Summary
The SMN complex is essential for the biogenesis of small nuclear ribonucleoproteins (snRNPs), the major constituents of the spliceosome. Deficiency in functional SMN protein causes spinal muscular atrophy (SMA), a common motor neuron degenerative disease of severity commensurate with SMN levels, and correspondingly, snRNP assembly decrease. We developed a high throughput screen for snRNP assembly modifiers and discovered that reactive oxygen species (ROS) inhibit SMN complex activity in a dose-dependent manner. ROS-generating compounds, e.g, the environmental toxins menadione and β-lapachone (in vivo IC50=0.45 μM) also cause intermolecular disulfide crosslinking of SMN. Both the oxidative inactivation and SMN crosslinking can be reversed by reductants. We identified two cysteines that form SMN-SMN disulfide crosslinks, defining specific contact points in oligomeric SMN. Thus, the SMN complex is a redox-sensitive assemblyosome and a novel ROS target, suggesting that it may play a role in oxidative stress pathophysiology, which is associated with many degenerative diseases.
Introduction
Genetic lesions in the survival of motor neurons gene (SMN1) that decrease the levels of functional SMN protein are the cause of spinal muscular atrophy (SMA), a common motor neuron degenerative disease and a leading genetic cause of infant mortality (Cifuentes-Diaz et al., 2002; Lefebvre et al., 1995; Talbot and Davies, 2001; Wirth et al., 2006). SMN, as an oligomeric protein, nucleates the SMN complex, which includes at least seven additional proteins called Gemins (Liu et al., 1997; Meister et al., 2002; Yong et al., 2004). SMN is ubiquitously expressed, present in all tissues and is essential for viability of all cells in eukaryotes (Schrank et al., 1997), yet SMN deficiency selectively affects motor neurons (Monani, 2005). Although a specific function for the SMN complex in motor neurons cannot be ruled out as the reason for the pathology of SMA, a definitive body of evidence indicates that the SMN complex has a fundamental role in cellular RNA metabolism, particularly in the biogenesis of RNA-protein complexes (RNPs) and post-transcriptional gene regulation (Zhang et al., 2008). The best characterized function of the SMN complex is in the assembly of small nuclear ribonucleoprotein particles (snRNPs) (Meister et al., 2001; Meister et al., 2002; Pellizzoni et al., 2002; Yong et al., 2004).
snRNPs are the major components of the spliceosome, the machinery that carries out pre-mRNA splicing (Nilsen, 2003; Tarn and Steitz, 1997; Will and Luhrmann, 2001). Each of the snRNPs, U1, U2, U4, U5, U4atac, U11 and U12, is comprised of one snRNA, a heptameric ring structure of Sm proteins (the Sm core), and several U snRNP-specific proteins (Kambach et al., 1999). The key step in snRNP biogenesis, the assembly of the common Sm proteins B/B′, D1, D2, D3, E, F, and G into a stable Sm core on a conserved uridine-rich sequence of the snRNA (the Sm site), occurs in the cytoplasm after the newly transcribed snRNAs are exported from the nucleus. Sm cores, which are remarkably stable, can form spontaneously in vitro with Sm proteins and RNA alone (Raker et al., 1999; Sumpter et al., 1992), but lack the necessary specificity for snRNAs. In cells, illicit Sm core formation is prevented and stringent specificity towards snRNAs is conferred by the SMN complex (Pellizzoni et al., 2002), which binds directly to specific sequences in the snRNAs (the snRNP code) (Golembe et al., 2005) and to the Sm proteins and carries the snRNP assembly reaction. SMN complex-mediated snRNP assembly is ATP-dependent, and can be recapitulated in vitro with cell extracts (Meister et al., 2001; Pellizzoni et al., 2002). Quantitative measurements of snRNP assembly (Wan et al., 2005) revealed that the extent of Sm core assembly strictly depends on the amount of SMN protein and is modulated by components of the SMN complex (Battle et al., 2006; Feng et al., 2005; Wan et al., 2005). Indeed, cells of SMA patients have lower snRNP assembly capacity corresponding directly to the lower amount of SMN protein they contain (Wan et al., 2005). These findings provided evidence for a biochemical deficit in cells of SMA patients, and demonstrated that the activity of the SMN complex in snRNP assembly is relevant to the pathogenesis of SMA. Other observations further supported the link between the deficiency in the biogenesis of snRNPs and the pathogenesis of SMA (Winkler et al., 2005).
Small molecules have been critically important for understanding the mechanism and regulation of diverse cellular processes, including those involving protein-nucleic acid interactions such as transcription and translation. Here we describe a high throughput assay to study the regulation of the SMN complex and the mechanism of snRNP assembly. Using this assay for high throughput screening (HTS), we identified small molecules that selectively inhibit the SMN complex, and their characterization revealed that reactive oxygen species (ROS) cause intermolecular disulfide crosslinking of SMN and inactivation of the SMN complex, both of which are prevented by reducing agents. Mutational analysis of oxidized SMN identified the positions of disulfide crosslinks and indicated contact points in SMN-SMN oligomers, demonstrating that ROS provide a powerful tool for investigating the structure of the SMN complex. ROS have been linked to numerous diseases, particularly neurodegenerative diseases and aging. Our findings reveal SMN as a novel target for ROS and indicate that oxidative stress causes a functional inactivation of the SMN complex similar to that caused by SMN protein deficiency. ROS may thus mimic or exacerbate the SMA phenotype and it is likely that the redox-sensitive SMN complex plays a potentially significant role in oxidative stress physiology and associated pathologies.
