Abstract
Ca2+-sensing receptors (CaSRs) represent a class of receptors that respond to changes in the extracellular Ca2+ concentration ([Ca2+]o) and activate multiple signaling pathways. A major barrier to advancing our understanding of the role of Ca2+ in regulating CaSRs is the lack of adequate information about their Ca2+-binding locations, which is largely hindered by the lack of a solved three-dimensional structure and rapid off rates due to low Ca2+-binding affinities. In this paper, we have reported the identification of three potential Ca2+-binding sites in a modeled CaSR structure using computational algorithms based on the geometric description and surface electrostatic potentials. Mutation of the predicted ligand residues in the full-length CaSR caused abnormal responses to [Ca2+]o, similar to those observed with naturally occurring activating or inactivating mutations of the CaR, supporting the essential role of these predicted Ca2+-binding sites in the sensing capability of the CaSR. In addition, to probe the intrinsic Ca2+-binding properties of the predicted sequences, we engineered two predicted continuous Ca2+-binding sequences individually into a scaffold protein provided by a non-Ca2+-binding protein, CD2. We report herein the estimation of the metal-binding affinities of these predicted sites in the CaSR by monitoring aromatic-sensitized Tb3+ fluorescence energy transfer. Removing the predicted Ca2+-binding ligands resulted in the loss of or significantly weakened cation binding. The potential Ca2+-binding residues were shown to be involved in Ca2+/Ln3+ binding by high resolution NMR and site-directed mutagenesis, further validating our prediction of Ca2+-binding sites within the extracellular domain of the CaSR.
Temporal and spatial changes of the Ca2+ concentration in the extra- and intracellular environments of cells affect the regulation of numerous cellular processes by modulating the activity of Ca2+ receptors and/or Ca2+-binding proteins (1–3). Multiple Ca2+-binding proteins with differing affinities have been identified in a variety of cellular compartments in all eukaryotic cells (4–6). Ca2+-binding proteins have Ca2+ affinities that vary by 106-fold or more depending upon their locations and functions (7, 8). Intracellularly, Ca2+-binding trigger proteins, such as calmodulin with its conserved EF-hand Ca2+-binding sites, have Ca2+-binding affinities in the submicromolar range (9). They respond to changes in the cytosolic free Ca2+ concentration ([Ca2+]i) and regulate numerous cellular events and processes (10, 11). Extracellularly, Ca2+ also functions as a first messenger to direct numerous intracellular functions (12–14). The Ca2+-sensing receptor (CaSR)4 that was initially cloned from the parathyroid gland (15) is a sensor of the extracellular Ca2+ concentration ([Ca2+]o) that transforms the [Ca2+]o stimulus into a variety of intracellular responses to regulate multiple signaling pathways (including activation of phospholipases C, A2, and D) and inhibition of cAMP formation (16, 17). This receptor, along with the metabotropic glutamate receptors (mGluRs), γ-aminobutyric acid type B receptors, and receptors for pheromones, amino acids, and sweeteners, belongs to family C of the G protein-coupled receptor superfamily. Most members in this family have the capacity to sense [Ca2+]o (18, 19). More than 100 mutations and polymorphisms have been identified in the CaSR that either inactivate (have a reduced sensitivity to [Ca2+]o) or activate (have an enhanced sensitivity to [Ca2+]o) the receptor and that cause familial hypocalciuric hypercalcemia and neonatal severe hyperparathyroidism (in the former) and autosomal dominant hypoparathyroidism (in the latter case) (20, 21).
The CaSR and other members of family C of the G protein-coupled receptor consist of a large extracellular domain (ECD), a transmembrane domain with seven transmembrane segments, and an intracellular C-tail segment (22, 23). Based on the functional responses at the cellular level, the ECD regions have been proposed to contain the major Ca2+-binding sites and to respond to [Ca2+]o for both the mGluRs and CaSRs. The Hill coefficient suggests that 3–5 Ca2+ ions bind cooperatively to the CaSR (22, 24). In addition to Ca2+, the CaSR also responds to Mg2+ (25), polyamines, and amino acids, such as l-Phe. Similar to the capacities of the mGluRs to respond to both l-Glu and high [Ca2+]o, the Ca2+-induced activation of the CaSR is potentiated by l-amino acids, particularly aromatic amino acids (26).
Progress in understanding the mechanism mediating [Ca2+]o regulation is largely hampered by a lack of knowledge regarding the Ca2+-binding sites in the CaSR. To date, the identity of the Ca2+-binding sites in the CaSR and related G protein-coupled receptors still remains unknown. X-ray structural determination by Kunishima et al. (27) reveals that the ECD of mGluR1 contains a Venus flytrap module. Key residues involved in glutamate binding were located at the interface between the two lobes (LB1 and LB2). However, no bound Ca2+ has been observed in these structures with or without the ligand glutamate (28). Major challenges in probing Ca2+-binding sites in this class of receptors include the difficulties in crystallization, the rapid off rates because of the low affinity of the Ca2+ binding and the existence of multiple conformations that are in equilibrium with one another (29). Indeed, successful crystallization of the CaSR ECD has not been reported despite a decade or more of effort directed to this end. Furthermore, methods for direct measurement of Ca2+ binding to the CaSR have not yet been established (30, 31), possibly because of its large size, multiple binding sites, and conformational flexibility.
In this study, to reveal the potential Ca2+-sensing locations in the CaSR, we first created model structures of the CaSR based on its structural homology to mGluR1. We then predicted several possible Ca2+-binding sites using our established computational algorithms based on the common structural properties of Ca2+-binding sites in proteins. We have shown that removing the predicted ligand residues in the full-length CaSR results in significant changes in the intracellular responses to [Ca2+]o. Two predicted continuous Ca2+-binding sites (e.g. sites with ligand-binding sites within short continuous primary sequences (<30 residues)) were further validated by inserting the sequences individually into a non-Ca2+-binding host protein, CD2. The resulting engineered proteins (designated CD2-CaSR1 and CD2-CaSR2) possess the ability to bind Ca2+ and Ln3+, in which the predicted Ca2+-binding residues were shown to be involved in Ca2+/Ln3+ binding by high resolution NMR and site-directed mutagenesis, further validating our prediction of Ca2+-binding sites within the ECD of the CaSR.
