
Tc1/mariner | Possibly similar to other Tc1/mariner transposons |
TA | Highly efficient gene trapping in tissue culture cells. Gene hits dominantly in introns |
Cultured vertebrate cell lines, zebrafish embryos |
|
Minos Drosophila hydei |
Tc1/mariner | Possibly similar to other Tc1/mariner transposons |
TA | No preference for genes. Gene hits dominantly in introns |
Cultured human cells, mouse tissues, Ciona intestinalis |
|
Hsmar1 Reconstructed from human |
Tc1/mariner | Possibly similar to other Tc1/mariner transposons |
TA | Similar to SB | Cultured human cells and zebrafish embryos |
|
Passport Pleuronectes platessa (plaice) |
Tc1/mariner | Possibly similar to other Tc1/mariner transposons |
TA | May have a preference for transcription units |
Mammalian and avian cell culture |
|
piggyback Trichoplusia ni (cabbage looper moth) |
piggyBac | Efficiency drops above 9.1 kb in pronucleus-injected mice |
TTAA | Preference for transcription units |
Mammalian cell culture including mouse ES cells, mouse tissues |
|
Tol1 Oryzias latipes (medaka fish) |
hAT | >20 kb was seen to move, but at reduced efficiency |
8-bp heterogenic sequences |
Unknown | Mammalian cell culture and zebrafish embryos |
|
Tol2 Oryzias latipes (medaka fish) |
hAT | >10 kb transposons jump efficiently in human cells and zebrafish embryos |
8-bp heterogenic sequences |
May prefer the 5’ regions of genes |
Cultured vertebrate cell lines including mouse ES cells, zebrafish, Xenopus and chicken embryos |
|
Ac/Ds Zea mays (maize) |
hAT | At least 6.5 kb in zebrafish embryos |
8-bp heterogenic sequences |
May have a preference for transcription units |
Mammalian cell culture and zebrafish embryos |
|
Harbinger3_DR Reconstructed from Danio rerio (zebrafish) |
PIF/Harbinger | Not tested experimentally |
Preferentially inserts into a 15-bp consensus target sequence |
Unknown | Cultured human and zebrafish cells |
|
ORFeus Synthetic mouse L1 retrotransposon |
LINE1 | 5’-truncations are frequent |
preference for AT- rich sequences |
~30% of insertions in genes |
Mouse and human cells |
RERERENCES
- 1.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Feng Q, Moran JV, Kazazian HH, Jr, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 3.Mathias SL, Scott AF, Kazazian HH, Jr, Boeke JD, Gabriel A. Reverse transcriptase encoded by a human transposable element. Science. 1991;254:1808–1810. doi: 10.1126/science.1722352. [DOI] [PubMed] [Google Scholar]
- 4.Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 5.Cost GJ, Feng Q, Jacquier A, Boeke JD. Human L1 element target-primed reverse transcription in vitro. Embo J. 2002;21:5899–5910. doi: 10.1093/emboj/cdf592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.An W, et al. Conditional activation of a single-copy L1 transgene in mice by Cre. Genesis. 2008;46:373–383. doi: 10.1002/dvg.20407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rushforth AM, Saari B, Anderson P. Site-selected insertion of the transposon Tc1 into a Caenorhabditis elegans myosin light chain gene. Mol Cell Biol. 1993;13:902–910. doi: 10.1128/mcb.13.2.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zwaal RR, Broeks A, van Meurs J, Groenen JT, Plasterk RH. Target-selected gene inactivation in Caenorhabditis elegans by using a frozen transposon insertion mutant bank. Proc Natl Acad Sci U S A. 1993;90:7431–7435. doi: 10.1073/pnas.90.16.7431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bessereau JL, et al. Mobilization of a Drosophila transposon in the Caenorhabditis elegans germ line. Nature. 2001;413:70–74. doi: 10.1038/35092567. [DOI] [PubMed] [Google Scholar]
- 10.Spradling AC. A Practical Approach. 1986. P element-mediated transformation in Drosophila; pp. 175–198. IRL Press. [Google Scholar]
- 11.Cooley L, Kelley R, Spradling A. Insertional mutagenesis of the Drosophila genome with single P elements. Science. 1988;239:1121–1128. doi: 10.1126/science.2830671. [DOI] [PubMed] [Google Scholar]
- 12.Ivics Z, Hackett PB, Plasterk RH, Izsvak Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- 13.Ding S, et al. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell. 2005;122:473–483. doi: 10.1016/j.cell.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 14.Drabek D, et al. Transposition of the Drosophila hydei Minos transposon in the mouse germ line. Genomics. 2003;81:108–111. doi: 10.1016/s0888-7543(02)00030-7. [DOI] [PubMed] [Google Scholar]