Results
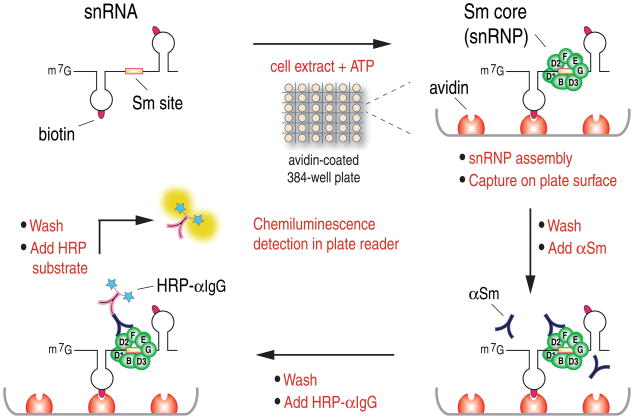
High throughput screening for small molecule modulators of SMN complex activity in snRNP assembly
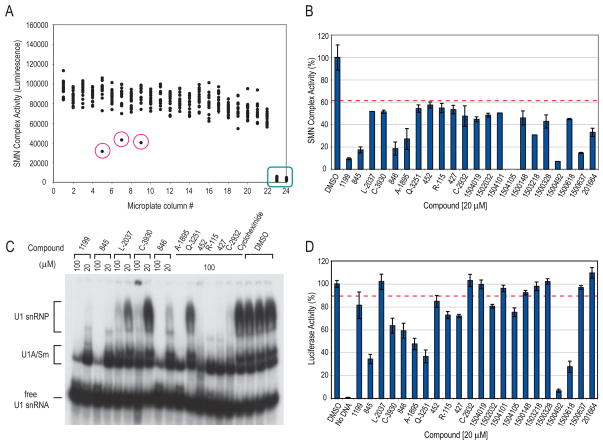
We have previously described a quantitative snRNP (Sm core) assembly assay based on quantitation of biotinylated snRNAs on which Sm cores assembled and subsequently captured on magnetic beads in 96-well plates (Wan et al., 2005). In order to perform HTS for small molecule modulators of SMN complex activity, we developed a different snRNP assembly assay that can be performed in 384- or 1536-well plate format. The scheme of this assay, illustrated in Figure 1, consists of incubating snRNAs, which are produced and labeled with biotins by in vitro transcription, with cell extracts in the presence of ATP for Sm core assembly to occur. The reactions are performed in avidin-coated microplates, which capture the RNAs onto the plate surface by avidin-biotin binding. After washing at high stringency with a buffer that spares only the highly stable Sm cores, the amount of Sm cores that assemble on the captured RNAs is determined with anti-Sm antibody (Y12), which is then detected with horseradish peroxidase (HRP)-conjugated secondary antibody. The HRP provides enzymatic amplification of a chemiluminescent substrate and the signal in each well is measured in a plate reader. Readings of a representative 384-well plate are shown in Figure 2A. Each dot represents the luminescence signal from a single well, which contains a standard assembly reaction with HeLa cell extract and U4 snRNA in the presence of a compound. The ratio of signal to background (assembly reactions without cell extracts, indicated in the green box) obtained in this assay is approximately 30, the standard deviation (SD) from all the assay wells is 13.17%, and the Z′ factor is 0.54. Compounds that have significant effects can easily be identified (dots circled in red), demonstrating that this assay is sensitive and robust, and therefore suitable for high throughput screens. Subsequent technical improvements, including direct conjugation of HRP to Y12, resulted in further improved assay parameters and simplified assay procedures. Initial screening was performed on a collection of ~5,000 bioactive compounds at a final concentration of 20 μM in triplicate. In this collection, 22 compounds strongly inhibited the activity of the SMN complex (>3SD; Figure 2B), but none were found that significantly increased it. The inhibitory activities of these compounds were confirmed and validated in magnetic beads snRNP assembly assay (Wan et al., 2005). Further validation of the HTS method was provided using gel mobility shift assay, a standard method for studying snRNP assembly (Kleinschmidt et al., 1989; Raker et al., 1999), with both U1 and U4 snRNAs (Figure 2C and data not shown).
Figure 1.
Experimental scheme of the high throughput SMN complex activity assay for detection of in vitro assembled snRNPs in 384-well microplate format (see text for details)
Figure 2. High throughput screening for small molecule modulators of the activity of the SMN complex in snRNP assembly.
(A)Scatter plot of a representative 384-well microplate from a chemical library screening. Each dot represents the signal from one well containing a standard assembly reaction mixture in the presence of individual compounds at 20 μM. Green box indicates reactions lacking cell extracts, representing non-specific background of the assay. Red circles indicate wells containing compounds that significantly decreased SMN complex activity.
(B) High throughput screening and selection of potential inhibitors of SMN complex activity. ~5,000 compounds were screened in triplicate at 20 μM. As shown, 22 compounds were selected as potential inhibitors based on the criteria that they inhibited SMN complex activity by more than 3 times the standard deviation (>3SD) of the assay, as indicated by the red dotted line. The compounds are labeled according to library nomenclature. Error bars represent SDs from triplicate samples.
(C) Validation of potential SMN complex inhibitors by gel mobility shift assay using [32P]UTP-labeled U1 snRNA mixed with HeLa total cell extracts in the presence of 20 or 100 μM compound, or cycloheximide or DMSO controls.
(D) Assessment of selectivity of potential SMN complex inhibitors by in vitro transcription and translation assay. Reactions were set up using luciferase DNA as a reporter in the presence of either 20 μM compound or DMSO control. Luciferase activities of the in vitro produced proteins were measured and compared to DMSO control (100% activity). The SD of this assay is 3.5%. Error bars represent SDs from triplicate measurements of each compound. Red dotted line indicates 3SD.
To assess the selectivity of the compounds and eliminate general inhibitors of ATP-dependent processes and RNA-protein interactions, we set up a counter-screening assay. For this, we carried out in vitro transcription and translation of luciferase DNA in rabbit reticulocyte lysate in the presence of 20 μM compound or DMSO control, and measured the amount of luciferase activity produced from the translated proteins. As shown in Figure 2D, 13 out of the 22 compounds identified from the primary screen also inhibited transcription and translation by >3SD of the assay, suggesting that these are not selective inhibitors of snRNP assembly.
β-lapachone is a potent inhibitor of SMN complex-dependent snRNP assembly
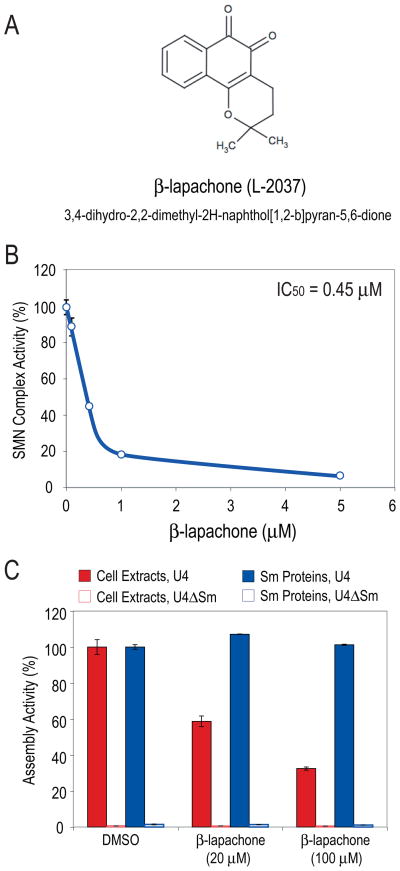
We next tested the in vivo effects of the remaining compounds on HeLa cells. Several of the compounds were extremely toxic to cells even at low concentrations and were not studied further. For the rest, extracts from cells treated for 6 hours with sub-toxic concentrations of each compound were prepared and their snRNP assembly activities were measured. Here, we describe one of these, β-lapachone (3,4-dihydro-2,2-dimethyl-2H-naphthol[1,2-b]pyran-5,6-dione; Figure 3A, labeled as L-2037), that showed a particularly strong inhibition of about 90%, but did not significantly inhibit the transcription and translation of a transfected luciferase reporter. Inhibition of snRNP assembly by >80% was observed within 30 minutes of β-lapachone treatment (data not shown), suggesting that its inhibition of the SMN complex is likely due to a direct effect of the compound. The inhibition of the activity of the SMN complex in treated cells was dependent on β-lapachone concentration with a calculated IC50 of 0.45 μM (Figure 3B). IC50 estimated from dose-response measurements in vitro using extracts of untreated cells was ~30 μM (data not shown). Thus, β-lapachone inhibits the activity of the SMN complex both in cell extracts and in living cells, showing higher potency in vivo than in vitro.
Figure 3. β-lapachone potently and selectively inhibits the SMN complex-mediated snRNP assembly in vitro and in cells.
(A) Chemical structure of β-lapachone.
(B) Concentration-dependent inhibition of SMN complex activity by β-lapachone in cells. HeLa cells were treated with various concentrations of β-lapachone or with DMSO (control) for 1 hour. SMN complex activity in extracts from treated cells was measured using magnetic beads snRNP assembly assay and compared to DMSO-treated control cells (100% activity). IC50 was calculated from the dose-reponse graph. Error bars represent SDs from 3 independent measurements.
