Abstract
The genome of the extremely radiation resistant bacterium Deinococcus radiodurans encodes 21 Nudix hydrolases of which only two have been characterized in detail. Here we report the activity and crystal structure for DR_0079, the first Nudix hydrolase observed to have a marked preference for cytosine ribonucleoside 5’-diphosphate (CDP) and cytosine ribonucleoside 5’-triphosphate (CTP). After CDP and CTP the next most preferred substrates for DR_0079, with a relative activity of < 50%, were the corresponding deoxyribose nucleotides, dCDP and dCTP. Hydrolase activity at the site of the phosphodiester bond was corroborated using 31P NMR spectroscopy to follow the phosphorus resonances for three substrates, CDP, IDP, and CTP, and their respective hydrolysis products, CMP + Pi, IMP + Pi, and CMP + PPi. Nucleophilic substitution at the β-phosphorus of CDP and CTP was established, using 31P NMR spectroscopy, by the appearance of an upfield shifted Pi resonance and line-broadened PPi resonance, respectively, when performing the hydrolysis in 40% H218O enriched water. Optimum activity for CDP was at pH 9.0 – 9.5 with the reaction requiring divalent metal cation (Mg2+ > Mn2+ > Co2+). The biochemical data is discussed with reference to the crystal structure for DR_0079 that was determined in the metal-free form at 1.9 Å resolution. The protein contains nine β-strands, three α-helices, and two 310-helices organized into three subdomains; an N-terminal β-sheet, a central Nudix core, and a C-terminal helix-turn-helix motif. As observed for all known structures of Nudix hydrolases, the α-helix of the ‘Nudix box’ is one of two helices that sandwich a ‘four-strand’ mixed β-sheet. To identify residues potentially involved in metal and substrate binding, NMR chemical shift mapping experiments were performed on 15N-labelled DR_0079 with the paramagnetic divalent cation Co2+ and the non-hydrolyzable substrate thymidine-5’-O-(α,β-methylenediphosphate) and the results mapped onto the crystal structure.
Keywords: chemical shift mapping, phosphorus-31 NMR, function screening, Nudix hydrolase, cytidine 5`diphosphate
Nudix hydrolases are widely distributed among species, having been found in the genomes of all three kingdoms and in organisms as diverse as viruses and humans (1). They are identified by the highly conserved sequence of amino acids, GX5EX7REUXEEXGU (where U = aliphatic and hydrophobic amino acid, X = any amino acid). This conserved sequence, called the ‘Nudix box’, forms part of the substrate-binding and catalytic site for the hydrolysis of nucleoside diphosphates linked to some other moiety, x, from whence the acronym ‘Nudix’ arose (2, 3). The glutamic acid residues in the core of the Nudix box, REUXEE, play a critical role in binding one to three divalent cations. At least one cation is essential for hydrolase activity and under physiological conditions the most relevant cation is likely Mg2+ (3). Nudix substrates include capped mRNA (4), dinucleotide coenzymes, nucleotide sugars, nucleotide alcohols, dinucleotide polyphosphates, and both canonical and oxidized (deoxy)ribonucleoside triphosphates ((d)NTPs) (5). Non-nucleotide Nudix substrates, such as thiamine pyrophosphate (6) and diphosphoinositol polyphosphates (7), have also been identified. By hydrolyzing such compounds the Nudix proteins effect the elimination of potentially toxic endogenous nucleotide metabolites from the cell and regulate the intercellular concentration of nucleotide cofactors and signaling molecules (3, 8, 9). For example, the prototypical member of the Nudix superfamily is the Escherichia coli MutT protein. MutT preferably hydrolyzes 7,8-dihydro-8-oxoguanosine triphosphate (8-oxo-d(GTP), a promutagenic compound generated during normal cellular metabolism and upon exposure to oxidative stress (10), into non-mutagenic nucleoside monophosphate and inorganic phosphate (3, 11) by catalyzing the nucleophilic substitution of H2O at the β-phosphorus (12).
Structures have been determined for a number of the proteins in the Nudix superfamily using both NMR-based and crystallographic methods and include: Escherichia coli MutT (nucleoside triphosphate pyrophosphohydroylase) (13), Caenorhabditis elegans and Lipinus angustifolius Ap4A hydrolase (14, 15), E. coli, Mycobacterium tuberculosis, and Homo sapien ADP-ribose pyrophosphatase (16–18), E. coli dihydroneopterin triphosphatase (19), and Deinococcus radiodurans coenzyme A pyrophosphatase (20). These structures have also been determined in a variety of states, such as with and without bound divalent cation and/or substrate. While the characterized Nudix proteins hydrolyze different substrates, they all conserve the Nudix box located on one of two α-helices that sandwich a central mixed β-sheet core (1, 21).
Recently, a new group of substrates for some members of the Nudix hydrolase superfamily has been identified. Fischer et al. (22) first reported that the D. radiodurans protein DR_0975 had a marked degree of specificity for ribonucleoside and deoxyribonucleoside 5’-diphosphates ((d)NDPs). Previously, Nudix hydrolase activity towards (d)NDPs had only been reported for one Nudix hydrolase, human NUDT5 ADP-sugar pyrophosphatase (23). Subsequently, activity towards (d)NDPs was reported for the ytkD gene product from Bacillus subtilis (8), and Orf17 (NtpA) (24) and orf153 (25) from E. coli. Indeed, E. coli MutT itself has recently been shown to be active upon 8-oxo-dGDP although its efficiency on the diphosphate is 4-fold smaller than on 8-oxo-dGTP (26). The moiety “x” linked to the nucleoside diphosphate is hydrogen or Me2+ and the hydrolysis products are a nucleoside monophosphate ((d)NMP) and inorganic phosphate (Pi) according to the following reaction:
Here we report that another D. radiodurans Nudix hydrolase, DR_0079, exhibits a marked degree of specificity for ribonucleoside and deoxyribonucleoside 5’-diphosphates. Unlike DR_0975 however, DR_0079 has a preference for cytidine diphosphate (CDP) and cytidine triphophate (CTP). The solution structure for D. radiodurans DR_0079 was previously determined using NMR-based methods (27). To complement this solution structure we report here the first crystal structure of a Nudix hydrolase with a marked specificity for CDP and CTP. Using this new crystal structure, the previously assigned amide resonances in the 1H-15N HSQC spectrum (28), and insights obtained from new biochemical studies, chemical shift perturbation experiments (29, 30) were performed on 15N-labelled DR_0079 with CoCl2 and the non-hydrolyzable substrate thymidine-5’-O-(α,β-methylenediphosphate) (TMP-CP) to map the potential metal-binding and nucleotide-binding surface, respectively.
EXPERIMENTAL PROCEDURES
All chemicals and enzymes were purchased from the Sigma Chemical Company (St. Louis, MI) except when indicated.