MATERIALS AND METHODS
Computational Prediction of Ca2+-binding Sites from a Model Structure
A sequence alignment of the ECD region (1–540 residues) of the human CaSR and mouse mGluR was carried out using the ClustalW program (32). Structural modeling of the CaSR was performed using SWISS-MODEL (33, 34) and MODELLER software (35) based on the structures of mGluR1 (Protein Data Bank (PDB) codes 1EWT (27) and 1ISR (28)). Thereafter, the putative Ca2+-binding sites in the CaSR were predicted using the program MetalFinder as described previously (36). Asp was used as the anchor ligand, because it is the most frequently used amino acid residue in Ca2+-binding sites. The oxygen atoms from backbone carbonyls and the side chains of Asp, Glu, Asn, Gln, Ser, Thr, and Tyr were used as potential Ca2+ ligand residues. In the computational calculation, an increased Ca-O distance (1.0–4.0 Å) and O-Ca-O angles (±60°) were used to compensate for the uncertainties in the model structures (37–39). The resulting potential sites identified by the algorithm were scored and ranked based on the ligands. Those that used all native, existing residues of the CaSR sequence as the ligands received the highest scores. The predicted sites that required mutations to similar residue types received higher scores than others. Similar residue types refer to the following pairs: Asn-Asp, Gln-Glu, Asp-Glu, and Ser-Thr. In contrast, the requirement of mutating non-Ca2+-binding ligands suggests that the location does not bind Ca2+ in the native CaSR. Finally, the electrostatic potentials were calculated using the program DelPhi (40, 41), and the model structures with the hydrogen atoms were built in by SYBYL. For the DelPhi calculations, interior and exterior dielectric constants of 2 and 80, respectively, were used. The salt concentration was 0, and the linear solution of the Poisson Boltzmann equation was imposed until convergence was reached. The geometrically predicted locations with 2–4 negatively charged ligands, strong negative surface potential, and functionally necessary residues were more likely to bind Ca2+. The model structures of CD2 variants with grafted potential Ca2+-binding sites of the CaSR were generated by SWISSMODEL (33, 34).
Measurement of [Ca2+]i in the Cell Population by Fluorimetry
The [Ca2+]i responses of wild type and mutant CaSRs were assessed as described by Bai et al. (42). In brief, the HEK293 cells transfected with the CaSR or its mutant cDNAs were loaded with Fura-2/AM. The remaining extracellular Fura-2/AM was washed out before the cells were transferred to a fluorescence cuvette. The emission at 510 nm was measured with excitation at 340 and 380 nm under varying [Ca2+]o (0.5–20.5 mm). The ratio of the fluorescence intensities was used to derive [Ca2+]i. All measurements were carried out in triplicate. Data are expressed with the S.E. of the mean as the index of dispersion.
Protein Engineering, Expression, and Purification
The CaSR sequences Gly222–Ile235 (GIEKFREEAEERDI) and Gly383–Ile408 (GHEESGDRFSNSSTAFRPLCTGDENI) were individually inserted between Ser52 and Gly53 of CD2 in the plasmid pGEX-2T (denoted as CD2-CaSR1 and CD2-CaSR2, respectively) by PCR using an established protocol (63). Two mutants, CD2-CaSR1-E228A/E229A and CD2-CaSR2-E378A/E379A, were also made by site-directed mutagenesis. Three glycines at the N terminus and two at the C terminus of the inserted sequences were thereafter inserted using a similar procedure (denoted as CD2-CaSR1-5G and CD2-CaSR2-5G). The mutants of the engineered proteins were produced using standard PCR methods. All sequences were verified by automated sequencing on an ABI PRISM-377 DNA sequencer (Applied Biosystems) at the Advanced Biotechnology Core Facilities of Georgia State University.
The proteins were expressed as glutathione S-transferase fusion protein using Escherichia coli BL21 (DE3) cells in LB medium with 100 mg/liter of ampicillin at 37 °C. For 15N isotopic labeling, 15NH4Cl was supplemented as the sole source of nitrogen in the minimal medium. Isopropyl-β-d-thiogalactopyranoside at 100 μm was added when the A600 reached 0.6 to induce protein expression for another 3–4 h. The cells were collected by centrifugation at 5000 × g for 30 min. The purification procedures followed the protocols for glutathione S-transferase fusion protein purification using glutathione-Sepharose 4B beads (GE Healthcare). The glutathione S-transferase tag of the proteins was cleaved on the beads by thrombin. The eluted engineered CD2 variants were further purified using Superdex 75 and Hitrap SP columns (GE Healthcare). The purified proteins were confirmed by matrix-assisted laser desorption ionization time-of-flight mass spectrometry at the Advanced Biotechnology Core Facilities of Georgia State University. The protein concentrations were determined using an ε280 of 11,700 m–1 cm–1 (43).
Electrospray Ionization Mass Spectrometry (ESI-MS)
ESI-MS was performed using a Waters Micromass Q-TOF micromass spectrometer. The data were acquired in positive ion mode by syringe pump infusion of the protein solutions at a flow rate of 7 μl/min. The protein sample stock (~1 mm) in 10 mm Tris, pH 7.4, was diluted 100-fold into water. Metal ions were added in 5-fold molar excess relative to the protein concentration.
Circular Dichroism
The circular dichroism spectra were recorded from 190 to 260 nm on a Jasco-810 spectropolarimeter at ambient temperature using a quartz cell of 1 mm path length with protein concentrations ranging from 15 to 20 μm in 10 mm Tris-HCl, pH 7.4. All spectra were the average of 10 scans with a scan rate of 100 nm/min. The spectra were converted to mean residue molar ellipticity after subtracting the spectrum of the buffer as the blank.
Fluorescence Spectroscopy
A PTI lifetime fluorimeter was used to record the fluorescence spectra at ambient temperature using a 1-cm path length cell. Intrinsic tryptophan emission spectra were recorded from 300 to 400 nm with the excitation wavelength at 282 nm. The slit widths were set at 4 and 8 nm for excitation and emission, respectively. Protein samples (1.5–3.0 μm) were in 20 mm PIPES-10 mm KCl at pH 6.8. For Tyr/Trp-sensitized Tb3+ fluorescence energy transfer (Tb3+-FRET) experiments, emission spectra were collected from 500 to 600 nm with the excitation at 282 nm with slit widths for excitation and emission set at 8 and 12 nm, respectively. A glass filter with cutoff of 320 nm was used to circumvent secondary Rayleigh scattering. The Tb3+ titration was performed by gradually adding different volumes of Tb3+ stock solutions (1 mm) into the protein samples (2.5 μm) in 20 mm PIPES, pH 6.8, with 10–150 mm KCl. For the Ca2+ competition studies, a solution containing 30 μm Tb3+ and 1.5 μm protein was set as the starting point. The stock solution of 10–100 mm CaCl2 with the same concentration of Tb3+ and protein was gradually added in the initial mixture. The fluorescence intensity was normalized by subtracting the contribution of the base-line slope using logarithmic fitting. The Tb3+-binding affinity of protein was obtained by fitting normalized fluorescence intensity data using the equation,
| (Eq. 1) |
where f is the fractional change, Kd is the dissociation constant for Tb3+, and [P]T and [M]T are the total concentrations of protein and Tb3+, respectively.
The Ca2+ competition data were first analyzed to derive the apparent dissociation constant by Equation 1. By assuming that the sample is saturated with Tb3+ at the starting point of the competition, the Ca2+-binding affinity is further obtained by using the equation,
| (Eq. 2) |
where Kd,Ca and Kd,Tb are the dissociation constants of Ca2+ and Tb3+, respectively. Kapp is the apparent dissociation constant.
NMR Spectroscopy
NMR spectra were collected on a 600-MHz NMR spectrometer. Two-dimensional total correlation spectra were collected using a water-gated total correlation spectroscopy pulse sequence with an isotropic mixing time of 75 ms at 25 °C. The spectrum width was 13.3 parts/million at both dimensions with a complex data point of 4096 at the first dimension and 400 increments at the second dimension. Two-dimensional (15N,1H)-heteronuclear single quantum correlation (HSQC) NMR spectra were collected with 4096 complex data points at the 1H dimension and 128 increments at the 15N dimension. Samples contained the protein at 1 mm in 20 mm PIPES-150 mm KCl, 10% D2O at pH 6.8. All of the NMR data were processed using FELIX (Accelrys) on a Silicon Graphics computer.