(C) β-lapachone selectively inhibits SMN complex-mediated Sm core assembly. Assembly reactions were performed using either cell extracts or purified native snRNP proteins lacking the SMN complex (Sm proteins). Both samples were adjusted to contain a similar amount of Sm proteins. Magnetic beads snRNP assembly assay was carried out with U4 or control U4ΔSm snRNA in the presence of either 20 or 100 μM β-lapachone or DMSO control. Sm core assembly on U4 snRNA in the presence of DMSO was considered 100% activity. The error bars represent SDs from 3 independent measurements.
The persistence of the inhibition in extracts prepared after β-lapachone was washed away from the treated cells suggested that it either caused a modification of one or more components in the snRNP assembly pathway or interfered with protein-protein or protein-RNA interactions. We first asked if β-lapachone inhibits the interactions among the Sm proteins or their capacity to bind snRNAs. Sm cores can form spontaneously in vitro from purified Sm proteins and exogenous snRNAs without auxiliary factors (Raker et al., 1999; Sumpter et al., 1992). This Sm core assembly, unlike assembly in cell extracts and in vivo, is independent of the SMN complex and does not require ATP. It reflects the high propensity of the Sm proteins to form heptameric cores on many Sm site-resembling sequences, but lacks the strict specificity that the SMN complex confers towards its appropriate substrates, the snRNAs (Pellizzoni et al., 2002). We therefore compared the effect of β-lapachone on snRNP assembly using either cell extracts, which contain the SMN complex and all the components required for Sm core assembly, or a purified fraction of native snRNP proteins which is highly enriched in Sm proteins but lacking the SMN complex components (Raker et al., 1999; Sumpter et al., 1992). As shown in Figure 3C, both of these preparations assembled Sm cores on the U4 snRNA but not on the control U4ΔSm RNA which lacks the Sm site. As expected, β-lapachone inhibited assembly in the cell extracts, which is SMN complex-directed, however, it had no effect on Sm core formation from the purified snRNP proteins. This indicates that the target of β-lapachone inhibition is not the Sm-Sm protein interactions or the Sm-snRNA binding, but rather is likely the assembly machinery, the SMN complex.
β-lapachone induces intermolecular disulfide bonds in SMN
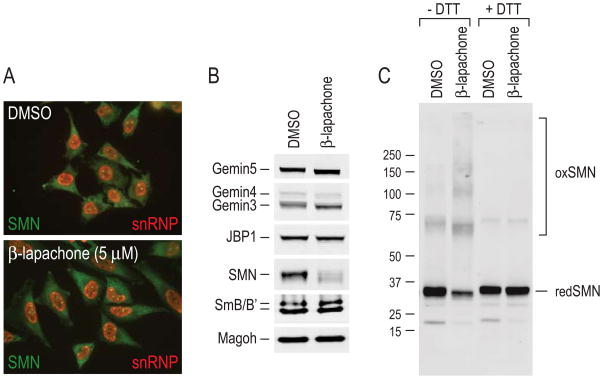
To further investigate the mechanism of action of β-lapachone, we asked whether it affected the cellular localization or composition of the SMN complex and of proteins involved in the snRNP biogenesis pathway. Typically, by immunofluorescence microscopy, SMN is found throughout the cytoplasm and in the nucleus where it is concentrated in Gems (Liu and Dreyfuss, 1996). However, no detectable difference was observed in SMN signal intensity or localization between β-lapachone treated and control cells (Figure 4A). There was also no obvious change in the amount or localization of Sm proteins, most of which are found in snRNPs (Fury and Zieve, 1996) and are detected by Y12 antibody (Figure 4A).
Figure 4. SMN protein is oxidized to form intermolecular disulfide bonds upon β-lapachone treatment.
(A) Indirect immunofluorescence staining of SMN (2B1; green) and snRNPs (Y12; red) in HeLa PV cells treated 3 hours with 5 μM β-lapachone or DMSO control.
(B) HeLa total cell extracts prepared from cells treated 3 hours with 5 μM β-lapachone or DMSO control were resolved by SDS-PAGE and analyzed by quantitative Western blotting, using JBP1 and Magoh as loading controls. Extracts were prepared and mixed with sample buffer without DTT. The membrane was cut into strips for probing of each protein at the corresponding molecular mass.
(C) β-lapachone causes intermolecular disulfide crosslinking of SMN. Total cell extracts from HeLa cells stably expressing Flag-Gemin2 (Yong et al., 2002) were used for in vitro assembly reactions in the presence of either 100 μM β-lapachone or DMSO control. The SMN complex was isolated by anti-Flag immunoprecipitation, mixed with sample buffer without or with DTT and resolved by SDS-PAGE. Western blot analysis was performed on the entire membrane with anti-SMN antibody 62E7. Molecular mass markers in kDa are indicated on the left. “redSMN” indicates monomer SMN migrating at normal molecular mass and “oxSMN” indicates disulfide-crosslinked SMN upon oxidation.
β-lapachone is a redox-active ortho-quinone (Boveris et al., 1978; Fernandez Villamil et al., 2004). We therefore considered the possibility that the inhibition of the activity of the SMN complex by β-lapachone may be due to its activity as a generator of redox cycles, a process in which the quinone is reduced and reactive oxygen species (ROS), superoxide anion (O2−) and hydrogen peroxide (H2O2) are produced. β-lapachone cannot directly react with sulfhydryl groups by 1,4-Michael addition since it is fully-substituted, but the ROS it generates might oxidize sulfhydryls which might then form disulfides or other cysteine modifications in protein components of the SMN complex. To test this, extracts from β-lapachone-treated cells or DMSO-treated control cells were prepared in gel electrophoresis sample buffer without the reducing agent dithiothreitol (DTT) to preserve potential disulfides, resolved by SDS-PAGE, and analyzed by quantitative Western blots. To simultaneously monitor different SMN complex components and associated proteins, the blots were cut into strips corresponding to the known molecular mass of the various proteins at their monomer size, and each was then probed separately. Strikingly, while there was no detectable change in signal intensities of the other proteins examined, the intensity of the SMN band was consistently and significantly lower in β-lapachone treated cells prepared without DTT (~60% decrease compared to DMSO controls; Figure 4B). Because the decrease in monomeric-size SMN most likely resulted from ROS-induced disulfide crosslinking, we performed Western blots on SMN complexes purified from β-lapachone- or DMSO-treated cell extracts prepared either with or without DTT, and probed the entire length of the membrane with anti-SMN antibody. As shown in Figure 4C, in the absence of DTT, the band at ~35 kDa, corresponding to monomeric SMN (redSMN), is decreased upon β-lapachone treatment and several higher molecular weight SMN-reactive forms (oxSMN), corresponding in sizes to SMN dimer and possibly larger forms are observed. When the same samples were prepared with DTT, all the higher molecular weight SMN-containing bands disappeared and the same amount of SMN migrated at the monomer size in both β-lapachone-treated and control extracts. These findings indicate that upon β-lapachone treatment, the SMN protein experienced an oxidative environment and became crosslinked via disulfide bonds. We note that even in the DMSO control, there was a detectable low level of SMN crosslinking. The β-lapachone-induced crosslinking appeared to be selective to SMN; it was not observed for the other proteins that were probed for in the same experiment, including several Gemins, the methyltransferase JBP1, Magoh and Sm proteins (Figure 4B), as well as several other proteins tested, e.g., hnRNP A1 and the fragile X mental retardation protein FMR1 (data not shown).