Cloning, Expression, and Purification
The cloning, expression, and purification protocol for 15N-labelled DR_0079 has previously been reported (28, 31). The procedure for preparing SeMet-labelled DR_0079 was similar using a protocol that inhibited the methionine biosynthesis pathway (32). In brief, this method involved growing the cells at 37°C to mid-log phase (OD600nm ~0.8) in M9 minimal medium supplemented with 34 µg/mL kanamycin (RPI Corporation, Prospect, IL), 30 µg/mL chloramphenicol, 120 µg/mL MgSO4, 11 µg/mL CaCl2, 10 ng/mL Fe2Cl3, 50 µg/mL NaCl, and 4 mg/mL glucose and then lowering the temperature to 25°C. At this point lysine (0.1 µg/mL), phenylalanine (0.1 µg/mL), threonine (0.1 µg/mL), isoleucine (0.05 µg/mL), valine (0.05 µg/mL) and selenomethionine (SeMet; 0.06 µg/mL) (Acrös Organics, Geel, Belgium) were added followed by the induction of protein expression ~15 minutes later with isopropyl β-D-1-thiogalactopyranoside (0.026 µg/mL) (RPI Corporation, Prospect, IL). Approximately 4 – 6 hours later the cells were harvested by mild centrifugation and then frozen at −80°C. From this point forward the protocol was identical to the previously described protocols except that the last step, gel filtration chromatography on a Superdex75 HiLoad column (GE Healthcare, Piscataway, NJ), used either buffer to grow crystals (100 mM NaCl, 20 mM Tris, 1 mM dithiothreitol (DTT), pH 7.1) to conduct the NMR studies (NMR buffer: 100 mM KCl, 20 mM potassium phosphate, 2 mM DTT, pH 7.1) or enzyme assay experiments (50 mM Hepes, 100 mM NaCl, pH 7.0).
CoCl2 and TMP-CP chemical shift mapping
Chemical shift mapping experiments (25°C) with CoCl2 were conducted with DR_0079 following the procedure previously described for MgCl2 and MnCl2 (27). Titration experiments were performed in a Shigemi tube (Tokyo, Japan) on a 250 mL sample of 0.6 mM (150 nmole) 15N-labelled DR_0079 in NMR buffer. Stock solution of CoCl2 (50 mM) was added to the protein sample in 1.5 µL (75 nmole) aliquots up to a 2-fold molar excess of the metal ion. Following mild agitation, two-dimensional 1H-15N HSQC spectra, at a 1H resonance frequency of 900 MHz (Varian Inova-900), were collected after the addition of each aliquot at metal:protein ratios of 0.5, 1.0, and 2.0 to 1.0.
To probe the substrate-binding surface the hydrolytically stable substrate thymidine-5’-O-(α,β-methylenediphosphate), sodium salt (TMP-CP) (Biolog Life Science Institute, Bremen, Germany), was titrated into a solution of DR_0079 containing 100 mM MgCl2 in the NMR buffer. TMP-CP (10 mM) was added to 400 mL of DR_0079 solution (0.3 mM, 120 nmole) in 12 mL aliquots (120 nmole) up to a four-fold molar excess of the substrate. Following mild agitation, two-dimensional 1H-15N HSQC spectra, at a 1H resonance frequency of 600 MHz (Varian Inova-600), were collected after the addition of each TMP-CP aliquot.
Enzymatic assays
Using (d)NDPs as substrates, DR_0079 produces the products (d)NMPs and orthophosphate (Pi). This enzymatic activity was determined by measuring the Pi release with the Malachite green reagent (33). The activity against (d)NDP substrates was measured in a reaction mixture (200 µL) containing 50 µM substrate, 50 mM Tris-HCl (pH 9.0), 7.5 mM MgCl2, and 0.7 µg DR_0079. As proposed previously for Nudix hydrolases (34), assays involving (d)NTPs included 10 µunits of inorganic pyrophosphatase to release Pi from any pyrophosphate (PPi) produced by the enzyme. After 15 min of incubation at 37°C the reaction was terminated by addition of 50 µL of Malachite green reagent (33). The mixture was developed for 3 min at room temperature and then absorption at 630 nm was measured with a Quant Universal Microplate Spectrophotometer (Bio-Tek Instruments, Inc.). The above reactions with CDP and CTP were performed in the presence and absence of inorganic pyrophosphatase to determine if PPi or Pi was a product of the hydrolysis reaction. The optimum pH for the enzymatic reaction was determined using the method described above, CDP as the substrate, and three different buffers: Tris-HCl (pH 7.5 – 8.5), CHES-K (pH 8.6 – 10.0), and CAPS-K (pH 9.7 – 11.0). The optimal metal ion requirements for the hydrolysis of CDP was determined with the above assay (pH 9), using 5 mM Mg2+ or 0.5 mM of other divalent cations (Mn2+, Co2+, Ni2+, Zn2+, Ca2+, or Cu2+). A 10-fold lower divalent cation concentration was necessary for the latter set of cations because at the 5 mM level significant inhibition of activity was observed. Kinetic parameters were determined by non-linear curve fitting using GraphPad Prism software (v 4.00 for Windows; GraphPad Software, San Diego, CA).
Enzyme activity was also assayed using 31P NMR spectroscopy by following the phosphorus resonances of three substrates, CDP, IDP, and CTP, and their products. Six hundred µL of 5.0 mM CDP, CTP, and IDP (~3000 nmole) were prepared in buffer containing 50 mM Tris, 10 mM MgCl2, pH 9.1. After recording a 1D 31P NMR spectrum on a Varian 600-Inova spectrometer at 37°C, 9 µL of 1.2 mM DR_0079 (~10 nmole, crystallization buffer) was added to the nucleotide diphosphate. Additional 1D 31P spectra were then recorded immediately afterwards (~5 min delay) and at ~10 (NDP) or ~60 (CTP) min intervals. To determine which phosphorus was undergoing substitution in the reaction, the experiments with CDP and CTP were repeated in the presence of 40% H218O. The data was processed with Felix98 (MSI, San Diego, CA) and the 31P chemical shifts were referenced to DSS (DSS = 0 ppm) using indirect methods.
Crystallization
Crystals of SeMet-labeled DR_0079 were grown using vapor-diffusion, hanging drop, crystallization methods at room temperature (~22°C) using precipitants from Hampton Research (Aliso Viejo, CA). The best “coffin-like” crystals of SeMet-substituted DR_0079, identical in appearance to those reported for unlabelled DR_0079 (31), were harvested a couple days after mixing 2 µL of protein (24 mg/mL) with 2 µL of buffer containing 0.2 M calcium acetate hydrate, 0.1 M sodium cacodylate trihydrate, 18% (w/v) polyethylene glycol 8000, pH 6.5. In order to grow diffraction quality crystals it was critical that the protein concentration was above 20 mg/mL.
