Abstract
Physical factors drive evolution and play important roles in motility and attachment as well as in differentiation. As animal cells adhere to survive, they generate force and "feel" various mechanical features of their surroundings – with mechanosensation mechanisms based in part on force-induced conformational changes. Single molecule methods for in vitro nano-manipulation together with new in situ proteomic approaches that exploit mass spectrometry are helping to identify and characterize the molecules and mechanics of structural transitions within cells and matrices. Given the diversity of cell and molecular responses, networks of biomolecules with conformations and interactions sculpted by force seem more likely than singular mechanosensors. Elaboration of the proteins that unfold and change structure in the extracellular matrix and in cells is needed, particularly the force-driven kinetics, to understand the systems biology of signaling in development, differentiation, and disease.
Introduction
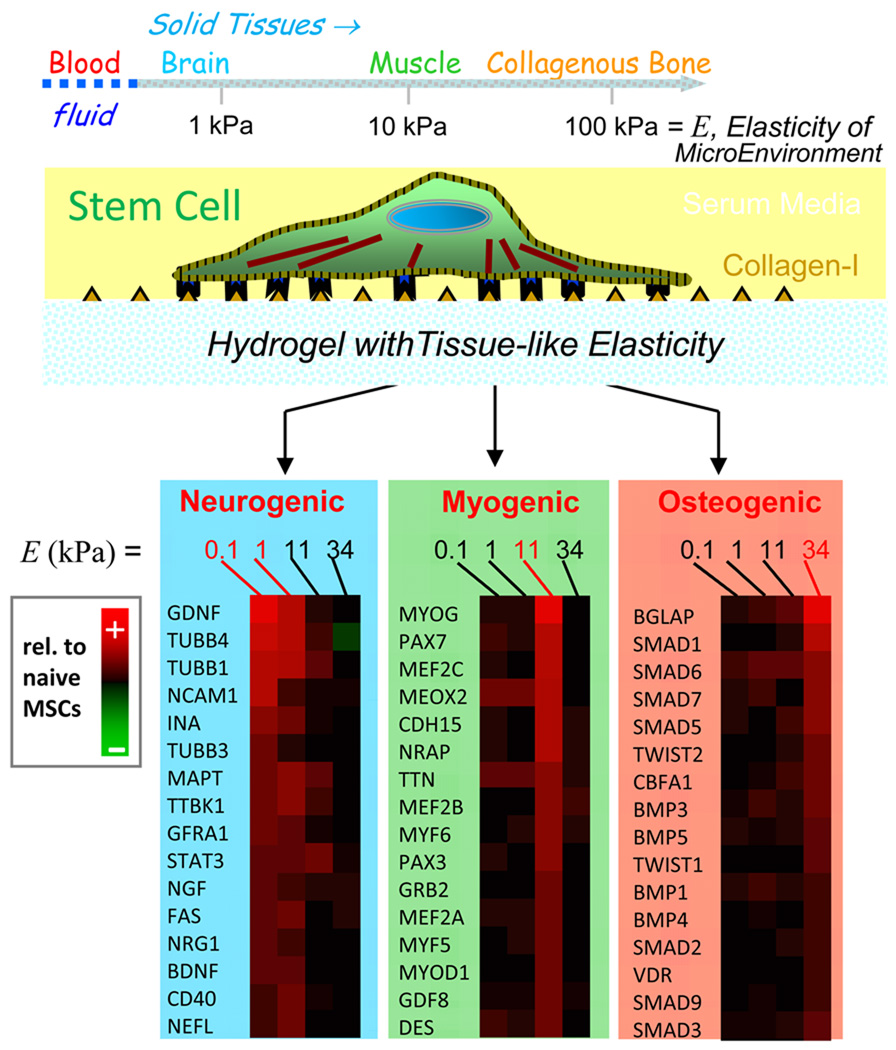
Evolutionary and developmental pressures such as outpacing pursuants, pumping essential fluids, or resisting mechanical stress, all physically select for beneficial changes in expressed genes. These examples involve forces of contact which must be sustained within a tissue by an ensemble of molecules that can also transduce – directly or indirectly – signals to the tissue-integrated cells. Since the beginnings of cell biology, various micro-tools have been developed to push or pull on cells and determine effective responses in terms of elastic, viscous, and yield parameters [1], though often without much molecular insight. Brownian and non-Brownian motions of particles within cells have also been tracked to analytically distinguish passive, thermally-scalable properties of cells from active, energy-driven characteristics [2]. The latter responses often involve signaling and exhibit molecular specificity: when a cell-matrix adhesion is stretched by an external force, for example, select proteins are post-translationally modified and actively enriched at the site of applied force [3]. When cells test the mechanics of their microenvironment through adhesive engagement and ATP driven contraction of actin-myosin ‘stress’ fibers [4], gene expression can change over hours to days, with recent examples including matrix elasticity directed lineage specification of stem cells (Fig.1) [5] and malignant transformations of breast epithelial cells on stiff substrates [6]. Regardless of process, however, the underlying molecular mechanics are in need of deeper study.
Figure 1.
Cells can ‘feel’ the physical properties of their microenvironment. In one recent example with mesenchymal stem cells, matrix elasticity is seen to direct lineage specification [5]. Inhibition of the cell’s contractile tension blocks mechanosensation.
Various signaling pathways have been described that transduce mechanical signals into biochemical responses which could lead to complex cell behaviors [7, 8]. Interactions within or between molecules that are directly modulated by force or by some other mechanical property of the microenvironment are likely to be critical in any putative mechanosensory circuit. Force-induced conformational changes have been thought to be important, but it is only relatively recent that such processes have been observed in vitro. Now there is also growing experimental evidence that force induced conformational changes indeed occur within matrix and cells [9].
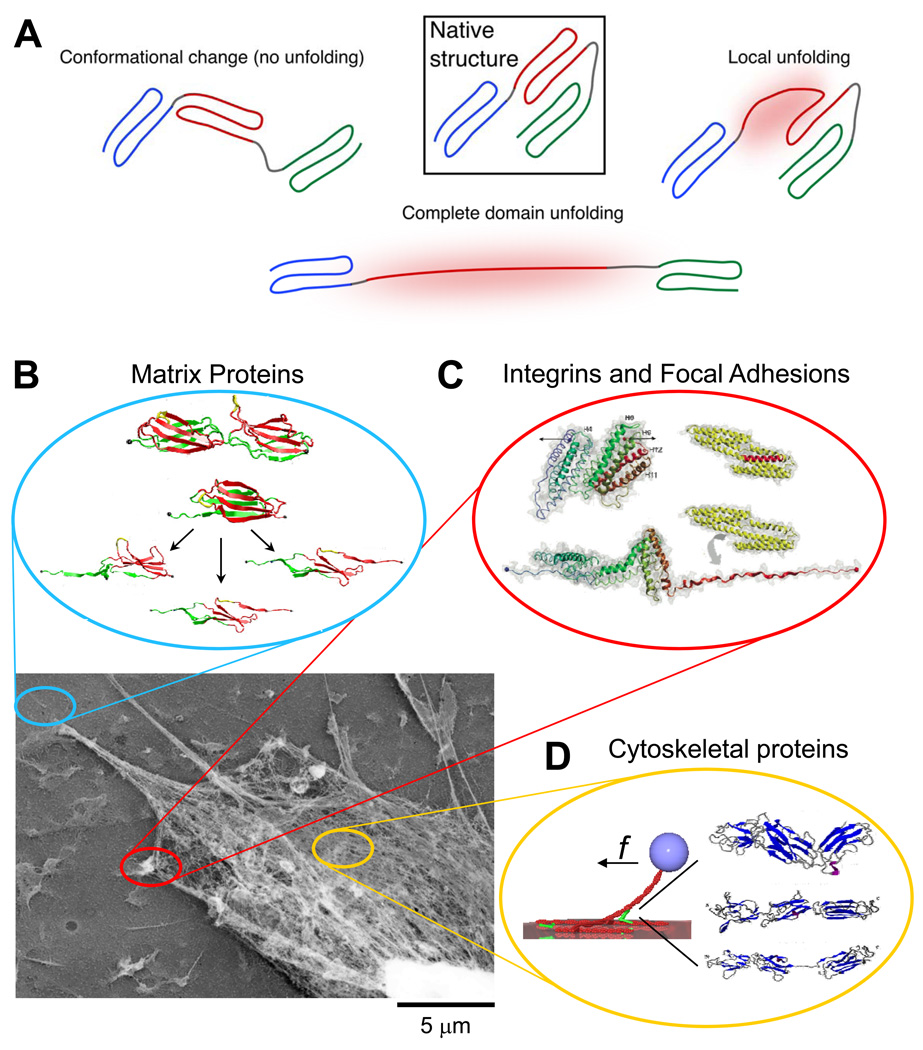
Of the various mechanisms that have been proposed for mechanosensing [7, 8, 10, 11], here we focus on tension-induced changes in protein structure (Fig. 2A). Forced unfolding can be localized to a part of a domain or loop, or it can involve complete domains; force can also re-orient domains or straighten unstructured regions such as hinges between domains. We review the experimental methods that have been used to probe transitions and also review how these observations fit into a broader cellular context. From outside a cell to inside, we first consider extracellular matrix, which defines a mechanical substrate for cells (Fig. 2B) and which interacts with cell surface receptors such as integrins to couple matrix to cell cytoskeleton (Fig. 2C). We then focus on the cytoskeleton itself, including its contractile myosin components with which a cell exerts pulling forces and probes its surroundings (Fig. 2D). More physical tools and biochemical insights are certainly needed, but it seems fair to suggest that ‘molecular mechanobiology’ is burgeoning.