ROS inhibit the SMN complex in vitro and in cells
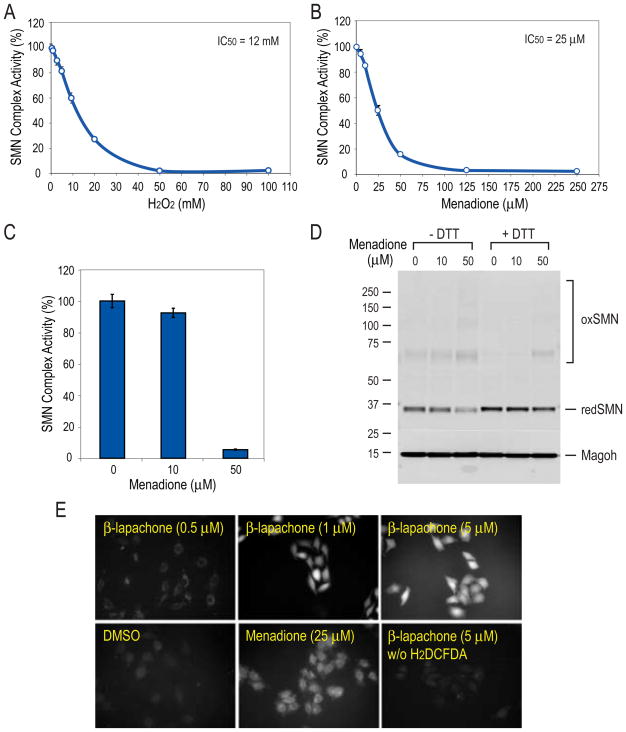
The data described above indicated that the activity of the SMN complex might be regulated by redox status and could be sensitive to other oxidative stress-inducing agents. We therefore examined the effect of other sources of ROS. H2O2 and cumene hydroperoxide, both of which are capable of oxidizing cysteines (Marnett et al., 2003), inhibited the activity of the SMN complex in vitro with IC50 values in the low millimolar range (Figure 5A and data not shown). Menadione (vitamin K3; 2-methyl-1,4-napthoquinone), a redox-active p-quinone, inhibited the activity of the SMN complex much more efficiently, yielding an IC50 of 25 μM (Figure 5B). Treatment of cells with menadione at concentrations commonly used to study oxidative stress also resulted in dose-dependent inhibition of SMN complex activity (Figure 5C) and a corresponding degree of SMN disulfide crosslinking (Figure 5D), similar to that observed with β-lapachone. These findings indicate that the SMN complex is susceptible to inactivation by ROS, and its activity can be modulated by the cellular redox state.
Figure 5. ROS reagents inhibit the activity of the SMN complex in vitro and in cells.
(A) Effect of H2O2 on the activity of the SMN complex in vitro. Magnetic beads snRNP assembly assay was carried out in the presence of increasing amounts of H2O2. IC50 was calculated from the dose-response curve. Error bars represent SDs from triplicate measurements.
(B) Effect of menadione on SMN complex activity in vitro. The same experimental procedure was carried out as in (A), except that menadione was used instead.
(C) Dose-dependent effect of menadione on SMN complex activity in cells. HeLa cells were treated with menadione at the indicated concentrations or with DMSO control for 1 hour. SMN complex activity in extracts from various treated cells was measured in comparison to DMSO control cell extract (100% activity) by magnetic beads snRNP assembly assay. Error bars represent SDs from 3 independent measurements.
(D) Extracts from (C) mixed with sample buffer without or with DTT were resolved by SDS-PAGE and analyzed by Western blot of the entire membrane with anti-SMN antibody 62E7. The molecular mass markers in kDa are indicated on the left. “redSMN” indicates monomer SMN migrating at normal molecular mass and “oxSMN” indicates disulfide-crosslinked SMN upon oxidation.
(E) β-lapachone and menadione generate ROS in live cells. HeLa cells were incubated 30 minutes with ROS indicator dye H2DCFDA (10 μM) or without dye as a control, then treated with compounds at indicated concentrations or DMSO as control. Fluorescence images were acquired 30 minutes after treatment.
Additional direct evidence for the ROS-generating activity of the compounds in live cells was obtained by staining cells with 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA), a nonfluorescent dye that becomes fluorescent upon exposure to ROS (Figure 5E). The fluorescence intensity of β-lapachone-treated cells was much higher than that of menadione-treated cells at similar concentrations, indicating that their potency as inactivators of SMN complex corresponds to their ROS-generating capacity.
DTT protects the SMN complex from ROS inactivation
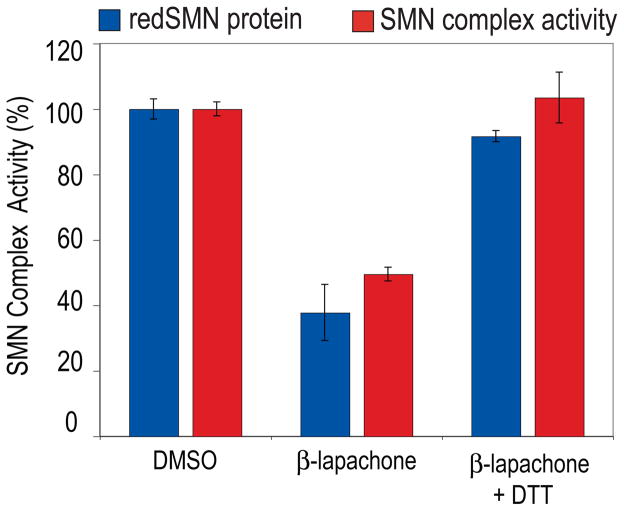
To further determine if the inhibition of the activity of the SMN complex by ROS is related to the oxidative modification of cysteines, reflected by disulfide protein crosslinking, we asked whether β-lapachone inhibition could be prevented by DTT, since DTT reversed SMN protein crosslinking (Figure 4C and 5D). As shown in Figure 6, DTT completely neutralized the inhibitory effect of β-lapachone. Furthermore, the extent of SMN oxidative crosslinking, determined by the corresponding decrease in unoxidized monomeric SMN (redSMN), is directly proportional to the inhibition of SMN complex activity. Similarly, another reductant and antioxidant N-acetyl-L-cysteine (NAC) also completely prevented the inhibition of β-lapachone, menadione and H2O2 (data not shown). This strongly suggests that ROS inhibit the activity of the SMN complex by causing oxidation of sulfhydryl groups of cysteines and inducing disulfide crosslinking, including in SMN itself.
Figure 6. DTT prevents the inhibition of the activity of the SMN complex by β-lapachone.
Cell extracts treated with 20 μM β-lapachone, or 20 μM β-lapachone together with 20 mM DTT, or DMSO control were analyzed by non-reducing Western blot. The relative levels of monomer SMN (“redSMN”) were calculated as the percentage of that in DMSO control and shown by the blue bar. Assembly activities of the SMN complex were measured by magnetic beads snRNP assembly assay using the same set of treated extracts and shown by the red bar. Error bars represent SDs from 3 independent experiments.