Data collection, structure determination and refinement
X-ray diffraction data for the DR_0079 crystals were collected at the National Synchrotron Light Source (NSLS), at Brookhaven National Laboratory, on the X29A beamline using an ADSC Q315 CCD detector. A Se peak SAD data set at 2.04 Å resolution (SeMet-labeled) and a native data set at 1.9 Å resolution (unlabeled) were collected on orthorhombic crystals of DR_0079 (31). The images were integrated and scaled with HKL2000. The heavy atom sites in the selenium-labeled DR_0079 were determined using the SHELX program suite and HKL2MAP. The Se peak wavelength data set, which contained useful anomalous signal out to 2.5 Å resolution, was used together with the program SOLVE to produce an excellent electron density map at 2.5 Å resolution. Fragments of the structure were built automatically into the 2.5 Å resolution map using RESOLVE, and the remainder of the structure was built manually using XtalView/Xfit and refined to 2.3 Å resolution. The model was further iteratively refined using a native data set extending out to 1.9 Å resolution and the refine.inp algorithm in CNS (http://cns.yale.edu/v1.1/) employing the maximum likelihood target using amplitudes. Assessment of the quality of stereochemistry of the final model, using the programs PROCHECK and MolProbity, indicated that the final model was a high quality representation of the crystal structure of DR_0079 and represented an improvement over the NMR-derived structure. MolProbity analysis showed that the overall protein geometry of the final model ranked in the 84th percentile (MolProbity score of 1.77) with a clash score for “all-atom contacts” of 18.92. PROCHECK analysis of the crystal data showed that 94% of the (ϕ,ψ) pairs for DR_0079 were found in the most favored regions and 6% within additionally allowed regions, a marked improvement over the 65 and 29% observed for these regions, respectively, in the average NMR structure (27). The data collection and structure refinement statistics are given in Table 1 and the coordinates have been submitted to the Protein Data Bank (PDB ID 2O5F).
Table 1.
Summary of data collection and structure refinement statistics for DR_0079.
| Data set | Se-Met | Native |
|---|---|---|
| Space Group | C2221 | C2221 |
| Unit Cell Parameters (Å,°) | a=35.46, b=157.62, c=127.03 | a=34.02, b=156.55, c=126.56 |
| α=β=γ=90.0 | α=β=γ=90.0 | |
| Matthew’s Coefficient | 2.3 | 2.2 |
| Percent Solvent (%) | 46.6 | 43.7 |
| X-ray Source | X29 | X29 |
| Temperature (K) | 100 | 100 |
| Resolution Limits (Å) | 50.0-2.04 (2.11-2.04) | 50.0-1.90 (1.97-1.90) |
| Detector Distance (mm) | 300 | 240 |
| Exposure Time (sec) | 2 | 3 |
| Oscillation angle (°) | 0.5 | 0.5 |
| No. Images | 720 | 360 |
| Total Angle (°) | 360 | 180 |
| Mosaicity (°) | 0.29-1.01 | 0.54–0.66 |
| Wavelength (Å) | 0.9792 | 1.1000 |
| No. Observations1 | 226118 (4306) | 170690 (14245) |
| No. Unique Reflections1 | 21753 (1305) | 27095 (2590) |
| Redundancy | 10.4 (3.3) | 6.3 (5.5) |
| Completeness | 91.4 (55.8) | 99.4 (97.8) |
| I/s (I) | 24.8 (2.01) | 49.6 (8.5) |
| Rsym (%) | 10.6 (39.4) | 7.1 (26.8) |
| Structure Refinement | ||
| PDB ID | 2O5F | |
| Rconv (%) | 23.3 | |
| Rfree (%) | 26.7 | |
| Protein atoms | 2538 | |
| Solvent atoms | 171 | |
| Protein residues | 316 | |
| Average B value, protein atoms (Å2) | 29.1 | |
| Average B value, main chain (Å2) | 26.7 | |
| Average B value, solvent atoms (Å2) | 35.5 | |
| Average B value, all atoms (Å2) | 29.5 | |
| R.m.s. deviations from ideal bond lengths (Å) | 0.006 | |
| R.m.s. deviations from ideal bond angles (Å) | 1.24 | |
| Structure Validation | ||
| Procheck | ||
| Ramachandran | ||
| Favored | 93.5 | |
| Allowed | 6.5 | |
| Generous | 0.0 | |
| Disallowed | 0.0 | |
| Molprobity | ||
| Clash Score – All atoms | 18.9 (33rd ) | |
| Clash Score – atoms B<40 | 17.1 (13th) | |
| MER score / (percentile) | 1.8 (84th) | |
Values in parentheses are for the highest resolution shell
RESULTS AND DISCUSSION
Overall structure of DR_0079
Two molecules of DR_0079 pack together in the asymmetric unit. The structure of each molecule in the asymmetric unit is similar with a backbone RMSD of 0.81 Å and an all heavy atom RMSD of 1.23 Å as calculated using the program SuperPose (35). We will refer to only one subunit, B, when discussing the crystal structure of DR_0079 primarily because electron density was missing for residues S54-F60 in subunit A.
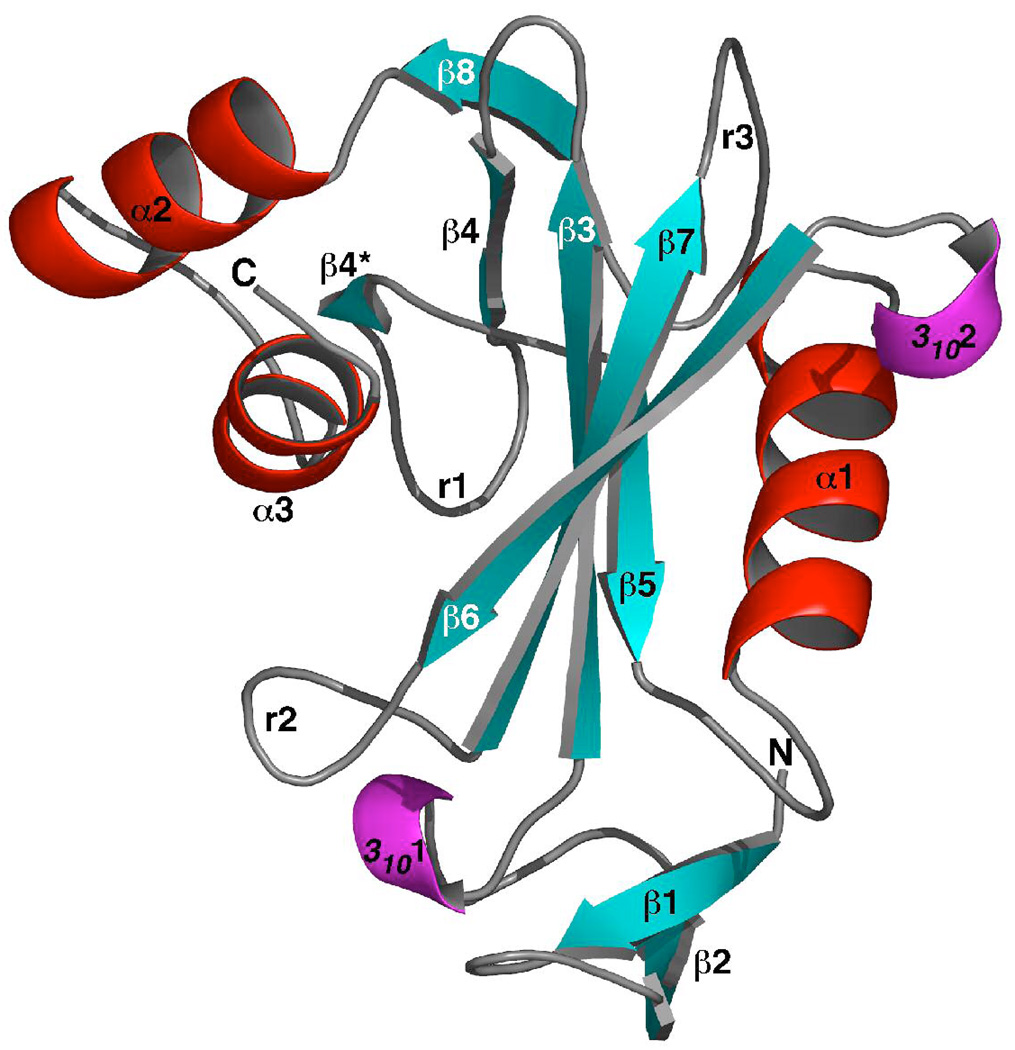
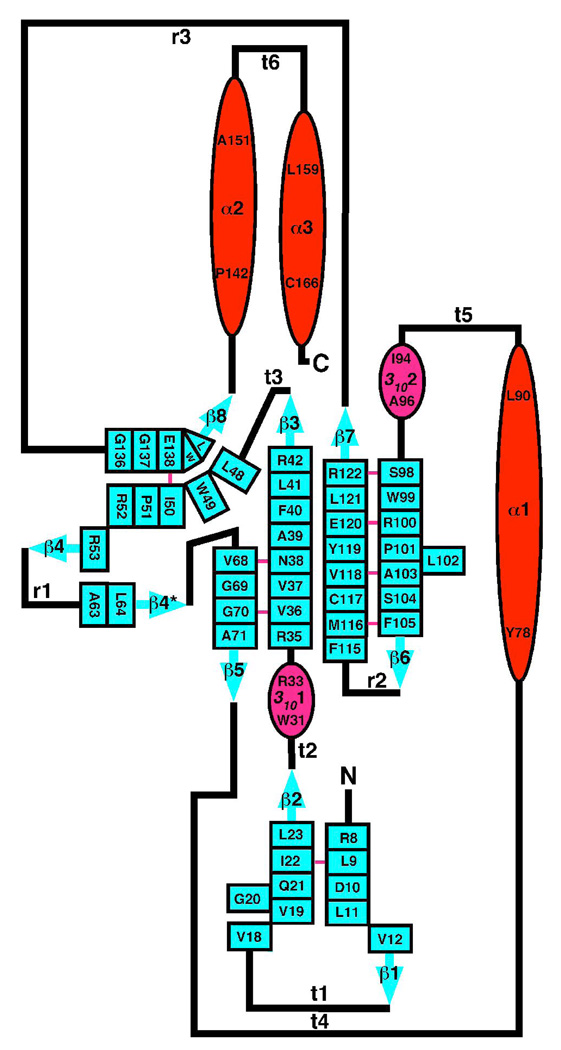