- 15.Miskey C, Izsvak Z, Plasterk RH, Ivics Z. The Frog Prince: a reconstructed transposon from Rana pipiens with high transpositional activity in vertebrate cells. Nucleic Acids Res. 2003;31:6873–6881. doi: 10.1093/nar/gkg910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miskey C, et al. The Ancient Mariner Sails Again: Transposition of the Human Hsmar1 Element by a Reconstructed Transposase and Activities of the SETMAR Protein on Transposon Ends. Mol Cell Biol. 2007 doi: 10.1128/MCB.02027-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawakami K, Shima A, Kawakami N. Identification of a functional transposase of the Tol2 element, an Ac-like element from the Japanese medaka fish, and its transposition in the zebrafish germ lineage. Proc Natl Acad Sci U S A. 2000;97:11403–11408. doi: 10.1073/pnas.97.21.11403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han JS, Boeke JD. A highly active synthetic mammalian retrotransposon. Nature. 2004;429:314–318. doi: 10.1038/nature02535. [DOI] [PubMed] [Google Scholar]
- 19.An W, et al. Active retrotransposition by a synthetic L1 element in mice. Proc Natl Acad Sci U S A. 2006;103:18662–18667. doi: 10.1073/pnas.0605300103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mates L, Izsvak Z, Ivics Z. Technology transfer from worms and flies to vertebrates: transposition-based genome manipulations and their future perspectives. Genome Biol. 2007;8 Suppl 1:S1. doi: 10.1186/gb-2007-8-s1-s1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Izsvak Z, Ivics Z, Plasterk RH. Sleeping Beauty, a wide host-range transposon vector for genetic transformation in vertebrates. J Mol Biol. 2000;302:93–102. doi: 10.1006/jmbi.2000.4047. [DOI] [PubMed] [Google Scholar]
- 22.Zayed H, Izsvak Z, Walisko O, Ivics Z. Development of hyperactive sleeping beauty transposon vectors by mutational analysis. Mol Ther. 2004;9:292–304. doi: 10.1016/j.ymthe.2003.11.024. [DOI] [PubMed] [Google Scholar]
- 23.Balciunas D, et al. Harnessing a high cargo-capacity transposon for genetic applications in vertebrates. PLoS Genet. 2006;2:e169. doi: 10.1371/journal.pgen.0020169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Urasaki A, Morvan G, Kawakami K. Functional dissection of the Tol2 transposable element identified the minimal cis-sequence and a highly repetitive sequence in the subterminal region essential for transposition. Genetics. 2006;174:639–649. doi: 10.1534/genetics.106.060244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cost GJ, Boeke JD. Targeting of human retrotransposon integration is directed by the specificity of the L1 endonuclease for regions of unusual DNA structure. Biochemistry. 1998;37:18081–18093. doi: 10.1021/bi981858s. [DOI] [PubMed] [Google Scholar]
- 26.Vigdal TJ, Kaufman CD, Izsvak Z, Voytas DF, Ivics Z. Common physical properties of DNA affecting target site selection of sleeping beauty and other Tc1/mariner transposable elements. J Mol Biol. 2002;323:441–452. doi: 10.1016/s0022-2836(02)00991-9. [DOI] [PubMed] [Google Scholar]
- 27.Liu G, et al. Target-site preferences of Sleeping Beauty transposons. J Mol Biol. 2005;346:161–173. doi: 10.1016/j.jmb.2004.09.086. [DOI] [PubMed] [Google Scholar]
- 28.Yant SR, et al. High-resolution genome-wide mapping of transposon integration in mammals. Mol Cell Biol. 2005;25:2085–2094. doi: 10.1128/MCB.25.6.2085-2094.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang W, et al. Chromosomal transposition of PiggyBac in mouse embryonic stem cells. Proc Natl Acad Sci U S A. 2008;105:9290–9295. doi: 10.1073/pnas.0801017105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilson MH, Coates CJ, George AL., Jr PiggyBac Transposon-mediated Gene Transfer in Human Cells. Mol Ther. 2007;15:139–145. doi: 10.1038/sj.mt.6300028. [DOI] [PubMed] [Google Scholar]
- 31.Fischer SE, Wienholds E, Plasterk RH. Regulated transposition of a fish transposon in the mouse germ line. Proc Natl Acad Sci U S A. 2001;98:6759–6764. doi: 10.1073/pnas.121569298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carlson CM, et al. Transposon mutagenesis of the mouse germline. Genetics. 2003;165:243–256. doi: 10.1093/genetics/165.1.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo G, Ivics Z, Izsvak Z, Bradley A. Chromosomal transposition of a Tc1/mariner-like element in mouse embryonic stem cells. Proc Natl Acad Sci U S A. 1998;95:10769–10773. doi: 10.1073/pnas.95.18.10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keng VW, et al. Region-specific saturation germline mutagenesis in mice using the Sleeping Beauty transposon system. Nat Methods. 2005;2:763–769. doi: 10.1038/nmeth795. [DOI] [PubMed] [Google Scholar]