ROS cause SMN-SMN disulfide crosslinking through at least two cysteines
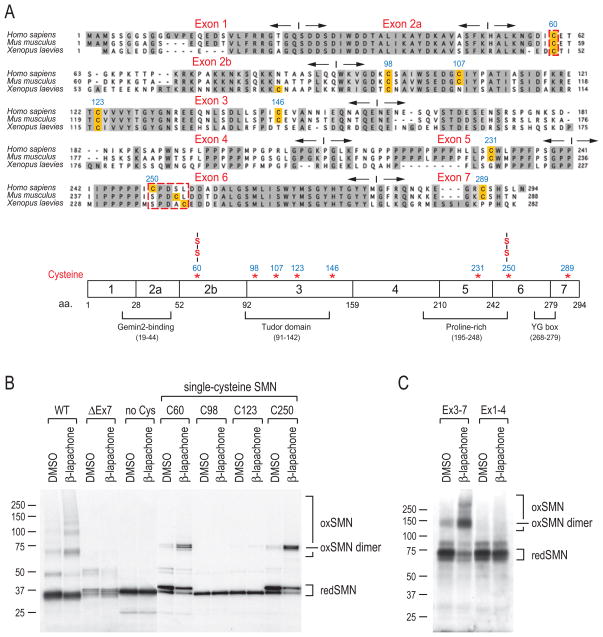
SMN can form homo-oligomers (Lorson et al., 1998; Pellizzoni et al., 1999) and it interacts with many other proteins (Yong et al., 2004). The apparent sizes of the ROS-induced disulfide-crosslinked forms correspond to SMN dimers or larger forms (Figure 4C and 5D), and we therefore asked if SMN-SMN crosslinks occur. To determine this, we generated constructs for full-length wild-type human SMN, amino- and carboxyl-terminal deletion mutants, and SMN in which all eight cysteines were mutated to alanines. The corresponding proteins were produced by transcription and translation in vitro with [35S]methionine. The samples were then treated with β-lapachone to generate ROS or with DMSO as a control, and resolved by SDS-PAGE without DTT. As shown in Figure 7B, wild-type SMN (WT) formed disulfide-crosslinked species similar in sizes to those observed in β-lapachone-treated cells and cell extracts (Figure 4C and 5D). This demonstrates that intermolecular SMN-SMN oxidative crosslinks occur. Although not precisely defined, sequences capable of homotypic interactions have been described in exons 6-7 and in exon 2b (Lorson et al., 1998; Pellizzoni et al., 1999). However, while SMN with an amino terminal deletion that includes sequences encoded by exons 1-2b (Figure 7C, labeled as Ex3-7) could still form a disulfide crosslinked dimer, deletion of the carboxyl terminal sequences encoded by exons 5-7 (Figure 7C, labeled as Ex1-4) completely abolished oxidative crosslinking. Additionally, no oxidative SMN-SMN crosslinking occurred in exon 7-deleted SMN (Figure 7B, labeled as ΔEx7), the predominant product of the remaining SMN2 gene in SMA patients, which is defective in oligomerization (Lorson et al., 1998; Pellizzoni et al., 1999). We conclude that SMN oligomerization, mediated by its carboxyl terminus, is required for SMN-SMN disulfide crosslinking.
Figure 7. ROS mapping of disulfide-crosslinked cysteines in SMN.
(A) Sequence alignment of human (Homo sapiens), mouse (Mus musculus), and frog (Xenopus laevies) SMN protein sequences. Conserved residues are shaded in gray. Cysteine residues are highlighted. Exons and their boundaries are indicated with opposite-directed arrows. The schematic diagram shows SMN protein domain organization and positions of cysteines. Two cysteines (C60 and C250) that form disulfide bridges are marked (-S-S-).
(B) Human SMN protein (WT, wild type; ΔEx7, exon7 deletion mutant; no Cys, mutation of all 8 cysteines to alanines; C60, C98, C123, and C250, mutation of 7 cysteines to alanines except for cysteine at positions 60, 98, 123, and 250, respectively) were produced by in vitro transcription and translation in the presence of 35S-Met and then treated with 40 μM β-lapachone or DMSO for 1 hour. Samples were mixed with sample buffer without DTT, and then analyzed by SDS-PAGE and autoradiography. Molecular mass markers in kDa are indicated on the left. Protein bands corresponding to monomer SMN (redSMN), disulfide-crosslinked SMN (oxSMN) and SMN dimer (oxSMN dimer) are indicated on the right.
(C) SMN amino terminus deletion (Ex3-7) and carboxyl terminus deletion (Ex1-4) mutants were tested for crosslinking, as described in panel (B). Note that these mutants were constructed in pcDNA3-myc-pyruvate kinase (PK, ~60kD) vector to facilitate detection of otherwise small fragments.
Among the eight cysteines in human SMN, four are evolutionarily conserved in vertebrates (Figure 7A). SMN in which all eight cysteines were mutated to alanines did not show oxidative crosslinking (Figure 7B), confirming that disulfides bridge SMN molecules. To identify the cysteines that can engage in SMN-SMN disulfide bond formation, we produced SMN constructs that mutated seven cysteines to alanines but retained each of the conserved cysteines, at positions 60, 98, 123, and 250, respectively. β-lapachone treatment of these mutants, each containing a single cysteine, showed that either C60 or C250 is sufficient to efficiently form SMN-SMN crosslinked dimers upon exposure to ROS. In contrast, the two most highly conserved cysteines, C98 and C123, which are buried in the structure of the Tudor domain (Selenko et al., 2001; Sprangers et al., 2003), did not (Figure 7B). Thus C60 and C250 must be juxtaposed to the corresponding cysteines in SMN-SMN oligomers.
Discussion
The high throughput assay we describe here provides a powerful tool to study the Sm core assembly process and the mechanism and regulation of the SMN complex. It is highly sensitive and robust, making it suitable for large-scale screens for chemical and genetic modifiers of the SMN complex. With slight modifications of the RNA, extract and reaction conditions, this assay can be broadly applied to many other reactions involving RNA-protein interactions. It obviates the need to analyze nucleic acid-protein complexes by gel electrophoretic mobility shifts and opens the way to rapidly identify effectors and dissect the pathway of RNP biogenesis. Although a genetically-determined decrease in functional SMN protein is manifested in loss of motor neurons, the activity of the SMN complex and the biogenesis of snRNPs are of fundamental importance to all eukaryotic cells (Schrank et al., 1997). Our rationale for developing this assay was that compounds that modulate the activity of the SMN complex would be useful both as research tools and as potential therapeutics for SMA. Compounds that increase or bypass the activity of the SMN complex, if such exist, as well as inhibitors of the SMN complex could point to how it is regulated and suggest therapeutic approaches. Using this HTS assay, we identified the first class of inhibitors of the SMN complex and of the snRNP biogenesis pathway, and this revealed a surprising and hitherto unknown aspect of the regulation of the SMN complex.
The most potent inhibitor we found, β-lapachone, is a naturally occurring naphthoquinone and a potent generator of a futile redox cycle that catalyzes the formation of ROS (Fernandez Villamil et al., 2004). β-lapachone has been shown to inhibit several enzymes, including NADH/NADPH oxidoreductase, topoisomerase I, HIV reverse transcriptase, telomerase and NF-κB (Pardee et al., 2002). The inhibition of NF-κB by β-lapachone can be reversed by DTT and involves critical sulfhydryl groups (Manna et al., 1999). Three lines of evidence indicate that the inhibition of the SMN complex by β-lapachone is the result of the ROS it generates. First, other structurally unrelated and diverse oxidants, including H2O2, cumene hydroperoxide as well as several environmental toxins known to generate ROS, menadione, 9,10-phenanthrenequinone and tetrachloro-1,2-benzoquinone, have the same effect. In contrast, the position isomer β-lapachone, which unlike α-lapachone has no appreciable redox cycling capability (Boveris et al., 1978), does not inhibit the SMN complex (Figure 5 and data not shown). Second, β-lapachone and the other oxidants cause formation of intermolecular disulfide bonds in SMN and these, as well as the inhibition of the activity of the SMN complex, are counter-acted or reversed by DTT (Figure 4, 5 and 6). Third, the potency of the compounds to induce SMN disulfide crosslinks and inactivate the SMN complex parallels their ROS-generating activity in live cells (Figure 5E).