Figure 1A is a molscript cartoon representation of the structure of DR_0079 and Figure 1B is a two-dimensional schematic of the secondary structure. The general secondary structure is similar to that observed in the NMR-based structure (27) aside from two short, three-residue, 310-helices (3101 and 3102) and an additional two-residue β-strand (β4*). The one additional β-strand has been labeled with an asterisk to facilitate comparison with the NMR-based structure. The molecule is composed of nine β-strands, three α-helics, and two 310-helices that can be grouped into three parts. Beta-strands one and two along with 3101 form an N-terminal β-sheet (1–34), alpha-helices three and four form a C-terminal helix-turn-helix motif (141–171), and the remainder of the molecule forms the Nudix core (35–140). Within this Nudix core is the ‘Nudix fold’, a mixed β-sheet sandwiched between the catalytic helix (α1) and the orthogonally-orientated N-terminal helix (α3). Such a general arrangement is observed in all Nudix proteins of known structure (1, 21). The mixed β-sheet of the Nudix fold is dominated by a ‘four’-strand mixed β-sheet with the central two β-strands parallel to each other. Such a general arrangement is also typical in all Nudix proteins of known structure. However, in DR_0079 one of the outer β-strands is discontinuous, composed of β5 and part of β4. In turn, β4 is the central β-strand in a small three strand β-sheet with β4* and β8 that is attached to the outer edge of the ‘four’-strand mixed β-sheet.
Fig. 1.
(A) Molscript ribbon representation of the crystal structure of DR_0079 in the metal-free form (PDB ID = 2O5F). The β-strands are colored blue, α-helices red, and 310-helices magenta. Residues 1 – 34 = N-terminal anti-parallel β-sheet; residues 35–140 = Nudix core; residues 141– 171 = C-terminal helix-turn-helix motif. (B) Secondary structure diagram of DR_0079. The α-helices are drawn as red ovals and 310-helices as magenta ovals with the residue number of the beginning and the end of each element shown. The β-strands are drawn as solid blue arrows with each residue indicated within a box. The β-sheets contain bulges at residues G20 and L102. A solid pink line between β-sheet residues indicates dual hydrogen bonds between two residues in an anti-parallel β-sheet.
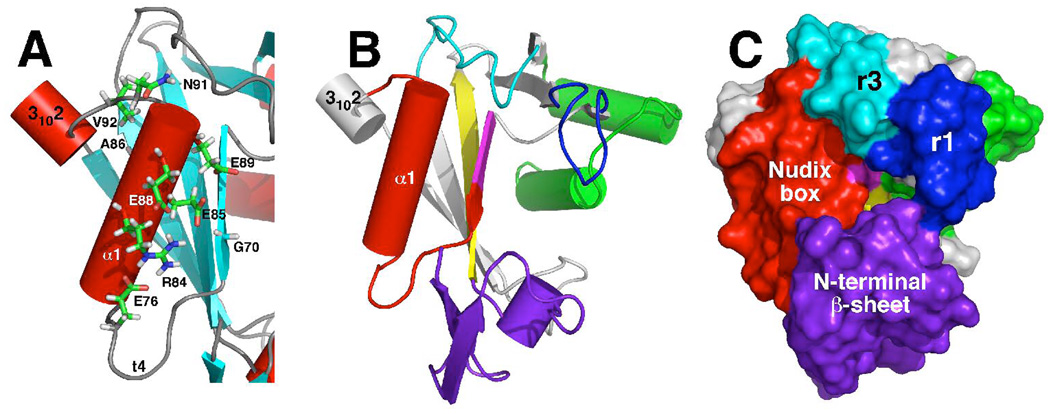
A characteristic feature of all Nudix proteins is the highly conserved 23-residue ‘Nudix box’, GX5EX7REUXEEXGU, where U is a bulky hydrophobic group and X is any residue (2, 3). The Nudix sequence forms a loop-helix-loop motif (36) that is responsible for coordinating the catalytically essential divalent cation (usually Mg2+) to the protein (13, 14, 16, 20). In DR_0079, the Nudix box is between G70-V92 and, as shown in Figure 2, it too adopts a loop-helix-loop structural motif. Of the 23 residues in the Nudix box, nine are highly conserved, G70X5E76X7R84E85A86XE88E89XN91V92 (the superscript refers to the residue number in the DR_0079 Nudix sequence) and Figure 2 illustrates the relative orientation of these highly conserved residues in the loop-helix-loop Nudix motif. As observed in other Nudix structures, all the polar conserved residues in the α-helix are orientated on the same side and oppose the central mixed β-sheet. The side chains of A86 and V92 form a small hydrophobic pocket and likely play a conserved role in stabilizing the loop-helix-loop motif. The similar three-dimensional orientation of the conserved residues in all known Nudix protein structures suggest that these residues play similar roles in all Nudix hydrolases – binding one or more divalent cation (37). Indeed, it has been established that binding of the catalytic divalent metal ion occurs through the side chains of the glutamic acid residues in the loop-helix-loop Nudix motif (13, 14, 16, 20, 38). The enzyme-bound metal, in turn, forms a second sphere complex with the diphosphate part of the substrate as part of the enzyme’s active site (13). While the diphosphate part of the Nudix substrate interacts with the loop-helix-loop Nudix motif and divalent metal cation, the other parts of the Nudix substrate, the nucleoside and a moiety ‘x’, interact with the side chains and motifs elsewhere in the protein (13–16, 21, 39). Differences in the shape and nature of the environment surrounding the Nudix box are responsible for the different substrate specificities of different Nudix hydrolases. As shown in Figure 2B, the immediate environment around the Nudix box are part of the N-terminal β-sheet, the backside of the mixed β-sheet, and loops r1 and r3. Note that no electron density was observed for residues S54-F60 in subunit A of the crystal structure and amide cross peaks were not observed for seven out of the 14 possible resonances in the 1H-15N HSQC spectrum of this region, suggesting that the r1 region may be especially disordered in solution (27, 40). Such intrinsically disordered regions are often sites of biological activity and become “ordered” only after binding their substrate (41).