RESULTS
Prediction of Ca2+-binding Sites in the CaSR
The ECD of the CaSR contains the majority of the Ca2+-sensing determinants, and glycosylation does not play a significant role in the activation of the receptor by high [Ca2+]o. Both mGluRs and CaSR respond to high [Ca2+]o and are potentiated (activated) by l-amino acids (26). Their ECDs share ~30% sequence identity and a highly similar arrangement of secondary structural elements to that of mGluR. Therefore, we performed sequence alignment of the ECD region (1–540 residues) of the human CaSR and mouse mGluR, and then model structures of the ECD of the CaSR were created based on three structures of mGluR1, the ligand-free form (PDB code 1EWT), the Glu- and Mg2+-bound forms (PDB code 1EWK) (27), and the Glu- and Gd3+-bound form (PDB code 1ISR) (28). Fig. 1 shows the model structure of the ECD of the CaSR with the Venus flytrap structure. Two subdomains, termed as lobe 1 (N terminus) and lobe 2 (C terminus), are linked by several loops. The structure is very similar to the model structures previously reported by Bai (22) and Silve et al. (44). This similarity of the models from different groups and methods strengthens the assumption that the model structures accurately represent the true structures of the CaSR.
FIGURE 1.
A, model structure of the Ca2+-sensing receptor based on the x-ray structure of the ECD domain of mGLuR1 (PDB code 1EWT), which contains a Venus flytrap motif that is conserved in more than 11 different proteins, including the bacterial periplasmic-binding proteins, maltose-binding proteins, and galactose-binding proteins. Two subdomains are connected by a flexible hinge region. Similar to other small molecules, such as maltose and galactose, Glu binds to the hinge region, resulting in a large conformational change with a hinge movement. B, electrostatic surface analysis on the model structure of the ECD of the CaSR. Circled positions are the predicted Ca2+-binding sites, which are rich in negatively charged residues. C, model structure of Ca2+-binding sites of sites 1–3. The proposed Ca2+-binding ligand residues are labeled.
We then predicted potential Ca2+-binding pockets in the modeled CaSR structure using our developed computational algorithms (“Materials and Methods”). Fig. 1 and Table 1 show three predicted potential Ca2+-binding pockets (sites 1–3) in the modeled CaSR after the evaluation of their local geometric properties and electrostatic potentials. site 1 is located in lobe 2. It contains five glutamate residues in a nine-residue sequence (Glu224–Glu232). Three positive residues (Lys225, Arg227, and Arg233) and one aspartate residue (Asp234) are either in or follow this sequence closely. Site 2, located in lobe 1, includes all of the predicted ligand residues in a 22-residue sequence (Glu378-Glu399), which are Glu378, Glu379, Thr396, Asp398, and Glu399. Predicted sites 1 and 2 do not have extensive interactions with other parts of the protein. Site 3 is formed by residues Ser147, Ser170, Asp190, Tyr218, and Glu297, which are located in the crevice between the two lobes. All three sites are located at flexible loops or helical regions and are largely exposed on the surface of the protein. These geometrically predicted sites overlap with the negatively charged “hot spots” predicted by the electrostatic potential (Fig. 1).
TABLE 1.
Predicted Ca2+ binding sites and their corresponding mutations in diseases. NA, not applicable
| Predicted sites | Residues | Disease-related mutations |
|
|---|---|---|---|
| Site mutations | Environmental mutations | ||
| Site 1 | Glu-224, -228, -229, -231, -232 | NSPHT: D215G ADH: E228Q | NSPHT: Y218G/S, R220W/Q, P221S, R227L/Q, E250K ADH: P221L, Q245R |
| Site 2 | Glu-378, Glu-379, Thr-396, Asp-398, Glu-399 | NA | FHH/NSPHT: C395R |
| Site 3 | Ser-147, Ser-170, Asp-190, Tyr-218, Glu-297 | FHH/NSPHT: S147A, S170A, D190K, Y218S, E297K | FHH/NSPHT: S137P, T138M, E191K, D215G, R220W/Q, P221S ADH: T151M |
Effect of Mutation of Putative Ca2+-binding Ligand Residues on the Biological Function of the CaSR
The CaSR responds to elevated levels of [Ca2+]o by activating phospholipase C, which leads to the production of inositol 1,4,5-trisphosphate and further results in transient increases in the cytosolic Ca2+ concentration. To investigate the role of the proposed Ca2+-binding sites in the biological function of the CaSR, we created several mutations in the predicted charged ligand residues in the full-length CaSR. Specifically, we used site-directed mutagenesis to mutate the following residues: E297I in predicted site 3 located at the crevice, E224I and E228I/E229I in site CaSR1 located in lobe 2, and E378I/E379I and E398I/E399I in site CaSR2 located in lobe 1 (Tables 1 and 2). The wild type CaSR and the respective mutants were expressed in HEK293 cells using previously reported methods (45). The effects of the mutations on the biological function of the CaSR are summarized in Table 2 and Fig. 2. As shown in Fig. 2, removal of putatively charged ligand residues at all three predicted Ca2+-binding sites led to significant alterations in either maximal Ca2+ response and/or sensitivity to [Ca2+]o of the CaSR. E224I and the double mutant E228I/E229I at predicted site 1 decreased the maximal Ca2+ response by 35–38% with a slight left shift in sensitivity to [Ca2+]o (EC50 ± 2.5 ± 0.3 and 2.8 ± 0.2 mm, respectively, versus 3.1 ± 0.2 mm for the wild type CaSR, n = 3, p < 0.05). The mutation E398I/E399I in the predicted site 2 also resulted in a 37% decrease in the maximal intracellular Ca2+ response. This mutant, however, exhibited a right-shifted EC50 of 4.4 ± 0.2 mm (p < 0.01). Interestingly, the E378I/E379I mutant at the same predicted site resulted in a 23% enhancement in the maximal response and a left-shifted EC50 (EC50 = 2.2 ± 0.1 mm, n = 3, p < 0.01). Mutation E297I in site 3 significantly impaired the sensitivity to the [Ca2+]o response with an EC50 of 9.6 ± 0.2 mm.
TABLE 2.
Summary of the maximal response and EC50 values of CaSR and its mutants
| Predicted site | Mutation | Maximal response | Ca2+, EC50 |
|---|---|---|---|
| % | mm | ||
| Site 1 | Wild type | 100 | 3.1 ± 0.2 |
| E224I | 65 ± 1a | 2.5 ± 0.3b | |
| E228I/E229I | 62 ± 6a | 2.8 ± 0.2 | |
| Site 2 | E378I/E379I | 123 ± 6b | 2.2 ± 0.1a |
| E398I/E399I | 63 ± 12b | 4.4 ± 0.2a | |
| Site 3 | E297I | 87 ± 6 | 9.6 ± 0.2a |
p < 0.01.
p < 0.05.
FIGURE 2. Effect of mutations in charged residues of the predicted Ca2+-binding sites on the responsiveness of the CaSR by monitoring intracellular Ca2+ using Fura-2.