- 35.Hamlet MR, et al. Tol2 transposon-mediated transgenesis in Xenopus tropicalis. Genesis. 2006;44:438–445. doi: 10.1002/dvg.20234. [DOI] [PubMed] [Google Scholar]
- 36.Nasevicius A, Ekker SC. Effective targeted gene 'knockdown' in zebrafish. Nat Genet. 2000;26:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- 37.Sinzelle L, et al. Generation of trangenic Xenopus laevis using the Sleeping Beauty transposon system. Transgenic Res. 2006 doi: 10.1007/s11248-006-9014-6. [DOI] [PubMed] [Google Scholar]
- 38.Sato Y, et al. Stable integration and conditional expression of electroporated transgenes in chicken embryos. Dev Biol. 2007;305:616–624. doi: 10.1016/j.ydbio.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 39.Dupuy AJ, et al. Mammalian germ-line transgenesis by transposition. Proc Natl Acad Sci U S A. 2002;99:4495–4499. doi: 10.1073/pnas.062630599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilber A, et al. RNA as a source of transposase for sleeping beauty-mediated gene insertion and expression in somatic cells and tissues. Mol Ther. 2006;13:625–630. doi: 10.1016/j.ymthe.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 41.Carlson CM, Frandsen JL, Kirchhof N, McIvor RS, Largaespada DA. Somatic integration of an oncogene-harboring Sleeping Beauty transposon models liver tumor development in the mouse. Proc Natl Acad Sci U S A. 2005;102:17059–17064. doi: 10.1073/pnas.0502974102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Friedrich G, Soriano P. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev. 1991;5:1513–1523. doi: 10.1101/gad.5.9.1513. [DOI] [PubMed] [Google Scholar]
- 43.Lukacsovich T, et al. Dual-tagging gene trap of novel genes in Drosophila melanogaster. Genetics. 2001;157:727–742. doi: 10.1093/genetics/157.2.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Collier LS, Carlson CM, Ravimohan S, Dupuy AJ, Largaespada DA. Cancer gene discovery in solid tumours using transposon-based somatic mutagenesis in the mouse. Nature. 2005;436:272–276. doi: 10.1038/nature03681. [DOI] [PubMed] [Google Scholar]
- 45.Dupuy AJ, Akagi K, Largaespada DA, Copeland NG, Jenkins NA. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature. 2005;436:221–226. doi: 10.1038/nature03691. [DOI] [PubMed] [Google Scholar]
- 46.Wu S, Ying G, Wu Q, Capecchi MR. Toward simpler and faster genome-wide mutagenesis in mice. Nat Genet. 2007;39:922–930. doi: 10.1038/ng2060. [DOI] [PubMed] [Google Scholar]
- 47.Skarnes WC, et al. A public gene trap resource for mouse functional genomics. Nat Genet. 2004;36:543–544. doi: 10.1038/ng0604-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen Y, et al. Genotype-based screen for ENU-induced mutations in mouse embryonic stem cells. Nat Genet. 2000;24:314–317. doi: 10.1038/73557. [DOI] [PubMed] [Google Scholar]
- 49.Lefebvre L, Dionne N, Karaskova J, Squire JA, Nagy A. Selection for transgene homozygosity in embryonic stem cells results in extensive loss of heterozygosity. Nat Genet. 2001;27:257–258. doi: 10.1038/85808. [DOI] [PubMed] [Google Scholar]
- 50.Luo G, et al. Cancer predisposition caused by elevated mitotic recombination in Bloom mice. Nat Genet. 2000;26:424–429. doi: 10.1038/82548. [DOI] [PubMed] [Google Scholar]
- 51.Yusa K, et al. Genome-wide phenotype analysis in ES cells by regulated disruption of Bloom's syndrome gene. Nature. 2004;429:896–899. doi: 10.1038/nature02646. [DOI] [PubMed] [Google Scholar]
- 52.Wang W, Bradley A. A recessive genetic screen for host factors required for retroviral infection in a library of insertionally mutated Blm-deficient embryonic stem cells. Genome Biol. 2007;8:R48. doi: 10.1186/gb-2007-8-4-r48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hansen GM, et al. Large-scale gene trapping in C57BL/6N mouse embryonic stem cells. Genome Res. 2008;18:1670–1679. doi: 10.1101/gr.078352.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Karran P. Mechanisms of tolerance to DNA damaging therapeutic drugs. Carcinogenesis. 2001;22:1931–1937. doi: 10.1093/carcin/22.12.1931. [DOI] [PubMed] [Google Scholar]