Fig. 2.
(A) Expansion of the loop-helix-loop Nudix box with the side chains of the nine highly conserved residues highlighted, labeled, and colored according to atom type. The regions that immediately surround the Nudix box are highlighted on the cartoon (B) and surface (C) structure of DR_0079. Red = Nudix box, purple = N-terminal β-sheet, blue = loop r1, cyan = loop r3, magenta = rest of β5, yellow = β3, and green = C-terminal helix-turn-helix.
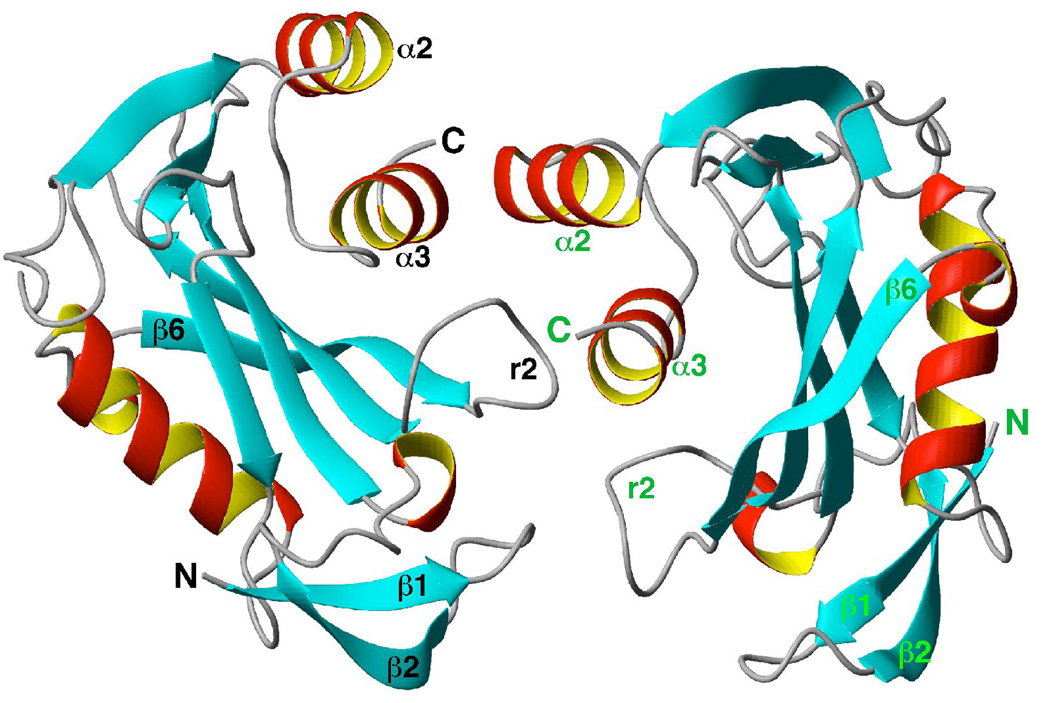
Two molecules of DR_0079 pack together in the crystalline state as shown in Figure 3. Hydrophobic intermolecular contacts at three regions, between α3 and α2, r2 and α3, and t1 and r2, are responsible for holding the dimer together in the asymmetric unit. Dimeric Nudix hydrolases with extensive domain swapping have previously been observed, both in solution and in the crystalline state (37). For the proteins with extensive domain swapping, oligomerization is necessary for the biological function of the protein. However, the dimer observed in the DR_0079 crystal structure is likely a crystal-packing artifact as NMR data and size exclusion chromatography strongly suggest that DR_0079 is a monomer in solution (27, 28).
Fig. 3.
Molscript ribbon representation of two molecules of DR_0079 in the asymmetric unit highlighting the intermolecular interface. The β-strands are colored blue, the alpha- and 310-helices red, and the turns and loops grey.
Enzymatic studies of DR_0079
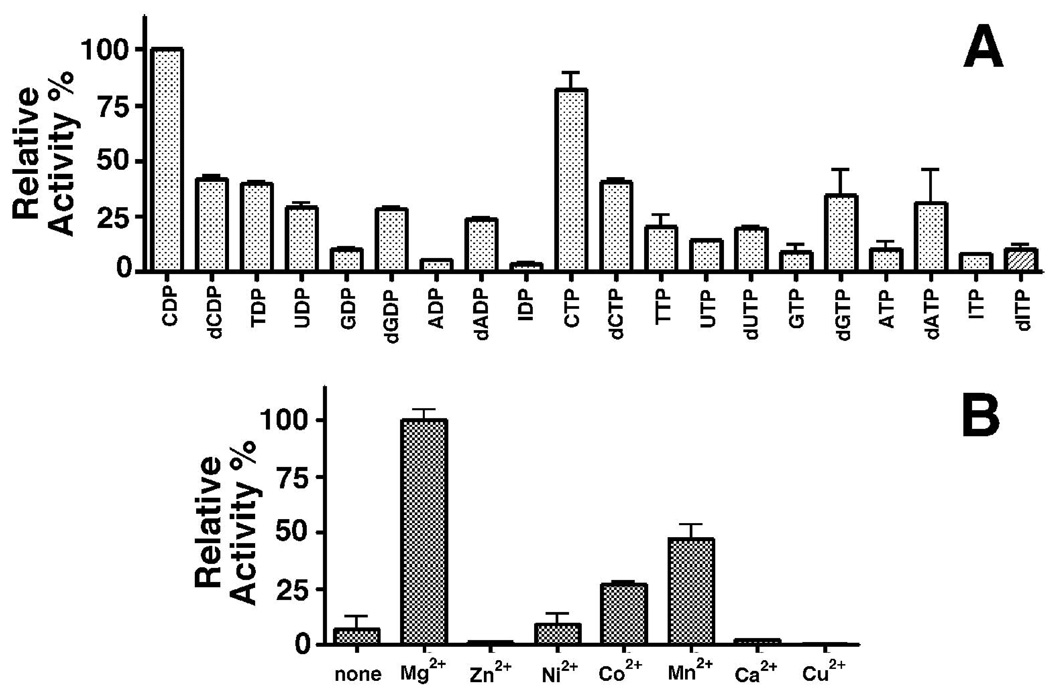
Purified DR_0079 was tested for enzymatic activity against a series of known Nudix hydrolase substrates (34) including (deoxy)nucleoside di- and triphosphates, nucleotide sugars and alcohols, and cofactors (FAD, NAD, CoA) in the standard assay at a fixed substrate concentration of 50 µM. As illustrated in Figure 4A, CDP and CTP were the best substrates for DR_0079 followed by dCDP and dCTP. Note that while cytidine nucleotides were the favored substrate for DR_0079, the enzyme was still promiscuous enough to show significant activity towards (d)GDP/GTP, (d)ADP/ATP, and (d)IDP/ITP (~ 20-fold difference between the poorest (IDP) and best (CDP) substrate). There was no detectable activity against GDP-glucose/mannose, UDP-glucose/galactose, ADP-glucose, CDP-choline, CDP-ethanolamine and CDP-glycerol (data not shown). Thus, DR_0079 is the first Nudix hydrolase selective for the cytidine nucleotides.