Figure 2.
(A) When force is applied to proteins their native structure can be perturbed. This can involve a conformational change in which only quaternary structure is perturbed or unfolding in which secondary or tertiary structure are disrupted locally or over an entire domain. Lower panel: Adherent cells attach to the extracellular matrix (ECM), spread, and apply contractile forces using acto-myosin stress fibers. The image in the lower left shows the detergent extracted cytoskeleton of a mesenchymal stem cell adhering to its substrate. This can in principle activate several force sensitive processes that may be mediated by protein unfolding. (B) ECM proteins such as fibronectin are extended, exposing cryptic binding sites that promote fiber assembly as well as cell adhesion (inset adapted from [23], permission pending). (C) Focal adhesions sense applied force and respond by recruiting additional proteins to shore up the cell-substrate interaction. A possible contributing mechanism is the force-induced exposure of vinculin binding sites in the talin rod domain, shown here to unfold under force in molecular dynamics simulations (adapted from [55], permission pending). (D) The cytoskeleton is actively contractile and these forces impact not only the ECM and focal adhesions, but also the cytoskeleton itself. The inset shows simulations of the forced unfolding of an actin binding protein called filamin (adapted from [76], permission pending). The schematic (adapted with permission from original by Hyungsuk Lee, Jorge M. Ferrer, and Matthew J. Lang) to the left shows how filamin unfolding and actin unbinding can be simultaneously studied in single molecule experiments [78].
Proteins Unfold under Force
Most proteins fold and fluctuate around an average three-dimensional structure that can be influenced by cell ‘signaling’ processes [12]. Unfolding through changes in temperature or solvent, with denaturants such as urea or guanidine hydrochloride, has taught us much about the determinants of protein structure. However, normal body temperatures tend to remain within tight limits in mammals and birds (especially in terms of the absolute temperature in Kelvin, K), and denaturants are not especially physiological. On the other hand, mechanical stresses are unavoidable in locomotion, flowing biofluids, and even just residing in the Earth’s gravitational field. Forced unfolding of single proteins was first shown about a decade ago [13, 14]. A ‘grip and break’ approach is perhaps the most intuitive way to think about disrupting any structure, but force is a vector (with magnitude and direction) that is difficult to apply to proteins tumbling in solution. Immobilization is required, as occurs with structural proteins within a cell or matrix, and there is also a need for nanoscale techniques to apply a force and monitor the effects of force at the molecular level.
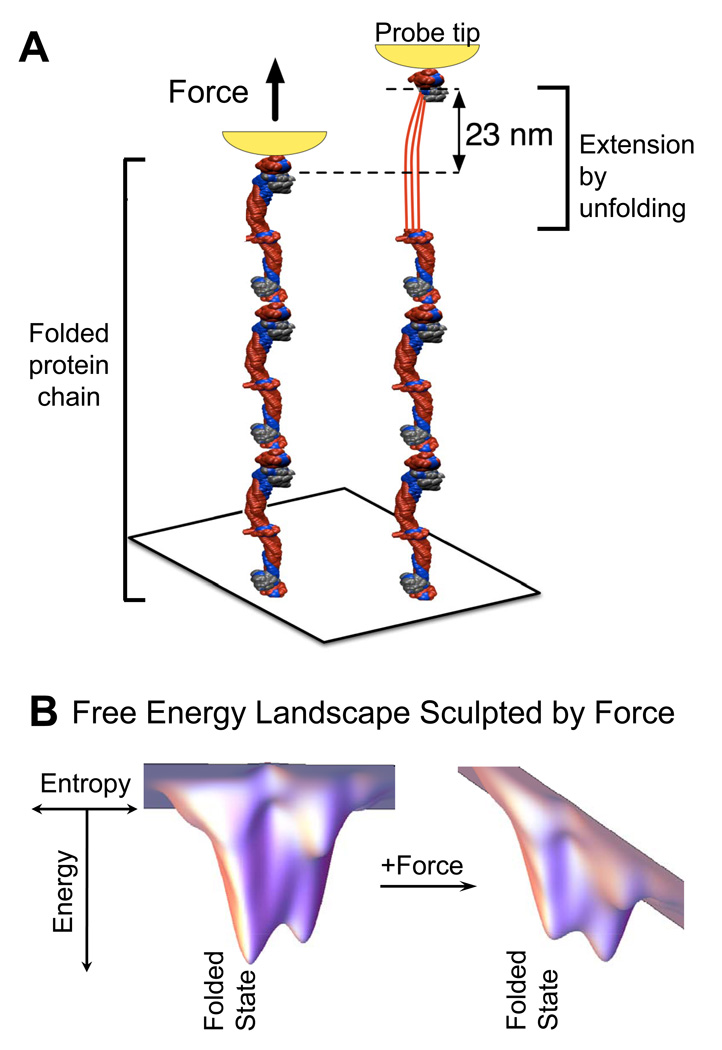
The most commonly used tools today for single molecule force spectroscopy are atomic force microscopes (AFM) (Fig. 3A) and optical tweezers as reviewed elsewhere [15]. Based on forced unfolding measurements of many proteins, forces in the range of f = 10 pN to 200 pN will unfold most proteins within a second or less. At lower forces, proteins unfold more slowly [16, 17], which one can understand from the simplest possible expression, widely attributed to Bell [16], for the rate k of reaction that is accelerated by force above a basal rate ko:
| Eq.1 |
Figure 3.
Nano-tools for protein folding and funnels. (A) Proteins can be extended and unfolded by force using a probe (shown in yellow) either in AFM or optical tweezers (adapted from [33], permission pending). (B) The energy of an extended protein conformation is typically higher than the folded native state and this is often conceptualized as an energy landscape in the form of a folding funnel where chain entropy dominates with the many unfolded states and energetic interactions pull the protein into one or a few well-defined structures. With force, the energy of more extended conformations is decreased, thereby accelerating unfolding.
The distance d is specific to a transition between protein states, and kBT (= 4 pN nm) is the standard Boltzmann constant multiplied by absolute temperature (~300 K in most biological systems). The exponential factor (i.e. Boltzmann factor) shifts and sculpts the free energy landscape which has the form of a folding funnel (Fig. 3B). Because cells are filled with myosin motors that each typically generate forces of ~1–10 pN in ~1–10 nm steps, unfolding reactions are expected to be accelerated and therefore more prominent within active cells.
With full unfolding of a typical repetitive domain within a multi-domain structural protein, extensions of 5–10 fold correspond to distensions of 20–40 nm, which is large compared to the dimensions of most other cytosolic proteins. While extension can serve as a strain-release mechanism that allows some biological scaffolds to distend without dissociating, for a growing list of proteins, unfolding seems to have additional functional consequences, such as exposure of previously hidden sites for binding and assembly.