Functional characterization of the wild type CaSRs and CaSRs with mutations in site 1 (A), site 2 (B), or site 3 (C) were performed using HEK293 cells transfected with the wild type and CaSR mutated in charged residues and are expressed as the normalized Ca2+ responses.
Our functional characterization of the putative ligand residues in the predicted Ca2+-binding sites of the CaSR were consistent with many mutations around the proposed Ca2+-binding sites, which are associated with clinical syndromes (autosomal dominant hypoparathyroidism and familial hypocalciuric hypercalcemia) due to either a decrease or an increase in the sensitivity of the respective receptors to [Ca2+]o. By monitoring the intracellular Ca2+ response of HEK293 cells transfected with the wild type and mutant receptors, mutations of Ser147, Ser170, Asp190, Tyr218, and E297K were shown to largely impair the activation of the human CaSR (45–47). Recently, Silve et al. (44) have shown that the missense mutations, E297K and Y218S, significantly reduce the maximum Ca2+-induced [3H]IP response. They postulate that the residues Ser170, Asp190, Gln193, Ser296, and Glu297 are critical for the CaSR Ca2+ binding and downstream biological functions, which is in excellent agreement with our prediction. Although these findings are not based on direct measurement of Ca2+ binding, they provide strong experimental evidence to support our model structures as well as the prediction of Ca2+-binding sites in the ECD of the CaSR.
Engineering Proteins by Grafting the Predicted Sequences into CD2
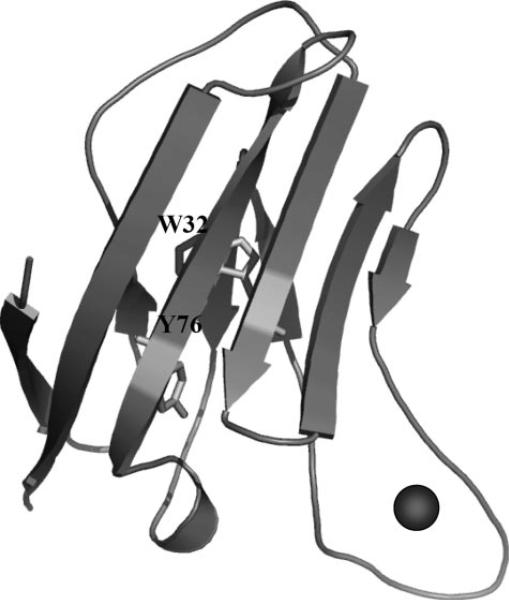
Investigation of the site-specific Ca2+-binding properties of the CaSR is one important step toward fully understanding the mechanism underlying its Ca2+-modulated functions. Our laboratory had previously established a grafting approach to investigate the site-specific metal-binding properties of calmodulin (CaM) by inserting the individual EF-loops of CaM into a host frame, CD2 (48). Because predicted Ca2+-binding sites 1 and 2 of the CaSR have contiguous stretches of amino acids, similar to the Ca2+-binding motifs in CaM, we extended the grafting approach to probe the site-specific Ca2+-binding affinity of these two sites. The continuous sequences were inserted into CD2 at position 52 between the strands C” and D, because our previous studies had shown that this position tolerates the insertion of EF-hand motifs from CaM. The distances between the two termini of the inserted Ca2+-binding sites in the model structures of the CaSR were within 15 Å. Accordingly, a total of 5–6 glycine linkers is sufficient to enable the grafted motifs to retain a native metal-binding conformation (48), and two more variants were thus engineered with 3 and 2 flanking Gly residues at the N and C termini of the CaSR sequence. Fig. 3 shows the modeled structure of CD2 with grafted Ca2+-binding sites from the CaSR. Trp32 and Tyr76 in the host proteins are ~12 Å distant from the grafted sites, which enables aromatic-sensitized energy transfer to the Tb3+ bound to the sites, providing a spectroscopic method to monitor the metal-binding process.
FIGURE 3. Modeled structure of CD2 with grafted Ca2+-binding sites in the CaSR.
Trp32 and Tyr76 in the host protein are ~12 Å away from the grafted Ca2+-binding sites.
The Conformation of Scaffold Protein Is Not Altered after Grafting Ca2+-binding Sites
To ensure that the grafted Ca2+-binding site has no major interaction with the host protein and that the host protein has not changed its native conformation, we carried out conformational analyses using various methods. As shown in Fig. 4A, the far UV CD spectra of all four engineered proteins shows a trough at 216 nm, indicating a typical β-sheet secondary structure, as in the wild type CD2. The deeper negative molar ellipticity in the short wavelength region compared with that of CD2 was consistent with the addition of loop/less structured sequences of the CaSR. In addition, the Trp fluorescence spectrum of wild type CD2 overlaps that of the engineered proteins, both of which exhibit two peaks at 314 and 335 nm, suggesting that the Trp environment remained unchanged after engineering of the protein (Fig. 4B). Furthermore, the majority of the NMR chemical shifts of the host frame in the engineered variants were not significantly different from those of CD2 (Fig. 4C), suggesting that the scaffold protein was minimally perturbed after the insertion of the foreign sequences. All of these results imply that the host scaffold protein retained its native structure, thereby ensuring a minimal contribution to the metal binding of the inserted sequences.
FIGURE 4. The secondary structures of wild type CD2 and engineered proteins.
A, far UV CD spectra of 15 μm wild type CD2, CD2-CaSR1, CD2-CaSR1–5G, CD2-CaSR2, and CD2-CaSR2–5G in 10 mm Tris, pH 7.4. B, Trp fluorescence spectra of wild type CD2 and the engineered proteins with grafted Ca2+-binding sites. C, comparison of chemical shifts of the α and backbone amide protons between wild type CD2 and the engineered protein CD2-CaSR1 with grafted Ca2+-binding sites from the CaSR.
The Grafted Sequences from CaSR Bind Cations
The Ca2+-binding capability of the inserted sequences was first revealed by NMR. Upon the addition of 10 mm Ca2+ under high salt conditions (150 mm KCl), several residues from the inserted sequences had altered resonances as shown in the HSQC spectra (Fig. 5). For example, Ca2+ binding resulted in chemical shift changes of at least two of the peaks arising from glutamates in CD2-CaSR1–5G (Fig. 5A). The addition of Mn2+ led to the disappearance of resonances from the inserted sequences and the CD2 host residues in proximity due to the line-broadening effect caused by the Mn2+ that substituted for the Ca2+ (data not shown). In contrast, wild type CD2 did not exhibit any detectable changes under identical conditions. Furthermore, the addition of La3+ to the CD2-CaSR2 led to significant changes of, for example, at least two resonances originating from the grafted sequence as well as Gly53 of CD2 (Fig. 5C). More convincingly, after substituting two proposed Ca2+-binding ligand residues within site 1 (Glu228/Glu229)or site 2 (Glu378/Glu379) with alanines, we could not detect such significant chemical shift changes upon the addition of metal ions (Figs. 5B and D), further supporting the involvement of these residues in the chelation of Ca2+ within each predicted metal-binding site. Mutagenesis studies on these putative Ca2+-binding ligands have also been shown to affect the biological function of CaSR (Fig. 2). Moreover, the mass peaks corresponding to the formation of 1:1 Ca2+-protein or Tb3+-protein complexes were observed in ESI-MS spectra in the presence of excess metal ions (Fig. 6). These results suggest that the inserted sequences from the CaSR have the capacity to bind Ca2+ and its trivalent analogs.