Fig. 4.
A) Relative substrate activity of DR_0079 towards a variety of nucleoside di- and triphosphate substrates. B) Nudix hydrolase activity of DR_0079 as a function of metal cation. The divalent cation requirements (metal profile) were determined towards CDP in the presence of 5 mM Mg2+ or 0.5 mM other divalent cation (Mn2+, Co2+, Ni2+, Zn2+, Ca2+, or Cu2+).
Like most known Nudix hydrolases (1, 22), DR_0079 had an alkaline pH optimum (pH 9.0 – 9.5) (data not shown) and required a divalent metal cation for activity. Figure 4B illustrates that Mg2+ was the most effective metal (optimal concentration 7 mM), whereas Mn2+ and Co2+ supported lower activity (47% and 27%, respectively). Negligible activity was observed in the presence of Ni2+, whereas Zn2+, Ca2+, and Cu2+ were inhibiting. Note that the result with Ca2+ corroborates with the crystal structure that showed no metal was bound to DR_0079 despite growing the crystals in presence of 0.1 M CaCl2. We also determined the kinetic parameters for the hydrolysis of various nucleotides by DR_0079. With all substrates, the protein showed classical Michaelis-Menten saturation kinetics with the lowest Km with dCTP (Km = 23.0 µM) and highest activity with CDP (kcat = 1.7 s−1) (Table 2). Thus, DR_0079 demonstrated the highest catalytic efficiency toward CDP and CTP and is the first characterized Nudix hydrolase with such a substrate specificity.
Table 2.
Kinetic parameters for DR_0079a
| Substrate |
Km, mM |
Vmax, µmol/min mg protein |
kcat, sec−1 |
kcat/Km, mM−1sec−1 |
|---|---|---|---|---|
| CDP | 0.082 ± 0.011 | 5.18 ± 0.17 | 1.67 | 20.37 |
| dCDP | 0.092 ± 0.010 | 3.52 ± 0.11 | 1.12 | 12.17 |
| TDP | 0.105 ± 0.023 | 3.18 ± 0.27 | 1.03 | 9.81 |
| UDP | 0.137 ± 0.033 | 4.19 ± 0.42 | 1.35 | 9.85 |
| GDP | 0.092 ± 0.026 | 1.21 ± 0.10 | 0.39 | 4.24 |
| dGDP | 0.051 ± 0.010 | 1.55 ± 0.09 | 0.51 | 10.0 |
| ADP | 0.217 ± 0.033 | 0.93 ± 0.13 | 0.29 | 1.34 |
| dADP | 0.145 ± 0.028 | 1.61 ± 0.09 | 0.52 | 3.59 |
| IDP | 0.046 ± 0.010 | 0.21 ± 0.04 | 0.06 | 1.30 |
| TTP | 0.138 ± 0.030 | 2.13 ± 0.15 | 0.69 | 4.97 |
| UTP | 0.124 ± 0.030 | 1.65 ± 0.12 | 0.53 | 4.28 |
| CTP | 0.034 ± 0.007 | 1.82 ± 0.10 | 0.59 | 17.22 |
| dCTP | 0.023 ± 0.004 | 0.92 ± 0.08 | 0.30 | 12.83 |
| ATP | 0.317 ± 0.090 | 0.91 ± 0.08 | 0.29 | 0.92 |
| dATP | 0.062 ± 0.009 | 0.95 ± 0.07 | 0.31 | 4.92 |
| GTP | 0.394 ± 0.089 | 0.99 ± 0.13 | 0.32 | 0.81 |
| dGTP | 0.083 ± 0.011 | 1.25 ± 0.10 | 0.40 | 4.82 |
| ITP | 0.147 ± 0.027 | 0.55 ± 0.05 | 0.18 | 1.19 |
| dITP | 0.088 ± 0.008 | 0.68 ± 0.05 | 0.22 | 2.5 |
All substrate concentrations were between 0.01 and 4.0 mM. Kinetic parameters were determined by non-linear curve fitting using GraphPad Prism software (v 4.00 for Windows, San Diego, CA)
The enzyme assays were first performed in the presence of inorganic pyrophosphatase to release Pi from any PPi produced by DR_0079 because Pi is the component that reacts with the Malachite green reagent (33). To determine if DR_0079 was hydrolyzing (d)NDPs to nucleosides plus PPi or to (d)NMPs plus Pi, the reaction with CDP was performed in the absence of inorganic pyrophosphatase and absorption at 630 nm measured. No change was observed indicating that (d)NMPs and Pi were generated by DR_0079 upon the hydrolysis of (d)NDPs. On the other hand, when the same reaction with CTP was performed in the absence of inorganic pyrophosphatase, no absorption at 630 nm was observed indicating that DR_0079 was converting (d)NTPs to (d)NMPs and PPi.
To further verify the catalytic activity of DR_0079, to positively identify the hydrolysis products, and to determine the site of nucleophilic substitution of water, the reaction with CDP, IDP, and CTP was followed by phosphorus-31 NMR spectroscopy. The phosphorus chemical shifts of the nucleoside mono-, di-, and tri-phosphates are different, and hence, the reaction can be monitored in real time by following the changes in the 31P NMR spectrum. The site of nucleophilic substitution can be determined by the small upfield shift (~0.02 ppm) in the phosphorus resonance upon the formation of a single 31P-18O chemical bond (12, 42). An additional advantage of using 31P NMR to follow hydrolysis by Nudix enzymes is that it eliminates the need for additional enzymes, such as inorganic pyrophosphatase, to release Pi.