Unfolding in the Extracellular Matrix Influences Assembly
Adherent cell types are viable only when they are attached to a solid substrate, which in vivo is the extracellular matrix (ECM) — a crosslinked hydrogel complex of protein and polysaccharide that is produced by either the cell it surrounds or specialized cells such as fibroblasts. ECM proteins are the most abundant proteins in animals and play critical mechanical roles for tissues by providing them with substantial tensile strength and, in the case of bone, substantial compressive strength results from calcification. The role of ECM in regulating cell behavior, and in particular how it can be modulated by force, is highlighted here.
Fibronectin is a well-studied ECM component that cells adhere to with surface receptors called integrins. Fibronectin consists of a series of immunoglobulin (Ig)-like domains that form a chain-like structure, and it further assembles into fibrous bundles and a network (Fig. 2B). Assembly depends on tension, whether it is produced by cells [18] or is applied externally [19, 20], and this has led to the suggestion of cryptic sites in fibronectin that are exposed by cell contractility [18]. The hypothesis that this exposure was due to a force-induced unfolding was plausible given that fibronectin unfolding under tension had already been predicted theoretically [21]. This early prediction was followed by molecular dynamics simulations [22, 23] and single molecule experiments on fibronectin domains from the related ECM protein tenascin [24]. These ‘force spectroscopy’ experiments also showed that the domains could refold in seconds after removal of force. Further evidence for some type of conformational change with the matrix under tension was provided by Foerster Resonance Energy Transfer (FRET) changes between fluorescent dyes attached to Cys and Lys residues in fibronectin [25, 26]. Although such Cys labeling tends to block disulfide formation that is essential to matrix crosslinking within the oxidative, extracellular space, the FRET results are quantitative and suggestive.
The results for fibronectin suggest that unfolding is activated by cell tension in the ECM, contributing to assembly, but fibronectin interacts with many other proteins including ECM components fibrin and collagen [27] as well as cell surface integrins [28]. Fibronectin’s multi-modular structure might therefore have evolved to serve as a multi-purpose mechanosensor in the ECM [29] with different binding events controlled by distinct mechanical stabilities of the different domains: at low forces, sites involved in fibril assembly of fibronectin could be exposed, while later, if the fibrils are put under higher tension, new binding sites could be exposed when the more stable domains are unfolded as suggested already by single molecule experiments [30]. A current challenge is to elaborate such mechanically propagating interactions.
Fibrin, fibronectin, and collagens (types 1 and 3) are linked physically through binding – perhaps with tension dependent mechanisms – and they are linked functionally during wound healing. An initial fibrin clot is replaced by ECM components that include fibronectin and then more permanent (and stiffer [31]) crosslinked collagen [32]. Fibrinogen binds several proteins in addition to fibronectin, and some of these interactions might also be regulated by force. The coiled-coils of fibrinogen have already been shown to unfold under force [33, 34]. More recent studies of fibrin gel stretching further indicate at small strains (<50%) that: (i) a volumetric phase transition occurs with a large negative compressibility that represents loss of water and association of unfolded protein, and (ii) a reversible order-disorder transition occurs as seen by small angle X-ray scattering [35]. Despite such macroscopic evidence of coiled-coil unfolding, it is not yet clear that binding sites found in fibrinogen’s globular end domains undergo conformational changes under force. Mechanically labile coiled-coils might protect the globular domains from unfolding and thus play the reverse role of unfolding in fibronectin. Another possibility is that force regulates the conformation of the C-terminal end of the γ-chain in fibrinogen, a natively unstructured region that contains an AGDV sequence involved in platelet adhesion [36, 37].
Collagen, like fibronectin, has a cryptic site that can be exposed by enzymatic degradation, and this has been shown to influence angiogenesis [38]. This exposure plays a role during enzymatic degradation of the matrix, but it is not known whether such sites can also be exposed by force. While the aligned nature of the collagen triple helix makes unfolding seem less likely, simulations have shown a variety of failure modes at high forces [39]. This could be relevant after traumatic injury, even if not under normal circumstances from cell-generated tension, but an active role of cells in any aspect of ECM remodeling should not be underestimated. Growth factors – so essential in development and differentiation – provide a valuable recent lesson in that many have long been known to bind ECM, but more recent experiments show that release of at least the canonical Transforming Growth Factor-β (TGF-β) is directly modulated by the elasticity of the matrix and by tension generated by cells [40].
Force Modulates Cell-Matrix Interactions
Cells have evolved many ways of modulating their physical interaction with each other and with the surrounding ECM. Most cells possess integrin receptors that bind to specific ECM peptides and mediate stable adhesion [28]. Forced unfolding with exposure (or protection) of these sequences is not the only way for mechanics to influence cell behavior; integrins and perhaps other focal adhesion proteins are also sensitive to force through changes in protein structure (Fig. 2C). Methods for interrogating forces from single adhesions on living cells [41] continue to be developed [42] and should provide more insight into the mechanics of adhesion that accompany these conformational changes, as they have for other processes that regulate adhesion, including mechanics [43].
With integrins, a conformational change from a more compact to a more extended conformation is thought to strengthen binding to ECM [44, 45]. Force sensitivity is suggested by recent work with fluid shear forces imposed on adherent cells [46]. With increasing stress, the average bond force per integrin increases, but this increase depended on exposure of a synergy site in fibronectin. This scenario is consistent with the formation of “catch-bonds” that exhibit an increased binding strength when put under mechanical load. This behavior has been directly observed at the single molecule level in other proteins [47–49], and similarly well-controlled single molecule experiments with integrins are to be anticipated.
In addition to regulating adhesion strength, integrin clustering acts as a signal involved in the formation of focal adhesions, which are currently estimated to contain around 160 component proteins [50]. When cells are stretched with a glass microneedle, they respond by locally recruiting focal adhesion proteins to shore up their attachment to substrate [3]. In this sense, focal adhesions are mechanosensitive complexes, however, changes in focal adhesion size and even the recruitment of specific proteins or the activation of particular kinases do not by themselves explain how mechanical cues are sensed since it is not yet clear how these processes could be modulated by force. Protein unfolding is one possibility.
Vinculin is one of the proteins recruited to focal adhesions when cells are stretched [51, 52] and talin, another focal adhesion component, is known to have a cryptic vinculin binding site [53]. Thus, a possible mechanism for the force dependent recruitment of vinculin to focal adhesions is the exposure of this site under force. Molecular dynamics simulations explored how this exposure could occur [54] and the hypothesis has recently gained experimental support with the finding that the rod domain of talin can be unfolded under moderate forces of ~10–100 pN in single molecule experiments [55]. Importantly, based on single molecule fluorescence during magnetic trapping and extension of proteins, vinculin is more likely to bind the talin rod domain when the rod domain is under tension. The number of binding events increased in constructs with tandem talin rod domains, while the low number of binding events to α-actinin was independent of the force, helping to rule out some possible experimental artifacts. These results are suggestive, but there are still several critical questions that remain unanswered. For example, what is the nature of the talin/vinculin binding? Simulations have provided insight [56], but experiments are lacking; rod domains with mutations in the binding sequence could help address whether the force-activated binding is similar to that observed in a talin construct that has been mutated to adopt the active conformation [57]. More importantly, a central open question is whether this mechanism is in fact operative in focal adhesions in cells. This question is now becoming addressable with new methods for the in situ labeling of exposed protein sites [9] as discussed in more detail below.
Given the complexity of the network of interactions in focal adhesions, it would be surprising if this were the only process regulated directly by force. The hypothesis that there are other critical binding interactions that are directly regulated by force, possibly through unfolding to expose cryptic sites, is made more attractive by studying the type of network motifs that are found in the ‘adhesome’. One common motif that appears 181 times by itself and is also frequently embedded in larger motifs consists of two enzymes that bind to the same scaffold protein [50]. Any time such a scaffold protein is incorporated structurally into a focal adhesion where it experiences an applied force, it could play a mechanosensitive role in which the affinity of one or both of its binding partners is altered when it completely or partially unfolds. There is recent evidence that this situation occurs for the mechanical activation of p130cas [58]. An important future task, perhaps best suited to higher throughput efforts, will be to determine which of the many candidate interactions in the adhesome are indeed directly modified by force. Interaction forces or ‘strengths’ will not provide immediate answers, however, because entropy dictates that more frequent structures or pathways will tend to be affected more often than the unitary Boltzmann factor of Eq.1 would predict. In other words, a more appropriate weight or probability P in comparing processes α and β that respectively exist Nα or Nβ times is:
| Eq.2 |
where f is the force applied to the protein a distance d along the reaction coordinate. Analogies to the width of a protein folding funnel (Fig. 3B), which represents the number of chain conformations, could perhaps be usefully formalized to multi-domain proteins in multi-protein network modules.