FIGURE 5. (1H,15N)-HSQC spectra of CD2-CaSR1, CD2-CaSR2, and their mutants.
A, Ca2+-induced chemical shift changes of two peaks arising from the four glutamates in the grafted Ca2+-binding sequence in CD2-CaSR1. This result provides direct evidence of Ca2+-induced movement of residues (Glu) within the grafted Ca2+-binding motif. Blue, CD2-CaSR1 with 1 mm EGTA; red, CD2-CaSR1 with 10 mm Ca2+. B, superimposed HSQC spectrum of CD2-CaSR1-E228A/E229A in the presence of 0.1 mm EGTA (blue) or 10 mm Ca2+ (red). No significant chemical shift changes were observed for the resonances from the inserted sequences (boxes). C, La3+-induced chemical shift changes of one of the linker residues, glycine 53 (left), and two resonances from the grafted CaSR2 sequence (right, boxed region). Meanwhile, the chemical shifts of resonances from the host framework (Ile14, Asn20, Asp25, Gly35, Glu41, Arg70, Ser82, and Thr83) remained almost unaltered. Blue, CD2-CaSR2 with 1 mm EGTA; red, CD2-CaSR2 with 1 mm La3+. D, overlaid HSQC spectrum of CD2-CaSR2-E378A/E379A in the presence of 0.1 mm EGTA (blue) or 1 mm La3+ (red). The addition of La3+ did not lead to chemical shift changes of glycine 53 or peaks originating from the inserted sequences (dashed boxes). The inset is another enlarged region of HSQC spectrum. The new resonance (dashed circle) emerged after mutagenesis was from one of the alanine residues in the inserted sequence. Addition of La3+ did not cause significant chemical shift change.
FIGURE 6. ESI-MS spectra of CD2-CaSR1 with metal ions.
The binding of Ca2+ (A) or Tb3+ (B) led to the emergence of additional peaks with molecular mass differences of +38 and +156, respectively.
The addition of Tb3+ into the engineered proteins (Fig. 7A) or vice versa resulted in large increases of Tb3+ fluorescence at 545 nm because of Trp-sensitized Tb3+-fluorescent resonance energy transfer (Tb3+-FRET) (36, 49), which was not observed for wild type CD2. The addition of Ca2+ into the Tb3+-protein mixture decreased the Tb3+ signal because of competition (Fig. 7B). The Tb3+- and Ca2+-binding affinities were derived from the Tb3+ fluorescence change and the metal competition, respectively. For CD2-CaSR1, the Tb3+- and Ca2+-binding dissociation constants obtained were 35 ± 3 and 890 ± 80 μm at low salt concentrations, respectively. At high salt concentrations (e.g. ~150 mm NaCl), as in the extracellular environment within which the extracellular domain of the native CaSR normally resides, the Ca2+-binding affinity decreased at least 10-fold (Table 3). For CD2-CaSR2, the Tb3+ and Ca2+ dissociation constants were 25 ± 2 μm and 4.3 ± 0.8 mm at low and 98 ± 7 μm and 18.6 ± 0.5 mm at high salt concentrations, respectively. The relatively stronger affinity for Tb3+ than for Ca2+ is consistent with other natural Ca2+-binding proteins and is partly because of the electrostatic nature of the metal binding (50). Further, the variants with or without flanking glycine linkers exhibit no significant differences in their metal-binding affinities, suggesting that the grafted Ca2+-binding sites are flexible and less sensitive to the host protein.
FIGURE 7. Probing metal binding with aromatic residue-sensitized Tb3+-FRET.
A, the enhancement of Tb3+ fluorescence of CD2-CaSR1 at 545 nm as a function of titrated Tb3+. A 1:1 binding mode was assumed for data fitting. B, Ca2+ competition assay. The addition of Ca2+ led to a decrease of Tb3+ fluorescence due to competitive binding of metal ions in CD2-CaSR1 that was preincubated with Tb3+. C, comparison of relative Tb3+ signal changes of wild type CD2-CaSR2 and its charged mutants.
TABLE 3.
Dissociation constants of CD2 variants with grafted Ca2+-binding sequences from the CaSR
| Variant | Tb3+ |
Ca2+ |
||
|---|---|---|---|---|
| 10 mm KCl | 150 mm NaCl | 10 mm KCl | 150 mm NaCl | |
| μ m | μ m | |||
| CD2-CaSR1 | 35 ± 3 | 144 ± 5 | 890 ± 80 | 4200 ± 400 |
| CD2-CaSR1-5G | 7 ± 1 | 94 ± 7 | 400 ± 240 | 5000 ± 700 |
| CD2-CaSR2 | 25 ± 2 | 98 ± 7 | 4300 ± 800 | 18600 ± 500 |
| CD2-CaSR2-5G | 19 ± 2 | 80 ± 3 | 2400 ± 1000 | 6100 ± 200 |
Site-directed mutagenesis that removed the predicted charged ligand residues resulted in a dramatic decrease of the Tb3+-FRET signal, suggesting attenuated metal-binding affinities. For example, the Tb3+ fluorescence enhancements of both mutants E378A/E379A and D398A/E399A are 70% lower than that of CD2-CaSR2. The signal of the mutant E231A/E232A is also 30% lower than that of CD2-CaSR. These results are consistent with our functional study of the mutations (Fig. 2 and Table 2) and further validate our prediction of Ca2+-binding sites in the ECD of the CaSR.
DISCUSSION
Predicting Ca2+ Binding in the CaSR and the Effect of Mutations on the Function of the CaSR
Ca2+ bound to proteins is predominantly chelated with oxygens with an average coordination of 6–7 (51–53). The binding of Ca2+ to multiple sites in a highly cooperative manner facilitates the response of a protein to small Ca2+ concentration changes (54–56), such as the binding of four Ca2+ ions to CaM. The Hill coefficient suggests that 3–5 Ca2+ ions bind cooperatively to the CaSR. However, the Ca2+-binding sites have not been identified.
Our laboratory has shown that naturally evolved Ca2+-binding sites can be identified and novel Ca2+-binding proteins can be designed using the pentagonal bipyramidal geometry with the side chains of the aforementioned residues and main chain carbonyls as potential ligands (53). In this study, taking advantage of the previously determined structures of mGLuR1 and the sequence homology of the CaSR to mGluR1, we predicted several Ca2+-binding sites in the modeled structure of the CaSR by applying our well developed computational algorithms. These sites possess high electrostatic potential and are located in the negatively charged environments (Fig. 1), which are preferred for Ca2+ binding (36). In addition, these putative Ca2+-binding sites are located in the flexible locations either in the crevices between two lobes (site 3) and/or at loop and exposed regions. This is consistent with a survey that revealed that almost all of the known Ca2+-binding sites are located in or adjacent to the flexible structures of their respective proteins, such as loop regions, turns, or the ends of α-helices and β-sheets (57).