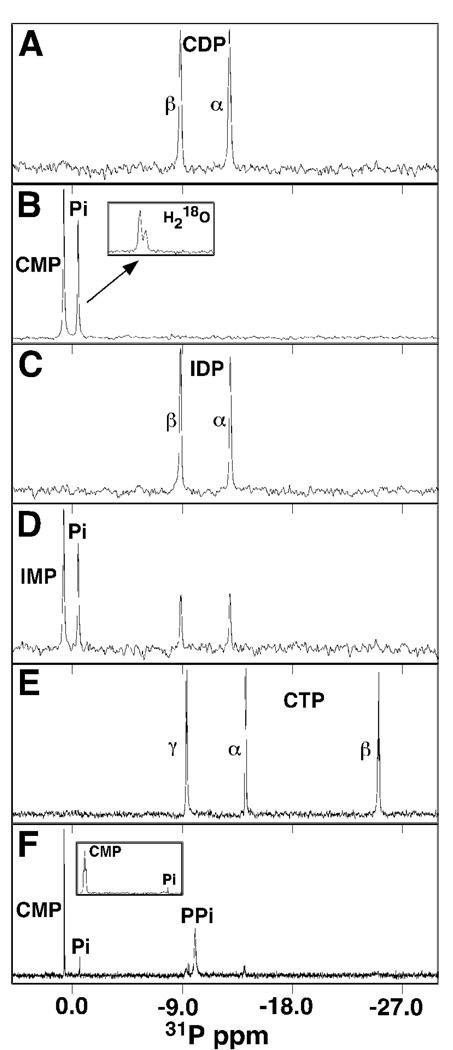
As shown in Figure 5A, CDP contains two 31P chemical shifts at −8.78 and −12.83 ppm that can be assigned to the β– and α– phosphorus atoms, respectively (43, 44). At the concentrations of DR_0079 used, the reaction was complete by the time the first 1D spectrum could be collected after its addition to the NMR tube as shown in Figure 5B. Two different resonances at 0.75 and 0.44 ppm were observed that correspond to CMP and Pi, respectively. To determine if the reaction was occurring via nucleophilic substitution of water on the α– or the β-phosphorus, the reaction was repeated in the presence of H218O-enriched (40%) water (12). As shown in the inset of Figure 5B, the orthophosphate (Pi) showed an additional upfield resonance (Δ = 0.02 ppm) upon completion of the reaction. Such an observation establishes that the reaction occurs via a nucleophilic substitution at the β-phosphorus of CDP with CMP displaced as the leaving group (12). While CDP is a good substrate for DR_0079, Figure 4A showed that IDP is a relatively poor substrate and this is evident in Figure 5D, the 1D 31P NMR spectrum fifty minutes after the addition of DR_0079 to IDP in an NMR tube. Four resonances are observed corresponding to unhydroylzed IDP (−8.83 and −12.88 ppm) and the hydrolysis products, IMP (0.77 ppm), and Pi (0.40 ppm).
Fig. 5.
One-dimensional 31P spectra of CDP, CTP, and IDP alone and at various time intervals after the addition of DR_0079 to the NMR tube. (A) CDP alone, (B) CDP + DR_0079 immediately after protein addition with the inset showing an expansion (0.4 ppm) of the Pi resonance in the presence of 40% H218O, (C) IDP alone, (D) CDP + DR_0079 after 50 min. (E) CTP alone, (F) CTP + DR_0079 at the completion of the reaction (~12 hours later) with the inset showing an expansion (0.5 ppm) of the downfield region. All spectra were recorded at a 1H resonance frequency of 600 MHz, 37°C, in buffer containing 50 mM Tris, 10 mM MgCl2, pH 9.1.
As shown in Figure 5E, CTP contains three 31P chemical shifts at −9.33, −14.14, and −25.04 ppm that can be assigned to the γ–, α–, and β–phosphorus atoms, respectively (12, 43). Figure 5F shows the spectrum 12 hours after the addition of DR_0079 when the reaction is almost complete. Most of the α–, β–, and γ–phosphorus signal for GTP disappear completely while two new major resonances, corresponding to CMP (0.69 ppm) and PPi (−10.02 ppm), appear. There is very little signal for Pi at 0.40 ppm. Indeed, this downfield region of the spectrum has been expanded in the inset of Figure 6F and the intergrated intensity of both resonances indicates the amount of Pi relative to CMP is approximately four percent. Hence, the spectrum in Figure 5F unambiguously shows that very little CTP is converted to CMP by sequential hydrolysis of individual phosphorus atoms. Instead, the primary reaction is the hydrolysis of the ester bond between the α– and β–phosphorus atoms of CTP. The repetition of this experiment in the presence of H218O-enriched (40%) water resulted in a broadening of the PPi resonance with no change in the CMP resonance (data not shown) indicating that the nucleophilic substitution of water was occurring on the β-phosphorus (12). Consequently, the hydrolysis of CTP by DR_0079 is likely occurring via a mechanism similar to the one reported for the hydrolysis of 8-oxo-dGTP by MutT (12), except the nucleotide binding pocket has a preference for cytosine di- and triphosphates over 7,8-dihydro-8-oxoguanosine triphosphate.
Fig. 6.
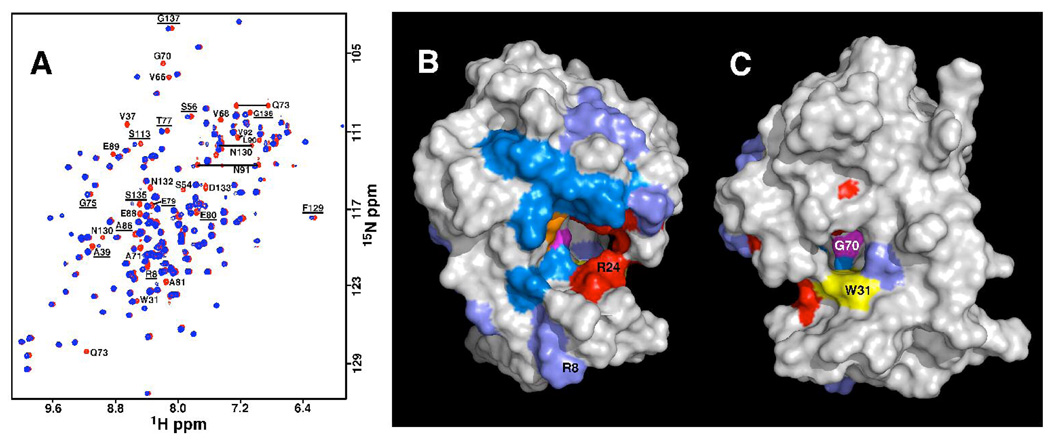
A) Overlay of the 1H–15N HSQC spectra of DR_0079 in the absence (red) and presence (blue) of ~1:1 molar ratio of CoCl2. The chemical shifts of backbone amide and identified side chain amine resonances that disappear or shift (underlined) upon the addition of CoCl2 are labeled. Spectra were collected at a 1H resonance frequency of 900 MHz, 25°C, in buffer containing 100 mM KCl, 20 mM potassium phosphate, 2 mM DTT, pH 7.1. B + C) Resonances for residues that disappear (marine blue) or move (slate) upon the addition of CoCl2 to DR_0079 are mapped onto the structure (2O5F) and shown in two orientations that differ by ~180°. The metal cation likely rests over G70 (magenta), the residue with the largest chemical shift perturbation with the addition of MgCl2 (27). Next to it, colored orange, is E85, the nearest glutamic acid side chain (the 1HN resonance of E85 overlaps with A160 and it is not possible to determine if this residue disappears with the addition of CoCl2). In the cleft there is a ring of lysine and arginine residues and these are colored red (R24, R53, K57, and K156). Directly behind the ring of positively charged residues is an aromatic tryptophan side chain (W31) that is colored yellow in orientation A. This tryptophan’s location in the cleft is more obvious in orientation B.