The cytoskeleton responds to and actively applies force
‘Integrin’ can be considered a linguistic contraction for tissue ‘integration’ in that focal adhesions provide a continuous, physical (if kinetic) link between the external ECM and the internal actin cytoskeleton. The cytoskeleton not only supports the cell and helps determine its shape, but also contains at least a few proteins that clearly respond to force and are likely involved in mechanosensing (Fig. 2D). The cytoskeleton also possesses a unique feature not present in the ECM or most focal adhesion components: it contains active assemblies that consume energy and are therefore capable of doing mechanical work [59]. This means that the cytoskeleton not only responds to force but also applies force to its surroundings [60]. This closes the mechanical loop described above and allows a cell to alter its surroundings by the forced-remodeling of proteins like fibronectin and also to probe the mechanics of its substrate [4].
One of the central components of the cytoskeleton is actin. Actin monomers polymerize to form filaments that are an important structural component of cells. For example, gels made from purified filamentous actin and actin crosslinkers recapitulate aspects of cellular mechanics [61]. Actin polymerization consumes ATP and, because it is directional, it can apply forces to the cell periphery that drive cell motility [62]. In order to control cell morphology and to help regulate this complex actin dynamics, a host of actin binding proteins regulate its assembly and interact with other pathways [63]. An interesting example that has received significant attention is filamin.
Filamin consists of a tandem array of Ig-like domains with an actin binding domain on one end and a homo-dimerization domain on the other. When mixed with filamentous actin, filamin forms cross-links between filaments and readily promotes gelation [64]. Actin-filamin gels are more resilient and stiffer than actin gels alone or gels formed with other actin binding proteins [65, 66] suggesting that one important role for filamin is in regulating the mechanical properties of the actin cytoskeleton. Indeed, several studies have found that filamin impacts cell mechanics [67–69] and filamin A expression has been shown to be required for active cell stiffening in response to substrate mechanics but not for passive stiffening in response to an external force [70]. For Dictyostelium discoideum filamin (ddFLN), this seems to be its main function as it is only known to bind to one other protein in addition to actin [71]. In contrast to ddFLN, human filamin has a more elaborate structure including hinge regions with important mechanical consequences and 24 instead of 4 Ig-like repeats. More than 20 binding partners have been identified for human filamin [72, 73]. These include proteins with a range of functions, including signaling. Particularly relevant here, filamin has also been shown to compete with talin in binding the cytoplasmic domain of integrin [74]. Thus there is a direct link between filamin and proteins known to be critical in mechanosensing. Given this connection with focal adhesions and the cytoskeleton as well as filamin’s structural similarity with Ig domains to proteins like fibronectin, it is natural to ask how filamin behaves under force and whether this behavior has functional consequences.
The mechanics of filamin have been measured at the single molecule level using AFM, and the Ig domains were found to unfold in an abrupt all-or-nothing manner at forces of order 100 pN [75], consistent with molecular dynamics simulations [76]. An important finding from work on ddFLN is that the force required to break the dimerization interaction is larger than the force required for unfolding [77], suggesting that filamin could unfold in vivo. However, these earlier AFM experiments could not address the strength of the filamin-actin interaction under load and therefore could not rule out the possibility that this interaction is the weakest link.
More recently, the mechanics of filamin have been measured in an elegant native-like system in which single filamin molecules are connected on one end to a fixed actin filament and on the other to a filament connected to a bead [78]. When the bead is trapped using optical tweezers and the sample stage is moved, the force required to either break the filamin-actin bond or to unfold one or more of the domains of filamin can be measured. One reason that this experiment is significant is that it gets directly to the issue of competition between alternative stress-release mechanisms. In this geometry at the rates studied, filamin was found to unbind most of the time, but unfolding was also observed. Similarity in the force required for unfolding and unbinding could have functional consequences in regulating cytoskeletal remodeling under force and highlights the importance of thinking about timing, rates, and kinetics. Furthermore, it is not known whether any of the proteins that bind the filamin repeats affect their stability or conversely whether applied stress can affect the binding affinity of the filamin binders. This question becomes especially interesting in light of recent work that suggests that filamin can serve to trap a transcription factor (i.e. PEBP2/CBF) in the cytoplasm [79], thereby regulating its action in the nucleus. If these results are confirmed, force-induced shifts in transcription factor binding could provide an elegant mechanism for mechanics to alter gene-expression, which is known to occur in (filamin-expressing) mesenchymal stem cells in response to substrate stiffness [5].
Mechanical regulation of a transcription factor has already been observed in a cardiomyocyte [80], which is one type of mesenchymal cell. A prototypical kinase domain in the sarcomeric protein titin can be activated by cell contractility to bind a signaling complex that ultimately regulates a transcription factor well-known to be regulated by the cytoskeleton, namely serum response factor (SRF). Molecular dynamics calculations [81] and single molecule AFM provide insight into a possible mechanism, suggesting that the kinase domain unfolds at lower force than individual immunoglobulin domains of titin [82] – although Eq.2 would suggest some of titin’s ~200 Ig domains are likely to unfold if the kinase domain unfolds. Mutations in the titin kinase domain interfere with the signaling pathway and lead to a hereditary muscle disease in humans [80], which implicates force-controlled unfolding of this domain and perhaps also some of titin’s Ig domains.
Another class of actin-binding proteins that have been relatively well studied also contain an actin-binding domain followed by a series of compact repeats, but instead of Ig-like domains of β-strands, they consist predominantly of α-helical bundles. The spectrin superfamily of proteins with their triple-helical bundle domains is prototypical: α-actinins are the shortest and ubiquitous isoforms, while spectrins – originally isolated from red blood cell membranes – are generally found at all cell membranes and are well known to be essential for membrane stability. Additional members of the family include dystrophin, which links the contractile apparatus of striated muscle to ECM, and nesprins, which link the nuclear envelope to the actin cytoskeleton. Dystrophin deficiency is the major cause of muscular dystrophy, with symptoms emerging at a few years of age when mechanical stresses of growth become critical. All proteins play mechanical roles in vivo, and a series of single molecule experiments have been directed at understanding the molecular bases of these functions [83–87]. All spectrin family proteins tested thusfar have been found to be remarkably labile, unfolding at forces around 15–30 pN even at the high loading rates typical in AFM experiments. Compared to β-sheet Ig-like domains, the force required to separate the helices of the spectrin bundles are relatively small and the helices themselves unfold easily because the hydrogen bonds holding them together are arranged in series in the direction of the applied force. In contrast, the bonds holding β-strands together are arranged in parallel and must be simultaneously sheared apart when the domains are pulled at their N- and C-termini [88]. Since proteins soften at higher temperatures [89, 90], spectrin domains will unfold at even lower forces in vivo. In fact, an exhaustive study of the thermal stability of the domains of spectrin in humans found that several domains have melting temperatures ≤ 37°C [91]; most single molecule experiments have been done at room temperature and will miss such effects. Strong effects of temperature (beyond kBT) and the stability of multiple serial versus parallel bonds are important to consider in future experiments with single molecules as well as in extrapolations or direct study of cellular structures.