Our model structure and predicted Ca2+-binding sites, especially site 3, are consistent with the reported functional studies. The x-ray structure of mGluR1 complexed with its ligand, glutamate, reveals that residues Ser164, Ser165, Thr188, Tyr236, and Glu292 in the protein form hydrogen bonds with the α-carboxyl group of the glutamate (58). These residues correspond to Ser147, Ser170, Asp190, Tyr218, and Glu297 in the CaSR, respectively. Two mutations, Y218S and E297K, markedly reduce the responsiveness and sensitivity of the receptor to [Ca2+]o (24). Furthermore, mutations around these predicted locations have been shown to alter the Ca2+ response of cells expressing the respective mutant receptors, supporting our prediction. For example, T151M in autosomal dominant hypoparathyroidism and R185Q in familial hypocalciuric hypercalcemia are adjacent to Ca2+ ligand residues in site 3. Additional mutations that alter the CaSR responses, such as Y218S/C, E297K, and R220S and R221S, are also either at or near the predicted ligand residues of site 3 (59, 60). In addition, Zhang et al. (24) have demonstrated that Ser170 is critical for functional modulation of the CaSR by l-Phe, in that the S170A mutation led to no significant change in the EC50 value of the protein for [Ca2+]o but a 1.6-fold increase in its response at 50 mm [Ca2+]o. Recently, Mun et al. (61) further showed that the double mutation T145A/S170T selectively disables l-amino acid sensing but does not change the [Ca2+]o-sensing capability of the CaSR, thereby suggesting that the two processes can be dissociated and that the l-amino acid is mainly bound near Ser170.
The predicted Ca2+-binding site in the crevice is likely to play a central role in modulating the function of the CaSR by Ca2+-induced conformational changes. Previous studies have shown that mutations in this site inactivate the protein (44). The mutations E297I and D215I provide additional support for this hypothesis. The glutamate-binding site in mGluR was also within the crevice of the protein, suggesting that the hinge that connects the two lobes directly responds to the stimuli-inducing receptor activation in the G protein-coupled receptor family C proteins.
The mutations outside of the crevice demonstrate the complexity of the regulatory mechanism of the CaSR. Removal of charged residues in both lobe 1 (E224I and E228I/E229I) and lobe 2 (E398I/E399I) decreased the maximal response levels but resulted in oppositely directed changes in the sensitivity of the receptor to [Ca2+]o. Perhaps the maximal responses and the sensitivity were determined by different locations of the protein via distinct mechanisms. In addition, loss of the charges on these residues could alter the interactions of these amino acids with charged residues elsewhere on the protein, potentially favoring conformations that either activate or inactivate the protein, depending on local geometrical factors. Because both mutations resulted in significantly weakened metal binding in the CD2-CaSR variants, the results suggest that Ca2+ binding at the predicted locations are necessary for maintaining the full function of the CaSR. Therefore, Ca2+ acts as both a stimulus and a regulator for the CaSR. The potential Ca2+ binding to Glu398/Glu399 might be crucial, because removal of these two charges causes a dual effect by decreasing both the maximal activity and the sensitivity to [Ca2+]o.
Grafting Approach for Probing Ca2+-binding Capability
To overcome the limitations of solely investigating the Ca2+-binding sites in native proteins, we established a grafting approach to dissect their site-specific properties. This approach has been used previously in the investigation of single EF-hand motifs in CaM (62, 63). CD2 has been shown to be a suitable host system, because it retains its native structure after insertion of the foreign sequences, both in the presence and absence of Ca2+ ions, so that the influence of the host protein on the inserted sites is minimized. The difficulties inherent in directly confirming the predicted Ca2+-binding ligands due to the multiple binding and conformational changes of the CaSR prompted us to investigate this issue indirectly using the grafting approach. In CD2-CaSR variants, additional residues and glycine linkers are at both termini of the predicted sequences to provide the conformational freedom needed for the formation of their native structures. In addition, the host protein retains the native structure of CD2, as indicated by CD, fluorescence, and NMR studies with and without cations. The addition of cations induced chemical shift changes of the resonances arising from the inserted sequences but not those from the host proteins, indicating that the binding is independent of the host protein and occurs at the inserted sequences.
The capability of the two predicted Ca2+-binding sites of the CaSR grafted into CD2 to bind Ca2+ and Ln3+ validated our computational identifications. In the CaSR model structures, the predicted sequences are in flexible loop regions. In CD2-CaSR variants, these sequences are also in flexible loops indicated by the far-UV CD spectra. The variants with or without glycine linkers exhibited similar Tb3+ and Ca2+-binding affinities, which also implies that these Ca2+-binding sites are highly flexible, and the contribution of the host protein frame to the metal binding is less likely to be significant. The probed site-specific Ca2+-binding affinities at 150 mm NaCl for CD2-CaSR1 and CD2-CaSR2 are 4.2 ± 0.3 and 18.6 ± 0.5 mm, respectively. These affinities are weaker than the EC50 value of the CaSR (Tables 2 and 3), which can be explained by several possible factors. First, the interaction of coupled metal-binding sites and cooperativity are likely to contribute to the overall sensing capacity of the intact CaSR to respond to [Ca2+]o. This is similar to the contribution of the cooperativity observed for CaM. This cooperativity could be the result of direct site-to-site interactions or through Ca2+-induced conformational change. The cooperativity is not necessarily positive in all cases. In fact, the opposite effects of the mutations at predicted sites 1 and 2 on the sensitivity of the protein to [Ca2+]o indicate that Ca2+ binding at various locations within the CaR ECD has diverse influences on the subsequent Ca2+ binding to other sites. If Ca2+ effectively cross-linked and neutralized negative charges in two different parts of the molecule (one of which was one of our binding sites), for example, which then caused the lobes to close, then loss of one of those negative charges could reduce the repulsion between the negative charges that was present in the absence of calcium and favor activation of the receptor at lower extracellular calcium. Second, other factors, such as dimer formation and the protein environment (e.g. hydrogen bonding and salt bridges), may also play a part in the affinity differences between the isolated sites and the full protein. In the engineered proteins, the host protein has minimal effects on metal binding. However, in the native CaSR, the surrounding residues will surely influence the electrostatic potentials at the Ca2+-binding sites, in a Ca2+-dependent or -independent manner. Third, it is possible that there are additional Ca2+ ligands for the sites that have not been identified in our prediction. Our work has opened up the opportunity for investigating the mechanism(s) underlying Ca2+-modulated function by determining Ca2+-binding properties in CaSRs at local levels.
In conclusion, several Ca2+-binding sites have been predicted in the model structure of the CaSR. Two continuous predicted sites have been grafted into the host protein CD2. The metal-binding studies on the engineered proteins using various spectroscopic methods validate the computational predictions and demonstrate the Ca2+-binding capability of the predicted sequences. Mutations that remove the predicted charged Ca2+ ligand residues lead to weakened metal binding in the engineered proteins and abnormal responses to [Ca2+]o in the full-length CaSR, supporting the predictions and indicating the importance of Ca2+ binding in regulating CaSR functions.
Acknowledgments
We thank Dr. April L. Ellis for the experimental procedures and techniques used in the study, Dr. Lisa Jones for the electrostatic calculation and Julian Johnson and Michael Kirberger for helpful suggestions. We thank Drs. Jun-tao Guo and Youxing Qu at University of Georgia at Athens, GA and Drs. Mei Bai and Stephen Quinn at Brigham and Women's Hospital for their generous help in structural modeling. We thank Dr. Siming Wang for MS analysis. We thank other members in the J. J. Y. laboratory for helpful discussions.