The sensitivity of the SMN complex to ROS inactivation is remarkable considering that transcription, splicing and translation are not inhibited under similar oxidative stress. Although the disulfide-crosslinked SMN demonstrates that SMN itself becomes oxidized, it is not yet known if this is the causative or the only target of ROS-mediated inactivation of the SMN complex. Other proteins involved in the snRNP assembly reactions may also be modified by ROS, causing inactivation. Nevertheless, ROS provide a powerful tool for studying the structure and domain interactions of SMN. Disulfide bonds can only form if the cysteines involved are in immediate juxtaposition (“zero-distance” crosslinking). The observation of SMN-SMN disulfide crosslinks, indicate that SMN homo-oligomers, previously shown in vitro (Lorson et al., 1998; Pellizzoni et al., 1999; Young et al., 2000), exist in cells. These homo-oligomers depend on sequences encoded in exons 6 and 7, near the carboxyl terminus, but which have not yet been fully characterized. The crosslinks of single-cysteine SMNs, C60 and C250, define these cysteines as specific contact points in exon 2b and exon 6, respectively. In contrast, the two most highly conserved SMN cysteines, C98 and C123, do not form intermolecular disulfides, consistent with their solvent-inaccessible positions in the interior of Tudor domain. The absence of C60-C60 contacts in C-terminal deleted SMN indicates that the exon 2b-2b interactions are either not sufficient for oligomerization or if they do occur, that the C-terminal domain influences the structure of the exon 2b-enoded peptide. ROS should facilitate further studies of the structure of SMN, about which little is known, including the arrangement of Tudor domains in SMN oligomers.
All cells are exposed to oxidative stress as an unavoidable consequence of the utilization of oxygen for energy production and other metabolic processes, as well as from exposure to environmental oxidants (Ischiropoulos and Beckman, 2003; Marnett et al., 2003; Valko et al., 2007). The physiologic process of oxygen reduction in mitochondria generates ROS which diffuse from mitochondria. ROS can also be generated by redox-active compounds, including numerous metabolites and xenobiotics e.g., quinones and polycyclic aromatic hydrocarbons, which can be converted to their corresponding redox-active quinones by aldo-keto reductases (Penning et al., 1999). Under normal conditions, cellular antioxidant defenses, comprised of small molecules (including glutathione and ascorbate) as well as enzymes (particularly superoxide dismutase, catalase, glutathione peroxidase and peroxyredoxins), are capable of neutralizing the hazardous ROS (Valko et al., 2007). However, when ROS exceed the antioxidant capacity, they react with proteins, DNA and lipids, and can impair numerous physiologic functions (Marnett et al., 2003; Valko et al., 2007). ROS readily react with thiols (-SH), including those in protein cysteines, to form sulfenic acid (-SOH), which can then readily react with available thiols either in glutathione or in an adjacent cysteine to form disulfides (Marnett et al., 2003). Disulfides (-S-S-) are reversible by cellular reducing systems that maintain the thiol-disulfide homeostasis. However, ROS can also over-oxidize cysteines by further reacting with sulfenic acid to form the corresponding sulfinic (-SO2H) and sulfonic (-SO3H) acids, which are not reversible and can result in permanent inactivation of proteins (Marnett et al., 2003). It is conceivable that the disulfide bonds that form in SMN cysteines protect it from irreversible oxidative damage. However, the possibility that irreversible inactivation of the SMN complex occurs upon excessive exposure to ROS, must also be considered. We have shown here that SMNΔEx7, the major protein product of SMN2, the only remaining gene in SMA patients, does not form reversible disulfide crosslinks upon ROS. Thus, SMNΔEx7 may be more susceptible to irreversible cysteine hyper-oxidation and permanent inactivation of its function.
SMN protein deficiency is deleterious to all cells, causing aberrations in gene expression and leading to SMA (Zhang et al., 2008). The dose-dependent ROS inactivation of the SMN complex causes a functional deficiency equivalent to that which occurs upon SMN protein deficiency. This suggests that cells that experience more oxidative stress, particularly if they already have decreased levels of SMN, as SMA patients do, may be especially vulnerable. Different cells experience oxidative stress to different degrees, depending on the balance of ROS and anti-oxidant defense they have. Neurons, including motor neurons, are amongst the most metabolically active cells in the body and their extremely high rate of oxygen consumption could generate relatively high levels of ROS (Barber et al., 2006; Ischiropoulos and Beckman, 2003). Oxidative stress has been suggested to play a major role in the etiology of many neurodegenerative diseases, including Alzheimer’s and Parkinson’s disease and in aging (Droge, 2003; Ischiropoulos and Beckman, 2003; Lin and Beal, 2006; Perry et al., 2002; Valko et al., 2007). Another motor neuron degenerative disease with similar clinical phenotypes to SMA is amyotrophic lateral sclerosis (ALS). Although the cause of ALS in the majority of patients is unknown, a small percentage of ALS cases are familial, resulting from mutations in the Cu/Zn superoxide dismutase (SOD1) (Beckman et al., 2001). The mechanism whereby SOD1 mutations cause motor neuron death is not certain, but considerable evidence suggest that excessive oxidative stress occurs and contributes to motor neuron injuries (Barber et al., 2006; Pacher et al., 2007; Rizzardini et al., 2005). Our findings that the SMN complex is highly susceptible to ROS and the role of ROS in neurodegenerative diseases suggest a potential mechanistic convergence of these diseases, particularly ALS with SMA, with the SMN complex as a plausible target. The vulnerability of the SMN complex to inactivation by ROS suggests that xenobiotics that could increase the exposure of the neuronal environment to oxidative stress, as well as anti-oxidants that may protect it from excessive ROS, deserve careful consideration, particularly for SMA patients.
Experimental Procedures
Chemical compounds
A library of ~5,000 pure bioactive chemicals that includes FDA-approved drugs, known inhibitors and activators of diverse enzymes and receptors, and pure natural compounds was assembled from several commercial sources (Microsource Diversity, Tocris, Lopac, Sigma-Aldrich and other suppliers) at 2 mM stock concentration for each compound dissolved in DMSO. For confirmation studies and treatment on cells, β-lapachone, H2O2, cumene hydroperoxide, and menadione were purchased from Sigma-Aldrich Chemical Co.