Towards a Characterization of the Cellular ‘Unfoldome’
Despite the many insights obtained to date in vitro, the difficulty of determining whether any domains are indeed wholly or partly unfolded in either cells or ECM highlights a crucial deficit in our knowledge. New methods are therefore being developed to assess the conformational state of proteins in cells and thus extend the knowledge gained in vitro closer to the relevant physiological environment. However, even with the relatively small number of cases that have been examined in some detail so far, it is clear that there is no singular protein transition or master mechanical switch at the heart of cellular mechanical responses. Instead, we are likely at the early stages of discovering what will prove to be a wide variety of force sensitive proteins and reactions. In terms of signaling networks, we have just begun to identify the machanosensitive nodes embedded in the cell’s broader system of signaling pathways. A more complete characterization of the proteins that are unfolded by external forces applied to cells or by their own myosin-driven contractility will help guide efforts to understand the molecular basis of mechanotransduction. Just as the study of focal adhesions will progress by understanding the interactions in the adhesome, the study of mechanotransduction more generally will benefit from a more complete characterization of the cellular ‘unfoldome,’ the set of proteins that can be unfolded as part of their physiological function. Although the focus of this review is on cell mechanics and transduction mechanisms, the unfoldome concept can be extended more broadly to other processes that induce changes in protein conformation or quaternary structure including heat shock and other pleiotropic perturbations.
It is possible in principle to characterize the unfoldome using experiments designed to monitor the conformation of a single protein of interest using, for example, antibodies against cryptic sites or FRET when the requisite double-labeling is possible. For example, an antibody that recognized an extended conformation of p130Cas in vitro also bound preferentially to the cell periphery where forces are expected to be highest in spreading cells [58]. However, the time and effort required to develop an assay for every protein to be investigated seems unlikely to provide significant coverage of the unfoldome in the near future. Methods are therefore required to search for a wider variety of proteins whose conformations are force sensitive.
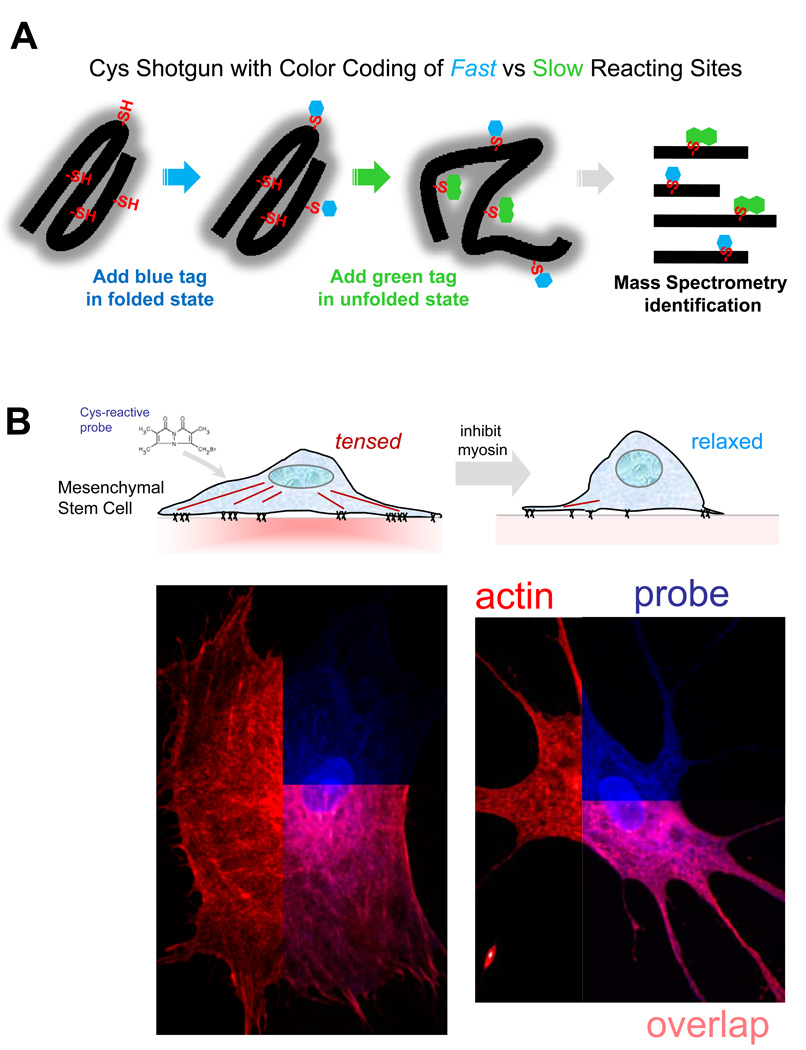
Advances in mass spectrometry based proteomics have set the stage to develop a method to screen the conformational state of a large number of proteins in living cells [9]. The experimental scheme uses cysteine accessibility as a probe (Fig. 4A): as a moderately hydrophobic amino acid, cysteines are often buried or partially buried within protein tertiary and/or quaternary structure and are thus not accessible to small molecules in the solvent. If a cysteine-reactive fluorescent dye is introduced into cells, only those cysteines that are surface-exposed will be labeled. If changing conditions alter the conformational state of a protein with a buried cysteine, a difference in dye labeling will be detected. Thus, the spatial distribution of unfolding can be monitored to some extent in cells using standard fluorescence microscopy. More importantly, mass spectrometry of tryptic fragments from cell lysates can identify not only the proteins that show a change in conformation, but even the precise cysteines that are exposed so that force sensitive domains can be mapped within proteins.
Figure 4.
(A) Cysteines are either buried within a protein fold or interaction site, or else they are exposed on the protein surface. The latter surface sites are rapidly labeled with one color before perturbation while the buried sites are then exposed and colored differently. This provides a ratiometric signal of unfolding and improves signal-to-noise ratio for exposed sites. (B) Addition of a membrane-permeable dye to adherent cells (in this case mesenchymal stem cells) labels cysteines depending on cell state, and relaxation of myosin (here with blebbistatin) leads to significant differences in protein labeling. Proteins that remain folded or assembled upon relaxation show a site-specific decrease in cysteine accessibility that is pinpointed using mass spectrometry (adapted from [9], permission pending).
When applied to the simplest possible cell, a red blood cell, this Cys shotgun approach identified a half dozen spectrin repeats that unfolded in response to physiological shear stress. Many sites in spectrin showed no change, and other membrane skeleton proteins such as actin and ankyrin also showed no detectable change. Cys shotgun can also be applied to adherent cells. Some proteins that unfold in response to cell-generated tension are expected to refold when that tension is released (Fig. 4B). This can be accomplished using a non-muscle myosin II inhibitor called blebbistatin. Cys shotgun labeling of mesenchymal stems cells attached to collagen-coated substrates in the presence or absence of blebbistatin gave a number of differential “hits” that included non-muscle myosin IIa (consistent with drug treatment preventing a conformational change in force generation), filamin, and the intermediate filament protein vimentin. The results were largely the same with embryonic cardiomyocytes that were labeled while beating spontaneously on a soft heart-like matrix or while static on a non-physiologically rigid matrix [92].
These initial studies serve as a proof-of-principle for Cys shotgun labeling of the unfoldome in cells and demonstrate the possibility of using such a proteomic-scale approach for the identification of protein conformational changes associated with a wide range of microenvironments or drug treatments. It will be important to explore mechanically perturbed proteins in other adherent cell types using blebbistatin and to examine cell responses to changes in applied strain. Cys shotgun investigations will also suggest promising targets and domains for more detailed study by other approaches. In combination with phosphorylation networks widely studied now by mass spectrometry, the approach is poised to clarify the breadth of interplay between molecular mechanics and signaling networks.
Conclusion
A wealth of information from in vitro studies now exists on the behavior of proteins and other macromolecules subjected to force. Some of the distended conformations already seem to have functional consequences ranging from the regulation of matrix assembly and cell adhesion to cytoskeleton dynamics. Proximal effects of mechanics also have ramifications for more complex cell processes that require changes in gene expression because these changes in protein conformation couple into other signaling pathways that can have a broad range of effects downstream. Despite some successes in identifying proteins involved in mechanosensing and an increasing understanding of their mechanism of action, it seems likely that there are many more to be discovered and that this search will be aided by methods that can broadly pinpoint relevant conformational changes amidst the complex background of mechanically silent cytosolic and cytoskeletal proteins—recognizing that there is no single cellular mechanosensor. Ultimately, a diverse network of proteins will likely respond in specific ways to different mechanical cues such as matrix stiffness, applied force, and fluid flow. As hinted at by the species differences in filamin, a variety of sensing mechanisms might reflect the importance of mechanical homeostasis in the evolution of larger multicellular organisms. New perspectives on human diseases – beyond heart disease [93], muscular dystrophy, and cancer [6] – are readily anticipated.