- 55.Geurts AM, et al. Conditional gene expression in the mouse using a Sleeping Beauty gene-trap transposon. BMC Biotechnol. 2006;6:30. doi: 10.1186/1472-6750-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dupuy AJ, Fritz S, Largaespada DA. Transposition and gene disruption in the male germline of the mouse. Genesis. 2001;30:82–88. doi: 10.1002/gene.1037. [DOI] [PubMed] [Google Scholar]
- 57.Horie K, et al. Efficient chromosomal transposition of a Tc1/mariner- like transposon Sleeping Beauty in mice. Proc Natl Acad Sci U S A. 2001;98:9191–9196. doi: 10.1073/pnas.161071798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Horie K, et al. Characterization of Sleeping Beauty transposition and its application to genetic screening in mice. Mol Cell Biol. 2003;23:9189–9207. doi: 10.1128/MCB.23.24.9189-9207.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kitada K, et al. Transposon-tagged mutagenesis in the rat. Nat Methods. 2007 doi: 10.1038/nmeth1002. [DOI] [PubMed] [Google Scholar]
- 60.Yae K, et al. Sleeping beauty transposon-based phenotypic analysis of mice: lack of Arpc3 results in defective trophoblast outgrowth. Mol Cell Biol. 2006;26:6185–6196. doi: 10.1128/MCB.00018-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Geurts AM, et al. Gene mutations and genomic rearrangements in the mouse as a result of transposon mobilization from chromosomal concatemers. PLoS Genet. 2006;2:e156. doi: 10.1371/journal.pgen.0020156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zagoraiou L, et al. In vivo transposition of Minos, a Drosophila mobile element, in mammalian tissues. Proc Natl Acad Sci U S A. 2001;98:11474–11478. doi: 10.1073/pnas.201392398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kawakami K, Noda T. Transposition of the Tol2 element, an Ac-like element from the Japanese medaka fish Oryzias latipes, in mouse embryonic stem cells. Genetics. 2004;166:895–899. doi: 10.1093/genetics/166.2.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kawakami K, et al. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev Cell. 2004;7:133–144. doi: 10.1016/j.devcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 65.Parinov S, Kondrichin I, Korzh V, Emelyanov A. Tol2 transposon-mediated enhancer trap to identify developmentally regulated zebrafish genes in vivo. Dev Dyn. 2004;231:449–459. doi: 10.1002/dvdy.20157. [DOI] [PubMed] [Google Scholar]
- 66.Davidson AE, et al. Efficient gene delivery and gene expression in zebrafish using the Sleeping Beauty transposon. Dev Biol. 2003;263:191–202. doi: 10.1016/j.ydbio.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 67.Sivasubbu S, et al. Gene-breaking transposon mutagenesis reveals an essential role for histone H2afza in zebrafish larval development. Mech Dev. 2006;123:513–529. doi: 10.1016/j.mod.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 68.Balciunas D, et al. Enhancer trapping in zebrafish using the Sleeping Beauty transposon. BMC Genomics. 2004;5:62. doi: 10.1186/1471-2164-5-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ostertag EM, Kazazian HH., Jr Twin priming: a proposed mechanism for the creation of inversions in L1 retrotransposition. Genome Res. 2001;11:2059–2065. doi: 10.1101/gr.205701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Collier LS, Largaespada DA. Transposons for cancer gene discovery: Sleeping Beauty and beyond. Genome Biol. 2007;8 Suppl 1:S15. doi: 10.1186/gb-2007-8-s1-s15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 72.Largaespada DA, Collier LS. Transposon-mediated mutagenesis in somatic cells: identification of transposon-genomic DNA junctions. Methods Mol Biol. 2008;435:95–108. doi: 10.1007/978-1-59745-232-8_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Su Q, et al. A DNA transposon-based approach to validate oncogenic mutations in the mouse. Proc Natl Acad Sci U S A. 2008;105:19904–19909. doi: 10.1073/pnas.0807785105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dupuy AJ, Jenkins NA, Copeland NG.Sleeping beauty: a novel cancer gene discovery tool Hum Mol Genet15200615R75–R79. [DOI] [PubMed] [Google Scholar]
- 75.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 76.Park IH, et al. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877–886. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Aitman TJ, et al. Progress and prospects in rat genetics: a community view. Nat Genet. 2008;40:516–522. doi: 10.1038/ng.147. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.