HTS assay for SMN complex activity in snRNP assembly
An automated and quantitative assay for in vitro snRNP assembly was developed for HTS using biotinylated U4 snRNA to capture snRNPs assembled from cell extracts onto 384-well neutravidin coated microplates. The plate map for the screen is shown in Figure 2A. Columns 1 and 2 contained controls corresponding to 100% assembly activity in the presence of DMSO. Library compounds were located in the 320 central wells, and columns 23 and 24 contained controls for non-specific background (no cell extract added). The assay was conducted as follows: 10 μl of reconstitution buffer containing 2.5 mM ATP, 0.25 μg/μl Escherichia coli tRNA, 0.2 U/μl RNasin RNase Inhibitor (Promega) and HeLa cytoplasmic extract (4 μg of total protein) was aliquoted into Reacti-Bind NeutrAvidin coated black 384-well microplates (Pierce) with a Multidrop (Thermo labsystems). Next, 5 μl of compounds in reconstitution buffer (final concentration 20 μM in < 0.1% DMSO), and 5 μl of biotin-labeled U4 snRNA (final concentration 5 nM) were added to each well using Biomek FX workstation (Beckman Coulter). The plates were centrifuged gently, and the assembly reactions were incubated for 1 hour at room temperature (RT). Subsequently, 20 μl of RSB-500 buffer (10 mM Tris/HCl pH 7.5, 500 mM NaCl, 2.5 mM MgCl2) containing 2 mg/ml heparin, 0.1% NP-40, 0.2 U/μl RNasin was added to each well, mixed by pipetting with the Biomek FX, and incubated for 1 hour at RT. The reaction mixtures were removed and 40 μl/well of Y12 antibody (diluted 1:1000 in RSB-500 containing 0.1% NP-40, 1 mg/ml BSA) was added with a Multidrop (Thermo labsystems). After 1 hour incubation at RT, the plates were washed 10 times with RSB-500 containing 0.1% NP-40 using an ELx405 microplate washer (Bio-Tek), followed by the addition of 40 μl/well of horseradish peroxidase (HRP)-conjugated AffiniPure goat anti-mouse IgG + IgM (diluted 1:10,000 in RSB-500 containing 0.1% NP-40, 1 mg/ml BSA; Jackson Laboratories) and incubation for 1 hour at RT. The plates were washed again 10 times and 40 μl of SuperSignal ELISA Femto Maximum Sensitivity enhanced chemiluminescence substrate working solution (Pierce) was added into each well. Luminescence signals were measured using an EnVision Reader (PerkinElmer) with standard luminescence settings. Percentage activity from each well of the plate was calculated from the equation:
Where S is sample well signal, N̄ is median non-specific background signal from wells in column 23, and S̄ is median sample signal from 320 central wells.
In vitro transcription and translation assay
In vitro transcription and translation reactions were set up in 384-well microplate using TNT Quick Coupled Transcription/Translation Systems (Promega) with luciferase DNA as a reporter in the presence of 20 μM compounds. The plate was centrifuged briefly and the reactions were incubated at 30°C for 1.5 hours. Subsequently, 20 μl of Luciferase Assay Reagent (Promega) was added into each well and mixed. Luminescence signals were measured immediately using an EnVision Reader (PerkinElmer) with standard luminescence settings.
Crosslinking assay of in vitro transcribed and translated SMN
In vitro transcription and translation reactions were performed with each plasmid in the presence of 35S-Methionine for protein labeling using T7 TNT Quick Coupled Transcription/Translation Systems (Promega). Subsequently, 0.7 μl of each translated product was added to in vitro snRNP assembly reaction mixture in the presence of 40 μM β-lapachone or DMSO as a control, incubated for 1 hour, and then mixed with sample buffer without DTT for analysis on 4–12% SDS-PAGE and detection by autoradiography.
Supplementary Material
Acknowledgments
We thank Drs. Trevor M. Penning, Harry Ischiropoulos, and Ian A. Blair for helpful discussions and Dr. Joan A. Steitz for providing the anti-Sm (Y12) monoclonal antibody. We are grateful to Congli Wang, Yoon Cho and Christopher Dengler for technical assistance and the members of our laboratory for stimulating discussions and comments on this manuscript. This work was supported by the Association Française Contre les Myopathies (AFM). G.D. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barber SC, Mead RJ, Shaw PJ. Oxidative stress in ALS: a mechanism of neurodegeneration and a therapeutic target. Biochim Biophys Acta. 2006;1762:1051–1067. doi: 10.1016/j.bbadis.2006.03.008. [DOI] [PubMed] [Google Scholar]
- Battle DJ, Lau CK, Wan L, Deng H, Lotti F, Dreyfuss G. The Gemin5 protein of the SMN complex identifies snRNAs. Mol Cell. 2006;23:273–279. doi: 10.1016/j.molcel.2006.05.036. [DOI] [PubMed] [Google Scholar]
- Beckman JS, Estevez AG, Crow JP, Barbeito L. Superoxide dismutase and the death of motoneurons in ALS. Trends Neurosci. 2001;24:S15–20. doi: 10.1016/s0166-2236(00)01981-0. [DOI] [PubMed] [Google Scholar]
- Boveris A, Docampo R, Turrens JF, Stoppani AO. Effect of β-lapachone on superoxide anion and hydrogen peroxide production in Trypanosoma cruzi. Biochem J. 1978;175:431–439. doi: 10.1042/bj1750431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes-Diaz C, Frugier T, Melki J. Spinal muscular atrophy. Semin Pediatr Neurol. 2002;9:145–150. doi: 10.1053/spen.2002.33801. [DOI] [PubMed] [Google Scholar]
- Droge W. Oxidative stress and aging. Adv Exp Med Biol. 2003;543:191–200. doi: 10.1007/978-1-4419-8997-0_14. [DOI] [PubMed] [Google Scholar]
- Feng W, Gubitz AK, Wan L, Battle DJ, Dostie J, Golembe TJ, Dreyfuss G. Gemins modulate the expression and activity of the SMN complex. Hum Mol Genet. 2005;14:1605–1611. doi: 10.1093/hmg/ddi168. [DOI] [PubMed] [Google Scholar]
- Fernandez Villamil S, Stoppani AO, Dubin M. Redox cycling of β-lapachone and structural analogues in microsomal and cytosol liver preparations. Methods Enzymol. 2004;378:67–87. doi: 10.1016/S0076-6879(04)78004-0. [DOI] [PubMed] [Google Scholar]
- Fury MG, Zieve GW. U6 snRNA maturation and stability. Exp Cell Res. 1996;228:160–163. doi: 10.1006/excr.1996.0311. [DOI] [PubMed] [Google Scholar]
- Golembe TJ, Yong J, Dreyfuss G. Specific sequence features, recognized by the SMN complex, identify snRNAs and determine their fate as snRNPs. Mol Cell Biol. 2005;25:10989–11004. doi: 10.1128/MCB.25.24.10989-11004.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ischiropoulos H, Beckman JS. Oxidative stress and nitration in neurodegeneration: cause, effect, or association? J Clin Invest. 2003;111:163–169. doi: 10.1172/JCI17638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambach C, Walke S, Young R, Avis JM, de la Fortelle E, Raker VA, Luhrmann R, Li J, Nagai K. Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell. 1999;96:375–387. doi: 10.1016/s0092-8674(00)80550-4. [DOI] [PubMed] [Google Scholar]
- Kleinschmidt AM, Patton JR, Pederson T. U2 small nuclear RNP assembly in vitro. Nucleic Acids Res. 1989;17:4817–4828. doi: 10.1093/nar/17.12.4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- Liu Q, Dreyfuss G. A novel nuclear structure containing the survival of motor neurons protein. EMBO J. 1996;15:3555–3565. [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Fischer U, Wang F, Dreyfuss G. The spinal muscular atrophy disease gene product, SMN, and its associated protein SIP1 are in a complex with spliceosomal snRNP proteins. Cell. 1997;90:1013–1021. doi: 10.1016/s0092-8674(00)80367-0. [DOI] [PubMed] [Google Scholar]