References
- 1.Pelling AE, Horton MA. An historical perspective on cell mechanics. Pflügers Arch. 2008;456:3–12. doi: 10.1007/s00424-007-0405-1. [DOI] [PubMed] [Google Scholar]
- 2.Wirtz D. Particle Tracking Microrheology of Living Cells: Principles and Applications. Ann. Rev. Biophys. 2009;38 doi: 10.1146/annurev.biophys.050708.133724. [DOI] [PubMed] [Google Scholar]
- 3.Riveline D, Zamir E, Balaban NQ, Schwarz US, Ishizaki T, Narumiya S, Kam Z, Geiger B, Bershadsky AD. Focal contacts as mechanosensors: externally applied local mechanical force induces growth of focal contacts by an mDia1-dependent and ROCK-independent mechanism. J. Cell Bio. 2001;153:1175–1186. doi: 10.1083/jcb.153.6.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Discher D, Janmey P, Wang Y. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310:1139–1143. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 5.Engler AJ, Sen S, Sweeney HL, Discher D. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 6.Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A, Reinhart-King CA, Margulies SS, Dembo M, Boettiger D, et al. Tensional homeostasis and the malignant phenotype. Cancer cell. 2005;8:241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 7.Orr AW, Helmke BP, Blackman BR, Schwartz MA. Mechanisms of mechanotransduction. Dev. Cell. 2006;10:11–20. doi: 10.1016/j.devcel.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 8.Chen CS. Mechanotransduction - a field pulling together? J. Cell Sci. 2008;121:3285–3292. doi: 10.1242/jcs.023507. [DOI] [PubMed] [Google Scholar]
- 9.Johnson CP, Tang HY, Carag C, Speicher DW, Discher D. Forced unfolding of proteins within cells. Science. 2007;317:663–666. doi: 10.1126/science.1139857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ingber DE. Cellular mechanotransduction: putting all the pieces together again. FASEB J. 2006;20:811. doi: 10.1096/fj.05-5424rev. [DOI] [PubMed] [Google Scholar]
- 11.Bershadsky A, Kozlov M, Geiger B. Adhesion-mediated mechanosensitivity: a time to experiment, and a time to theorize. Curr. Opin. Cell Bio. 2006;18:472–481. doi: 10.1016/j.ceb.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 12.Smock RG, Gierasch LM. Sending Signals Dynamically. Science. 2009;324:198–203. doi: 10.1126/science.1169377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rief M, Gautel M, Oesterhelt F, Fernandez JM, Gaub HE. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 1997;276:1109–1112. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- 14.Kellermayer MSZ, Smith SB, Granzier HL, Bustamante C. Folding-unfolding transitions in single titin molecules characterized with laser tweezers. Science. 1997;276:1112–1116. doi: 10.1126/science.276.5315.1112. [DOI] [PubMed] [Google Scholar]
- 15.Neuman KC, Nagy A. Single-molecule force spectroscopy: optical tweezers, magnetic tweezers and atomic force microscopy. Nat. Meth. 2008;5:491–505. doi: 10.1038/nmeth.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bell GI. Models for specific adhesion of cells to cells. Science. 1978;200:618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- 17.Evans EA, Calderwood DA. Forces and bond dynamics in cell adhesion. Science. 2007;316:1148–1153. doi: 10.1126/science.1137592. [DOI] [PubMed] [Google Scholar]
- 18.Zhong C, Chrzanowska-Wodnicka M, Brown J, Shaub A, Belkin AM, Burridge K. Rho-mediated contractility exposes a cryptic site in fibronectin and induces fibronectin matrix assembly. J. Cell Bio. 1998;141:539–551. doi: 10.1083/jcb.141.2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ejim OS, Blunn GW, Brown RA. Production of artificial-oriented mats and strands from plasma fibronectin - a morphological study. Biomaterials. 1993;14:743–748. doi: 10.1016/0142-9612(93)90038-4. [DOI] [PubMed] [Google Scholar]
- 20.Baneyx G, Vogel V. Self-assembly of fibronectin into fibrillar networks underneath dipalmitoyl phosphatidylcholine monolayers: Role of lipid matrix and tensile forces. Proc. Nat. Acad. Sci. USA. 1999;96:12518–12523. doi: 10.1073/pnas.96.22.12518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Erickson HP. Reversible unfolding of fibronectin type III and immunoglobulin domains provides the structural basis for stretch and elasticity of titin and fibronectin. Proc. Nat. Acad. Sci. USA. 1994;91:10114–10118. doi: 10.1073/pnas.91.21.10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krammer A, Lu H, Isralewitz B, Schulten K, Vogel V. Forced unfolding of the fibronectin type III module reveals a tensile molecular recognition switch. Proc. Nat. Acad. Sci. USA. 1999;96:1351–1356. doi: 10.1073/pnas.96.4.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao M, Craig D, Vogel V, Schulten K. Identifying Unfolding Intermediates of FN-III10 by Steered Molecular Dynamics. J. Mol. Biol. 2002;323:939–950. doi: 10.1016/s0022-2836(02)01001-x. [DOI] [PubMed] [Google Scholar]
- 24.Oberhauser AF, Marszalek PE, Erickson HP, Fernandez JM. The molecular elasticity of the extracellular matrix protein tenascin. Nature. 1998;393:181–185. doi: 10.1038/30270. [DOI] [PubMed] [Google Scholar]
- 25.Baneyx G, Baugh L, Vogel V. Fibronectin extension and unfolding within cell matrix fibrils controlled by cytoskeletal tension. Proc. Nat. Acad. Sci.USA. 2002;99:5139–5143. doi: 10.1073/pnas.072650799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith M, Gourdon D, Little W, Kubow K, Eguiluz RA, Luna-Morris S, Vogel V. Force-induced unfolding of fibronectin in the extracellular matrix of living cells. PLoS Biol. 2007;5:e268. doi: 10.1371/journal.pbio.0050268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hynes RO, Yamada KM. Fibronectins: multifunctional modular glycoproteins. J. Cell Bio. 1982;95:369–377. doi: 10.1083/jcb.95.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 29.Vogel V. Mechanotransduction involving multimodular proteins: converting force into biochemical signals. Ann. Rev. Biophys. Biomol. Struct. 2006;35:459–488. doi: 10.1146/annurev.biophys.35.040405.102013. [DOI] [PubMed] [Google Scholar]
- 30.Oberhauser AF, Badilla-Fernandez C, Carrion-Vazquez M, Fernandez JM. The mechanical hierarchies of fibronectin observed with single-molecule AFM. J. Mol. Biol. 2002;319:433–447. doi: 10.1016/S0022-2836(02)00306-6. [DOI] [PubMed] [Google Scholar]
- 31.Berry MF, Engler AJ, Woo YJ, Pirolli TJ, Bish LT, Jayasankar V, Morine KJ, Gardner TJ, Discher D, Sweeney HL. Mesenchymal stem cell injection after myocardial infarction improves myocardial compliance. Am. J. Phys. Heart and Circ.Phys. 2006;290:H2196–H2203. doi: 10.1152/ajpheart.01017.2005. [DOI] [PubMed] [Google Scholar]
- 32.Singer AJ, Clark RA. Cutaneous wound healing. New Eng. J. Med. 1999;341:738–746. doi: 10.1056/NEJM199909023411006. [DOI] [PubMed] [Google Scholar]
- 33.Brown AEX, Litvinov RI, Discher DE, Weisel JW. Forced unfolding of coiled-coils in fibrinogen by single-molecule AFM. Biophys. J. 2007;92:L39–L41. doi: 10.1529/biophysj.106.101261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lim BBC, Lee EH, Sotomayor M, Schulten K. Molecular basis of fibrin clot elasticity. Structure. 2008;16:449–459. doi: 10.1016/j.str.2007.12.019. [DOI] [PubMed] [Google Scholar]