Footnotes
This work was supported, in part, by National Institutes of Health Grants GM62999-1 and GM070555, National Science Foundation Grant MCB-0092486, American Heart Association Grant 0655168B (to J. J. Y.), and a Predoctoral Fellowship from the Brain and Behavior Program at Georgia State University (to Y. H.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The abbreviations used are: CaSR, Ca2+-sensing receptor; mGluR, metabotropic glutamate receptor; ECD, extracellular domain; ESI-MS, electrospray ionization mass spectrometry; PIPES, 1,4-piperazinediethanesulfonic acid; FRET, fluorescence energy transfer; HSQC, heteronuclear single quantum correlation; CaM, calmodulin.
REFERENCES
- 1.Carafoli E. Proc. Natl. Acad. Sci. U. S. A. 2002;99:1115–1122. doi: 10.1073/pnas.032427999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berridge MJ, Bootman MD, Lipp P. Nature. 1998;395:645–648. doi: 10.1038/27094. [DOI] [PubMed] [Google Scholar]
- 3.Maurer P, Hohenester E, Engel J. Curr. Opin. Cell Biol. 1996;8:609–617. doi: 10.1016/s0955-0674(96)80101-3. [DOI] [PubMed] [Google Scholar]
- 4.Michalak M, Robert Parker JM, Opas M. Cell Calcium. 2002;32:269–278. doi: 10.1016/s0143416002001884. [DOI] [PubMed] [Google Scholar]
- 5.Ikura M, Osawa M, Ames JB. BioEssays. 2002;24:625–636. doi: 10.1002/bies.10105. [DOI] [PubMed] [Google Scholar]
- 6.Feske S, Okamura H, Hogan PG, Rao A. Biochem. Biophys. Res. Commun. 2003;311:1117–1132. doi: 10.1016/j.bbrc.2003.09.174. [DOI] [PubMed] [Google Scholar]
- 7.Goll DE, Thompson VF, Li H, Wei W, Cong J. Physiol. Rev. 2003;83:731–801. doi: 10.1152/physrev.00029.2002. [DOI] [PubMed] [Google Scholar]
- 8.Teplyakov AV, Kuranova IP, Harutyunyan EH, Vainshtein BK, Frommel C, Hohne WE, Wilson KS. J. Mol. Biol. 1990;214:261–279. doi: 10.1016/0022-2836(90)90160-n. [DOI] [PubMed] [Google Scholar]
- 9.Kawasaki H, Nakayama S, Kretsinger RH. Biometals. 1998;11:277–295. doi: 10.1023/a:1009282307967. [DOI] [PubMed] [Google Scholar]
- 10.Ermak G, Morgan TE, Davies KJ. J. Biol. Chem. 2001;276:38787–38794. doi: 10.1074/jbc.M102829200. [DOI] [PubMed] [Google Scholar]
- 11.Zhang X, Joseph SK. Biochem. J. 2001;360:395–400. doi: 10.1042/0264-6021:3600395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Breitwieser GE, Miedlich SU, Zhang M. Cell Calcium. 2004;35:209–216. doi: 10.1016/j.ceca.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 13.Handford PA. Biochim. Biophys. Acta. 2000;1498:84–90. doi: 10.1016/s0167-4889(00)00085-9. [DOI] [PubMed] [Google Scholar]
- 14.Maki M, Kitaura Y, Satoh H, Ohkouchi S, Shibata H. Biochim Biophys Acta. 2002;1600:51–60. doi: 10.1016/s1570-9639(02)00444-2. [DOI] [PubMed] [Google Scholar]
- 15.Brown EM, Gamba G, Riccardi D, Lombardi M, Butters R, Kifor O, Sun A, Hediger MA, Lytton J, Hebert SC. Nature. 1993;366:575–580. doi: 10.1038/366575a0. [DOI] [PubMed] [Google Scholar]
- 16.Brown EM, MacLeod RJ. Physiol. Rev. 2001;81:239–297. doi: 10.1152/physrev.2001.81.1.239. [DOI] [PubMed] [Google Scholar]
- 17.Bai M. Int. J. Mol. Med. 1999;4:115–125. doi: 10.3892/ijmm.4.2.115. [DOI] [PubMed] [Google Scholar]
- 18.Jingami H, Nakanishi S, Morikawa K. Curr. Opin. Neurobiol. 2003;13:271–278. doi: 10.1016/s0959-4388(03)00067-9. [DOI] [PubMed] [Google Scholar]
- 19.Zonta M, Sebelin A, Gobbo S, Fellin T, Pozzan T, Carmignoto G. J. Physiol. (Lond.) 2003;553:407–414. doi: 10.1113/jphysiol.2003.046706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu J, Mora S, Weber G, Zamproni I, Proverbio MC, Spiegel AM. J. Bone Miner. Res. 2004;19:578–586. doi: 10.1359/JBMR.040106. [DOI] [PubMed] [Google Scholar]
- 21.Sayer JA, Pearce SH. Clin. Endocrinol. (Oxf.) 2003;59:419–421. doi: 10.1046/j.1365-2265.2003.01869.x. [DOI] [PubMed] [Google Scholar]
- 22.Bai M. Cell Calcium. 2004;35:197–207. doi: 10.1016/j.ceca.2003.10.018. [DOI] [PubMed] [Google Scholar]
- 23.Quinn SJ, Bai M, Brown EM. J. Biol. Chem. 2004;279:37241–37249. doi: 10.1074/jbc.M404520200. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Z, Sun S, Quinn SJ, Brown EM, Bai M. J. Biol. Chem. 2001;276:5316–5322. doi: 10.1074/jbc.M005958200. [DOI] [PubMed] [Google Scholar]
- 25.Vetter T, Lohse MJ. Curr. Opin. Nephrol. Hypertens. 2002;11:403–410. doi: 10.1097/00041552-200207000-00006. [DOI] [PubMed] [Google Scholar]
- 26.Francesconi A, Duvoisin RM. J. Neurosci. Res. 2004;75:472–479. doi: 10.1002/jnr.10853. [DOI] [PubMed] [Google Scholar]
- 27.Kunishima N, Shimada Y, Tsuji Y, Sato T, Yamamoto M, Kumasaka T, Nakanishi S, Jingami H, Morikawa K. Nature. 2000;407:971–977. doi: 10.1038/35039564. [DOI] [PubMed] [Google Scholar]
- 28.Tsuchiya D, Kunishima N, Kamiya N, Jingami H, Morikawa K. Proc. Natl. Acad. Sci. U. S. A. 2002;99:2660–2665. doi: 10.1073/pnas.052708599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang W, Wilkins AL, Li S, Ye Y, Yang JJ. Biochemistry. 2005;44:8267–8273. doi: 10.1021/bi050463n. [DOI] [PubMed] [Google Scholar]
- 30.Nagar B, Overduin M, Ikura M, Rini JM. Nature. 1996;380:360–364. doi: 10.1038/380360a0. [DOI] [PubMed] [Google Scholar]