- Lorson CL, Strasswimmer J, Yao JM, Baleja JD, Hahnen E, Wirth B, Le T, Burghes AH, Androphy EJ. SMN oligomerization defect correlates with spinal muscular atrophy severity. Nat Genet. 1998;19:63–66. doi: 10.1038/ng0598-63. [DOI] [PubMed] [Google Scholar]
- Manna SK, Gad YP, Mukhopadhyay A, Aggarwal BB. Suppression of tumor necrosis factor-activated nuclear transcription factor-kappaB, activator protein-1, c-Jun N-terminal kinase, and apoptosis by β-lapachone. Biochem Pharmacol. 1999;57:763–774. doi: 10.1016/s0006-2952(98)00354-2. [DOI] [PubMed] [Google Scholar]
- Marnett LJ, Riggins JN, West JD. Endogenous generation of reactive oxidants and electrophiles and their reactions with DNA and protein. J Clin Invest. 2003;111:583–593. doi: 10.1172/JCI18022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Buhler D, Pillai R, Lottspeich F, Fischer U. A multiprotein complex mediates the ATP-dependent assembly of spliceosomal U snRNPs. Nat Cell Biol. 2001;3:945–949. doi: 10.1038/ncb1101-945. [DOI] [PubMed] [Google Scholar]
- Meister G, Eggert C, Fischer U. SMN-mediated assembly of RNPs: a complex story. Trends Cell Biol. 2002;12:472–478. doi: 10.1016/s0962-8924(02)02371-1. [DOI] [PubMed] [Google Scholar]
- Monani UR. Spinal muscular atrophy: a deficiency in a ubiquitous protein; a motor neuron-specific disease. Neuron. 2005;48:885–896. doi: 10.1016/j.neuron.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Nilsen TW. The spliceosome: the most complex macromolecular machine in the cell? Bioessays. 2003;25:1147–1149. doi: 10.1002/bies.10394. [DOI] [PubMed] [Google Scholar]
- Pacher P, Beckman JS, Liaudet L. Nitric oxide and peroxynitrite in health and disease. Physiol Rev. 2007;87:315–424. doi: 10.1152/physrev.00029.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardee AB, Li YZ, Li CJ. Cancer therapy with β-lapachone. Curr Cancer Drug Targets. 2002;2:227–242. doi: 10.2174/1568009023333854. [DOI] [PubMed] [Google Scholar]
- Pellizzoni L, Charroux B, Dreyfuss G. SMN mutants of spinal muscular atrophy patients are defective in binding to snRNP proteins. Proc Natl Acad Sci U S A. 1999;96:11167–11172. doi: 10.1073/pnas.96.20.11167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellizzoni L, Yong J, Dreyfuss G. Essential role for the SMN complex in the specificity of snRNP assembly. Science. 2002;298:1775–1779. doi: 10.1126/science.1074962. [DOI] [PubMed] [Google Scholar]
- Penning TM, Burczynski ME, Hung CF, McCoull KD, Palackal NT, Tsuruda LS. Dihydrodiol dehydrogenases and polycyclic aromatic hydrocarbon activation: generation of reactive and redox active o-quinones. Chem Res Toxicol. 1999;12:1–18. doi: 10.1021/tx980143n. [DOI] [PubMed] [Google Scholar]
- Perry G, Cash AD, Smith MA. Alzheimer Disease and Oxidative Stress. J Biomed Biotechnol. 2002;2:120–123. doi: 10.1155/S1110724302203010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raker VA, Hartmuth K, Kastner B, Luhrmann R. Spliceosomal U snRNP core assembly: Sm proteins assemble onto an Sm site RNA nonanucleotide in a specific and thermodynamically stable manner. Mol Cell Biol. 1999;19:6554–6565. doi: 10.1128/mcb.19.10.6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzardini M, Mangolini A, Lupi M, Ubezio P, Bendotti C, Cantoni L. Low levels of ALS-linked Cu/Zn superoxide dismutase increase the production of reactive oxygen species and cause mitochondrial damage and death in motor neuron-like cells. J Neurol Sci. 2005;232:95–103. doi: 10.1016/j.jns.2005.02.004. [DOI] [PubMed] [Google Scholar]
- Schrank B, Gotz R, Gunnersen JM, Ure JM, Toyka KV, Smith AG, Sendtner M. Inactivation of the survival motor neuron gene, a candidate gene for human spinal muscular atrophy, leads to massive cell death in early mouse embryos. Proc Natl Acad Sci U S A. 1997;94:9920–9925. doi: 10.1073/pnas.94.18.9920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selenko P, Sprangers R, Stier G, Buhler D, Fischer U, Sattler M. SMN tudor domain structure and its interaction with the Sm proteins. Nat Struct Biol. 2001;8:27–31. doi: 10.1038/83014. [DOI] [PubMed] [Google Scholar]
- Sprangers R, Groves MR, Sinning I, Sattler M. High-resolution X-ray and NMR structures of the SMN Tudor domain: conformational variation in the binding site for symmetrically dimethylated arginine residues. J Mol Biol. 2003;327:507–520. doi: 10.1016/s0022-2836(03)00148-7. [DOI] [PubMed] [Google Scholar]
- Sumpter V, Kahrs A, Fischer U, Kornstadt U, Luhrmann R. In vitro reconstitution of U1 and U2 snRNPs from isolated proteins and snRNA. Mol Biol Rep. 1992;16:229–240. doi: 10.1007/BF00419662. [DOI] [PubMed] [Google Scholar]
- Talbot K, Davies KE. Spinal muscular atrophy. Semin Neurol. 2001;21:189–197. doi: 10.1055/s-2001-15264. [DOI] [PubMed] [Google Scholar]
- Tarn WY, Steitz JA. Pre-mRNA splicing: the discovery of a new spliceosome doubles the challenge. Trends Biochem Sci. 1997;22:132–137. doi: 10.1016/s0968-0004(97)01018-9. [DOI] [PubMed] [Google Scholar]
- Valko M, Leibfritz D, Moncol J, Cronin MT, Mazur M, Telser J. Free radicals and antioxidants in normal physiological functions and human disease. Int J Biochem Cell Biol. 2007;39:44–84. doi: 10.1016/j.biocel.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Wan L, Battle DJ, Yong J, Gubitz AK, Kolb SJ, Wang J, Dreyfuss G. The survival of motor neurons protein determines the capacity for snRNP assembly: biochemical deficiency in spinal muscular atrophy. Mol Cell Biol. 2005;25:5543–5551. doi: 10.1128/MCB.25.13.5543-5551.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will CL, Luhrmann R. Spliceosomal UsnRNP biogenesis, structure and function. Curr Opin Cell Biol. 2001;13:290–301. doi: 10.1016/s0955-0674(00)00211-8. [DOI] [PubMed] [Google Scholar]
- Winkler C, Eggert C, Gradl D, Meister G, Giegerich M, Wedlich D, Laggerbauer B, Fischer U. Reduced U snRNP assembly causes motor axon degeneration in an animal model for spinal muscular atrophy. Genes Dev. 2005;19:2320–2330. doi: 10.1101/gad.342005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth B, Brichta L, Hahnen E. Spinal muscular atrophy: from gene to therapy. Semin Pediatr Neurol. 2006;13:121–131. doi: 10.1016/j.spen.2006.06.008. [DOI] [PubMed] [Google Scholar]
- Yong J, Pellizzoni L, Dreyfuss G. Sequence-specific interaction of U1 snRNA with the SMN complex. EMBO J. 2002;21:1188–1196. doi: 10.1093/emboj/21.5.1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong J, Wan L, Dreyfuss G. Why do cells need an assembly machine for RNA-protein complexes? Trends Cell Biol. 2004;14:226–232. doi: 10.1016/j.tcb.2004.03.010. [DOI] [PubMed] [Google Scholar]
- Young PJ, Man NT, Lorson CL, Le TT, Androphy EJ, Burghes AH, Morris GE. The exon 2b region of the spinal muscular atrophy protein, SMN, is involved in self-association and SIP1 binding. Hum Mol Genet. 2000;9:2869–2877. doi: 10.1093/hmg/9.19.2869. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Lotti F, Dittmar K, Younis I, Wan L, Kasim M, Dreyfuss G. SMN Deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell. 2008;133:585–600. doi: 10.1016/j.cell.2008.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.