- 35.Brown AEX, Litvinov RI, Discher DE, Purohit P, Weisel JW. Science. 2009 doi: 10.1126/science.1172484. to appear. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Joel SB. Platelet-Fibrinogen Interactions. Ann. NY Acad. Sci. 2001;936:340–354. doi: 10.1111/j.1749-6632.2001.tb03521.x. [DOI] [PubMed] [Google Scholar]
- 37.Springer TA, Zhu J, Xiao T. Structural basis for distinctive recognition of fibrinogen gammaC peptide by the platelet integrin alphaIIbbeta3. J. Cell Bio. 2008;182:791–800. doi: 10.1083/jcb.200801146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu J, Rodriguez D, Petitclerc E, Kim JJ, Hangai M, Yuen SM, Davis GE, Brooks PC. Proteolytic exposure of a cryptic site within collagen type IV is required for angiogenesis and tumor growth in vivo. J. Cell Bio. 2001;154:1069. doi: 10.1083/jcb.200103111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buehler MJ. Nature designs tough collagen: explaining the nanostructure of collagen fibrils. Proc. Nat. Acad. Sci. USA. 2006;103:12285–12290. doi: 10.1073/pnas.0603216103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wipff P-J, Rifkin DB, Meister J-J, Hinz B. Myofibroblast contraction activates latent TGF- 1 from the extracellular matrix. J. Cell Biol. 2007;179:1311–1323. doi: 10.1083/jcb.200704042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Benoit M, Gabriel D, Gerisch G, Gaub HE. Discrete interactions in cell adhesion measured by single-molecule force spectroscopy. Nat Cell Biol. 2000;2:313–317. doi: 10.1038/35014000. [DOI] [PubMed] [Google Scholar]
- 42.Helenius J, Heisenberg C-P, Gaub HE, Muller DJ. Single-cell force spectroscopy. J Cell Sci. 2008;121:1785–1791. doi: 10.1242/jcs.030999. [DOI] [PubMed] [Google Scholar]
- 43.Schmitz J, Gottschalk KE. Mechanical regulation of cell adhesion. Soft Matter. 2008;4:1373–1387. doi: 10.1039/b716805p. [DOI] [PubMed] [Google Scholar]
- 44.Sims PJ, Ginsberg MH, Plow EF, Shattil SJ. Effect of platelet activation on the conformation of the plasma membrane glycoprotein IIb-IIIa complex. J. Biol. Chem. 1991;266:7345–7352. [PubMed] [Google Scholar]
- 45.Hato T, Pampori N, Shattil SJ. Complementary Roles for Receptor Clustering and Conformational Change in the Adhesive and Signaling Functions of Integrin alpha IIbbeta 3. J. Cell Biol. 1998;141:1685–1695. doi: 10.1083/jcb.141.7.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Friedland JC, Lee MH, Boettiger D. Mechanically activated integrin switch controls alpha5beta1 function. Science. 2009;323:642–644. doi: 10.1126/science.1168441. [DOI] [PubMed] [Google Scholar]
- 47.Marshall BT, Long M, Piper JW, Yago T, McEver RP, Zhu C. Direct observation of catch bonds involving cell-adhesion molecules. Nature. 2003;423:190–193. doi: 10.1038/nature01605. [DOI] [PubMed] [Google Scholar]
- 48.Yakovenko O, Sharma S, Forero M, Tchesnokova V, Aprikian P, Kidd B, Mach A, Vogel V, Sokurenko E, Thomas WE. FimH Forms Catch Bonds That Are Enhanced by Mechanical Force Due to Allosteric Regulation. J. Biol. Chem. 2008;283:11596–11605. doi: 10.1074/jbc.M707815200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Evans E, Leung A, Heinrich V, Zhu C. Mechanical switching and coupling between two dissociation pathways in a P-selectin adhesion bond. Proc. Nat. Acad. Sci. USA. 2004;101:11281–11286. doi: 10.1073/pnas.0401870101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zaidel-Bar R, Itzkovitz S, Ma'ayan A, Iyengar R, Geiger B. Functional atlas of the integrin adhesome. Nat. Cell Bio. 2007;9:858–867. doi: 10.1038/ncb0807-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sawada Y, Sheetz MP. Force transduction by Triton cytoskeletons. J. Cell Bio. 2002;156:609–615. doi: 10.1083/jcb.200110068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Galbraith CG, Yamada KM, Sheetz MP. The relationship between force and focal complex development. J. Cell Bio. 2002;159:695–705. doi: 10.1083/jcb.200204153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gingras AR, Ziegler WH, Frank R, Roberts GCK, Critchley DR, Emsley J. Mapping and consensus sequence identification for multiple vinculin binding sites within the talin rod. J. Biol. Chem. 2005 doi: 10.1074/jbc.M508060200. M508060200. [DOI] [PubMed] [Google Scholar]
- 54.Lee SE, Kamm RD, Mofrad MR. Force-induced activation of talin and its possible role in focal adhesion mechanotransduction. J. Biomech. 2007;40:2096–2106. doi: 10.1016/j.jbiomech.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 55.del Rio A, Perez-Jimenez R, Liu R, Roca-Cusachs P, Fernandez JM, Sheetz MP. Stretching single talin rod molecules activates vinculin binding. Science. 2009;323:638–641. doi: 10.1126/science.1162912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee SE, Chunsrivirot S, Kamm RD, Mofrad MR. Molecular dynamics study of talin-vinculin binding. Biophys. J. 2008;95:2027–2036. doi: 10.1529/biophysj.107.124487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Papagrigoriou E, Gingras AR, Barsukov IL, Bate N, Fillingham IJ, Patel B, Frank R, Ziegler WH, Roberts GC, Critchley DR, et al. Activation of a vinculin-binding site in the talin rod involves rearrangement of a five-helix bundle. EMBO J. 2004;23:2942–2951. doi: 10.1038/sj.emboj.7600285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sawada Y, Tamada M, Dubin-Thaler BJ, Cherniavskaya O, Sakai R, Tanaka S, Sheetz MP. Force sensing by mechanical extension of the Src family kinase substrate p130Cas. Cell. 2006;127:1015–1026. doi: 10.1016/j.cell.2006.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pellegrin S, Mellor H. Actin stress fibres. J. Cell Sci. 2007;120:3491–3499. doi: 10.1242/jcs.018473. [DOI] [PubMed] [Google Scholar]
- 60.Wang JH, Lin JS. Cell traction force and measurement methods. Biomech. Model Mechanobiol. 2007;6:361–371. doi: 10.1007/s10237-006-0068-4. [DOI] [PubMed] [Google Scholar]
- 61.Gardel ML, Nakamura F, Hartwig JH, Crocker JC, Stossel TP, Weitz DA. Prestressed F-actin networks cross-linked by hinged filamins replicate mechanical properties of cells. Proc. Nat. Acad. Sci. USA. 2006;103:1762–1767. doi: 10.1073/pnas.0504777103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rafelski SM, Theriot JA. Crawling toward a unified model of cell mobility: spatial and temporal regulation of actin dynamics. Ann. Rev. Biochem. 2004;73:209–239. doi: 10.1146/annurev.biochem.73.011303.073844. [DOI] [PubMed] [Google Scholar]
- 63.Revenu C, Athman R, Robine S, Louvard D. The co-workers of actin filaments: from cell structures to signals. Nat. Rev. Mol. Cell Biol. 2004;5:635–646. doi: 10.1038/nrm1437. [DOI] [PubMed] [Google Scholar]
- 64.Shizuta Y, Shizuta H, Gallo M, Davies P, Pastan I. Purification and properties of filamin, and actin binding protein from chicken gizzard. J. Biol. Chem. 1976;251:6562–6567. [PubMed] [Google Scholar]
- 65.Tseng Y, An KM, Esue O, Wirtz D. The Bimodal Role of Filamin in Controlling the Architecture and Mechanics of F-actin Networks. J. Biol. Chem. 2004;279:1819–1826. doi: 10.1074/jbc.M306090200. [DOI] [PubMed] [Google Scholar]
- 66.Janmey PA, Hvidt S, Lamb J, Stossel TP. Resemblance of actin-binding protein/actin gels to covalently crosslinked networks. Nature. 1990;345:89–92. doi: 10.1038/345089a0. [DOI] [PubMed] [Google Scholar]