- 31.Hu J, Spiegel AM. Trends Endocrinol. Metab. 2003;14:282–288. doi: 10.1016/s1043-2760(03)00104-8. [DOI] [PubMed] [Google Scholar]
- 32.Higgins DG. Methods Mol. Biol. 1994;25:307–318. doi: 10.1385/0-89603-276-0:307. [DOI] [PubMed] [Google Scholar]
- 33.Kopp J, Schwede T. Pharmacogenomics. 2004;5:405–416. doi: 10.1517/14622416.5.4.405. [DOI] [PubMed] [Google Scholar]
- 34.Schwede T, Kopp J, Guex N, Peitsch MC. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marti-Renom MA, Stuart AC, Fiser A, Sanchez R, Melo F, Sali A. Annu. Rev. Biophys. Biomol. Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 36.Yang W, Wilkins AL, Ye Y, Liu ZR, Li SY, Urbauer JL, Hellinga HW, Kearney A, van der Merwe PA, Yang JJ. J. Am. Chem. Soc. 2005;127:2085–2093. doi: 10.1021/ja0431307. [DOI] [PubMed] [Google Scholar]
- 37.Hellinga HW, Richards FM. J. Mol. Biol. 1991;222:763–785. doi: 10.1016/0022-2836(91)90510-d. [DOI] [PubMed] [Google Scholar]
- 38.Yang W, Jones LM, Isley L, Ye Y, Lee HW, Wilkins A, Liu ZR, Hellinga HW, Malchow R, Ghazi M, Yang JJ. J. Am. Chem. Soc. 2003;125:6165–6171. doi: 10.1021/ja034724x. [DOI] [PubMed] [Google Scholar]
- 39.Deng H, Chen G, Yang W, Yang JJ. Proteins. 2006;64:34–42. doi: 10.1002/prot.20973. [DOI] [PubMed] [Google Scholar]
- 40.Honig B, Nicholls A. Science. 1995;268:1144–1149. doi: 10.1126/science.7761829. [DOI] [PubMed] [Google Scholar]
- 41.Nicholls A, Honig B. J. Comput. Chem. 1991;12:435–445. [Google Scholar]
- 42.Bai M, Quinn S, Trivedi S, Kifor O, Pearce SH, Pollak MR, Krapcho K, Hebert SC, Brown EM. J. Biol. Chem. 1996;271:19537–19545. doi: 10.1074/jbc.271.32.19537. [DOI] [PubMed] [Google Scholar]
- 43.Driscoll PC, Cyster JG, Somoza C, Crawford DA, Howe P, Harvey TS, Kieffer B, Campbell ID, Williams AF. Biochem. Soc. Trans. 1993;21:947–952. doi: 10.1042/bst0210947. [DOI] [PubMed] [Google Scholar]
- 44.Silve C, Petrel C, Leroy C, Bruel H, Mallet E, Rognan D, Ruat M. J. Biol. Chem. 2005;280:37917–37923. doi: 10.1074/jbc.M506263200. [DOI] [PubMed] [Google Scholar]
- 45.Zhang Z, Qiu W, Quinn SJ, Conigrave AD, Brown EM, Bai M. J. Biol. Chem. 2002;277:33727–33735. doi: 10.1074/jbc.M200976200. [DOI] [PubMed] [Google Scholar]
- 46.Tu CL, Oda Y, Komuves L, Bikle DD. Cell Calcium. 2004;35:265–273. doi: 10.1016/j.ceca.2003.10.019. [DOI] [PubMed] [Google Scholar]
- 47.Chang W, Shoback D. Cell Calcium. 2004;35:183–196. doi: 10.1016/j.ceca.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 48.Ye Y, Lee HW, Yang W, Yang JJ. J. Inorg. Biochem. 2005;99:1376–1383. doi: 10.1016/j.jinorgbio.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 49.Ye Y, Shealy S, Lee HW, Torshin I, Harrison R, Yang JJ. Protein Eng. 2003;16:429–434. doi: 10.1093/protein/gzg051. [DOI] [PubMed] [Google Scholar]
- 50.Falke JJ, Drake SK, Hazard AL, Peersen OB. Q. Rev. Biophys. 1994;27:219–290. doi: 10.1017/s0033583500003012. [DOI] [PubMed] [Google Scholar]
- 51.Glusker JP. Adv. Protein Chem. 1991;42:1–76. doi: 10.1016/s0065-3233(08)60534-3. [DOI] [PubMed] [Google Scholar]
- 52.Dudev T, Chang LY, Lim C. J. Am. Chem. Soc. 2005;127:4091–4103. doi: 10.1021/ja044404t. [DOI] [PubMed] [Google Scholar]
- 53.Yang W, Lee HW, Hellinga H, Yang JJ. Proteins. 2002;47:344–356. doi: 10.1002/prot.10093. [DOI] [PubMed] [Google Scholar]
- 54.Akke M, Forsen S, Chazin WJ. J. Mol. Biol. 1995;252:102–121. doi: 10.1006/jmbi.1995.0478. [DOI] [PubMed] [Google Scholar]
- 55.Hiraoki T, Vogel HJ. J. Cardiovasc. Pharmacol. 1987;10(Suppl. 1):S14–S31. doi: 10.1097/00005344-198710001-00004. [DOI] [PubMed] [Google Scholar]
- 56.Malmberg NJ, Varma S, Jakobsson E, Falke JJ. Biochemistry. 2004;43:16320–16328. doi: 10.1021/bi0482405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pidcock E, Moore GR. J. Biol. Inorg. Chem. 2001;6:479–489. doi: 10.1007/s007750100214. [DOI] [PubMed] [Google Scholar]
- 58.Suzuki Y, Moriyoshi E, Tsuchiya D, Jingami H. J. Biol. Chem. 2004;279:35526–35534. doi: 10.1074/jbc.M404831200. [DOI] [PubMed] [Google Scholar]
- 59.Pearce SH, Trump D, Wooding C, Besser GM, Chew SL, Grant D, Heath DA, Hughes A, Paterson CR, Whyte MP, Thakker RV. J. Clin. Investig. 1995;96:2683–2692. doi: 10.1172/JCI118335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cetani F, Pardi E, Borsari S, Tonacchera M, Morabito E, Pinchera A, Marcocci C, Dipollina G. Clin. Endocrinol. (Oxf.) 2003;58:199–206. doi: 10.1046/j.1365-2265.2003.01696.x. [DOI] [PubMed] [Google Scholar]
- 61.Mun HC, Franks AH, Culverston E, Krapcho K, Nemeth EF, Conigrave AD. J. Biol. Chem. 2004;279:51739–51744. doi: 10.1074/jbc.M406164/200. [DOI] [PubMed] [Google Scholar]
- 62.Ye Y, Lee HW, Yang W, Shealy SJ, Yang JJ. J. Am. Chem. Soc. 2005;127:3743–3750. doi: 10.1021/ja042786x. [DOI] [PubMed] [Google Scholar]
- 63.Ye Y, Lee HW, Yang W, Shealy SJ, Wilkins AL, Liu ZR, Torshin I, Harrison R, Wohlhueter R, Yang JJ. Protein Eng. 2001;14:1001–1013. doi: 10.1093/protein/14.12.1001. [DOI] [PubMed] [Google Scholar]