- 67.Cunningham CC, Gorlin JB, Kwiatkowski DJ, Hartwig JH, Janmey PA, Byers HR, Stossel TP. Actin-binding protein requirement for cortical stability and efficient locomotion. Science. 1992;255:325–327. doi: 10.1126/science.1549777. [DOI] [PubMed] [Google Scholar]
- 68.Tandon R, Levental I, Huang C, Byfield FJ, Ziembicki J, Schelling JR, Bruggeman LA, Sedor JR, Janmey PA, Miller RT. HIV infection changes glomerular podocyte cytoskeletal composition and results in distinct cellular mechanical properties. Am J Physiol Renal Physiol. 2007;292:F701–F710. doi: 10.1152/ajprenal.00246.2006. [DOI] [PubMed] [Google Scholar]
- 69.Coughlin MF, Puig-de-Morales M, Bursac P, Mellema M, Millet E, Fredberg JJ. Filamin-A and Rheological Properties of Cultured Melanoma Cells. Biophys. J. 2006;90:2199–2205. doi: 10.1529/biophysj.105.061267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kasza KE, Nakamura F, Hu S, Kollmannsberger P, Bonakdar N, Fabry B, Stossel TP, Wang N, Weitz DA. Filamin A Is Essential for Active Cell Stiffening but not Passive Stiffening under External Force. Biophys. J. 2009;96:4326–4335. doi: 10.1016/j.bpj.2009.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Knuth M, Khaire N, Kuspa A, Lu SJ, Schleicher M, Noegel AA. A novel partner for Dictyostelium filamin is an alpha-helical developmentally regulated protein. J. Cell Sci. 2004:5013–5022. doi: 10.1242/jcs.01366. [DOI] [PubMed] [Google Scholar]
- 72.Popowicz GM, Schleicher M, Noegel A, Holak TA. Filamins: promiscuous organizers of the cytoskeleton. Trends Biochem. Sci. 2006;31:411–419. doi: 10.1016/j.tibs.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 73.Stossel TP, Condeelis J, Cooley L, Hartwig JH, Noegel A, Schleicher M, Shapiro SS. Filamins as integrators of cell mechanics and signalling. Nat. Rev. Mol. Cell Biol. 2001;2:138–145. doi: 10.1038/35052082. [DOI] [PubMed] [Google Scholar]
- 74.Kiema T, Lad Y, Jiang P, Oxley CL, Baldassarre M, Wegener KL, Campbell ID, Ylänne J, Calderwood DA. The Molecular Basis of Filamin Binding to Integrins and Competition with Talin. Mol. Cell. 2006;21:337–347. doi: 10.1016/j.molcel.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 75.Furuike S, Ito T, Yamazaki M. Mechanical unfolding of single filamin A (ABP-280) molecules detected by atomic force microscopy. FEBS Lett. 2001;498:72–75. doi: 10.1016/s0014-5793(01)02497-8. [DOI] [PubMed] [Google Scholar]
- 76.Kolahi KS, Mofrad MR. Molecular mechanics of filamin's rod domain. Biophys. J. 2008;94:1075–1083. doi: 10.1529/biophysj.107.118802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schwaiger I, Kardinal A, Schleicher M, Noegel AA, Rief M. A mechanical unfolding intermediate in an actin-crosslinking protein. Nat. Struct. Mol. Biol. 2004;11:81–85. doi: 10.1038/nsmb705. [DOI] [PubMed] [Google Scholar]
- 78.Ferrer JM, Lee H, Chen J, Pelz B, Nakamura F, Kamm RD, Lang MJ. Measuring molecular rupture forces between single actin filaments and actin-binding proteins. Proc. Nat. Acad. Sci. USA. 2008;105:9221–9226. doi: 10.1073/pnas.0706124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yoshida N, Ogata T, Tanabe K, Li S, Nakazato M, Kohu K, Takafuta T, Shapiro S, Ohta Y, Satake M, et al. Filamin A-Bound PEBP2 beta/CBF beta Is Retained in the Cytoplasm and Prevented from Functioning as a Partner of the Runx1 Transcription Factor. Mol. Cell. Biol. 2005;25:1003–1012. doi: 10.1128/MCB.25.3.1003-1012.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lange S, Xiang F, Yakovenko A, Vihola A, Hackman P, Rostkova E, Kristensen J, Brandmeier B, Franzen G, Hedberg B, et al. The Kinase Domain of Titin Controls Muscle Gene Expression and Protein Turnover. Science. 2005;308:1599–1603. doi: 10.1126/science.1110463. [DOI] [PubMed] [Google Scholar]
- 81.Gräter F, Shen J, Jiang H, Gautel M, Grubmüller H. Mechanically Induced Titin Kinase Activation Studied by Force-Probe Molecular Dynamics Simulations. 2005;88:790–804. doi: 10.1529/biophysj.104.052423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Puchner EM, Alexandrovich A, Kho AL, Hensen U, Schäfer LV, Brandmeier B, Gräter F, Grubmüller H, Gaub HE, Gautel M. Mechanoenzymatics of titin kinase. Proc. Nat. Acad. Sci. USA. 2008;105:13385–13390. doi: 10.1073/pnas.0805034105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rief M, Pascual J, Saraste M, Gaub HE. Single molecule force spectroscopy of spectrin repeats: Low unfolding forces in helix bundles. J. Mol. Bio. 1999;286:553–561. doi: 10.1006/jmbi.1998.2466. [DOI] [PubMed] [Google Scholar]
- 84.Law R, Carl P, Harper S, Dalhaimer P, Speicher DW, Discher DE. Cooperativity in Forced Unfolding of Tandem Spectrin Repeats. Biophys. J. 2003;84:533–544. doi: 10.1016/S0006-3495(03)74872-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Randles LG, Rounsevell RWS, Clarke J. Spectrin domains lose cooperativity in forced unfolding. Biophys. J. 2007;92:571–577. doi: 10.1529/biophysj.106.093690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Law R, Harper S, Speicher DW, Discher DE. Influence of Lateral Association on Forced Unfolding of Antiparallel Spectrin Heterodimers. J. Biol. Chem. 2004;279:16410–16416. doi: 10.1074/jbc.M313107200. [DOI] [PubMed] [Google Scholar]
- 87.Bhasin N, Law R, Liao G, Safer D, Ellmer J, Discher BM, Sweeney HL, Discher DE. Molecular extensibility of mini-dystrophins and a dystrophin rod construct. J. Mol. Bio. 2005;352:795–806. doi: 10.1016/j.jmb.2005.07.064. [DOI] [PubMed] [Google Scholar]
- 88.Lu H, Isralewitz B, Krammer A, Vogel V, Schulten K. Unfolding of Titin Immunoglobulin Domains by Steered Molecular Dynamics Simulation. Biophys. J. 1998;75:662–671. doi: 10.1016/S0006-3495(98)77556-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Law R, Liao G, Harper S, Yang G, Speicher DW, Discher DE. Pathway Shifts and Thermal Softening in Temperature-Coupled Forced Unfolding of Spectrin Domains. Biophys. J. 2003;85:3286–3293. doi: 10.1016/S0006-3495(03)74747-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schlierf M, Rief M. Temperature softening of a protein in single-molecule experiments. J. Mol. Bio. 2005;354:497–503. doi: 10.1016/j.jmb.2005.09.070. [DOI] [PubMed] [Google Scholar]
- 91.An X, Guo X, Zhang X, Baines AJ, Debnath G, Moyo M, Salomao M, Bhasin N, Johnson C, Discher D, et al. Conformational stabilities of the structural repeats of erythroid spectrin and their functional implications. J. Biol. Chem. 2006 doi: 10.1074/jbc.M513725200. M513725200. [DOI] [PubMed] [Google Scholar]
- 92.Engler AJ, Carag-Krieger C, Johnson CP, Raab M, Tang H-Y, Speicher DW, Sanger JW, Sanger JM, Discher DE. Embryonic cardiomyocytes beat best on a matrix with heart-like elasticity: scar-like rigidity inhibits beating. J. Cell Sci. 2008;121:3794–3802. doi: 10.1242/jcs.029678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Collet JP, Allali Y, Lesty C, Tanguy ML, Silvain J, Ankri A, Blanchet B, Dumaine R, Gianetti J, Payot L, et al. Altered Fibrin Architecture Is Associated With Hypofibrinolysis and Premature Coronary Atherothrombosis. Arterioscler. Thromb. Vasc. Biol. 2006;26:2567–2573. doi: 10.1161/01.ATV.0000241589.52950.4c. [DOI] [PubMed] [Google Scholar]