Co2+ chemical shift mapping
Previous 1H-15N HSQC experiments with 15N-labeled DR_0079 in the absence and presence of five-fold molar excess EDTA suggested that DR_0079 was prepared in the metal-free form (27). This conclusion is corroborated by the biochemical data presented here showing that a divalent cation must be added to the protein before statistically significant activity is observed. This is also corroborated in the X-ray determined structure presented here showing no divalent cation bound to the protein (although Mg2+ would be difficult to detect). Changes were previously observed in 1H-15N HSQC spectrum of DR_0079 in the presence of 100-fold molar excess MgCl2 while in the presence of 25-fold molar excess MnCl2 DR_0079 precipitated out of solution (27). In light of the data shown in Figure 4B, and because excess MgCl2 was necessary to observe significant chemical shift perturbations to the 1H-15N HSQC spectrum of DR_0079, the chemical shift perturbation experiments were repeated using CoCl2 in order to better characterize the metal binding site on the protein. This technique is based on the premise that protein-ligand interactions perturb the chemical environment of the nuclei at the interface of ligand binding. Because such perturbations are often accompanied by changes in the chemical shifts of the backbone 1HN and 15N resonances (30, 45), by identifying resonances that undergo a binding dependent chemical shift or intensity perturbation, it is possible to identify ligands that bind to a protein and to map the location of the ligand binding site on to the three-dimensional structure of the protein. The 1H-15N HSQC spectrum of DR_0079 should be more sensitive to chemical shift perturbations upon binding Co2+ instead of Mg2+ because Co2+ is paramagnetic. Consequently, in addition to any chemical shift perturbations due to structural changes effected by divalent cation binding, the unpaired electrons in Co2+ can also effect the surrounding nuclear environment in two other major ways. One is a through-bond scalar interaction, known as a contact shift, which can propagate a maximum of five bonds from the metal center. The second is a through-space dipolar interaction, known as a pseudocontact shift, that propagates to neighboring nuclei with a 1/r3 distance dependence (30, 46). Figure 6A overlays the 1H-15N HSQC spectra for DR_0079 in the presence (blue) and absence (red) of one molar equivalent of CoCl2 and shows that a substantial subset of backbone, amide cross peaks either disappear (16) or shift (12) in the presence of the divalent metal cation. In contrast, the average chemical shift perturbation of only nine residues was observed to shift more than 0.05 ppm with the addition of MgCl2 to DR_0079 (27). Of these Mg2+ perturbed chemical shifts, the most significantly effected residue was G70 with R8, E89, and D95 affected moderately. The small number of residues with chemical shift perturbations and the small magnitude of the perturbations suggested that the global structure of the protein was not significantly altered upon binding Mg2+, consistent with the minor structural changes reported for other Nudix proteins upon the addition of divalent cation (16, 20). Evidently, the larger number of perturbations observed upon binding Co2+ primarily reflect through-bond and through-space interactions that propagate from the metal’s center.
Figures 6B and 6C map the positions of the 1HN residues affected by the addition of CoCl2 on to the crystal structure of DR_0079. All the amide residues perturbed by the addition of Co2+ are clustered together on one face of the molecule at, or near, the loop-helix-loop Nudix motif. Divalent cations have been identified in many of the known Nudix structures with the side chain carboxyl group of one or more glutamic acid residues in the loop-helix-loop Nudix motif participating in the metal ligation (13, 14, 16, 20). It is likely that at least one of the glutamic acid residues in the Nudix box of DR_0079 is participating in metal ligation. These glutamic residues are most likely E85, the residue commonly observed in most crystal structures with a metal (38) and possibly E89, the second most commonly observed residue complexed to a metal in crystal structures (38) and the glutamic acid amide in the Nudix box with the second-most significant chemical shift perturbation in the presence of Mg2+ (E85 overlaps with A160 therefore it is not possible to identify chemical shift perturbations for this residue). Note that the residue disturbed most by Mg2+ binding was G70, and it also disappears in the presence of Co2+. Glycine-70 is the first highly conserved residue of the Nudix box and in other Nudix proteins, including DR_0079, this glycine residue is often preceded by a second glycine residue (20, 34). It has been suggested that these physically ‘small’ glycine residues may be necessary to allow the metal to sit on the Nudix fold (47). The observation that the chemical shift for G70 is most perturbed in the presence of Mg2+ (27), and disappears completely in the presence of Co2+, supports the hypothesis that the metal may indeed be sitting over this residue in DR_0079.
Chemical shift mapping of the substrate-binding surface with TMP-CP
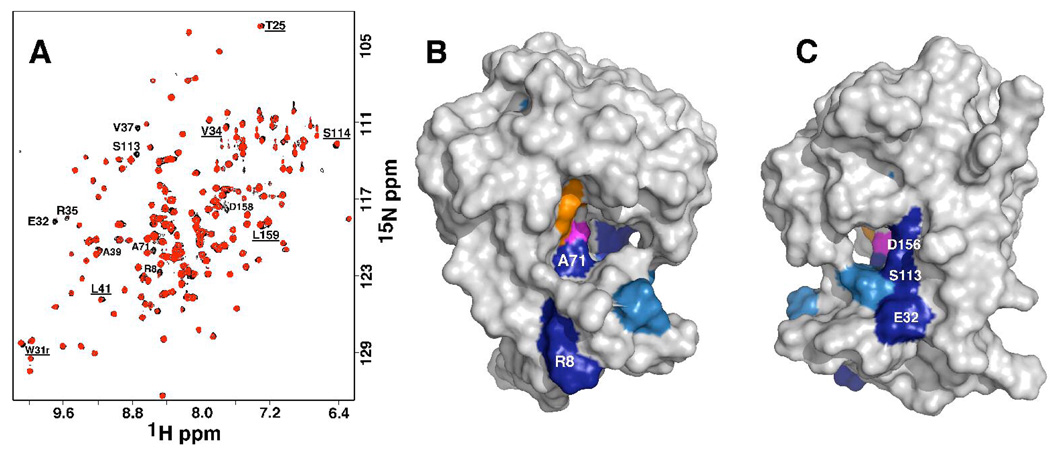
Substrate screening indicated that (deoxy)nucleoside diphosphates were the preferred substrate for DR_0079 with pyrimidine generally preferred over purine nucleotides. It is therefore not surprising that previous efforts to map the substrate-binding surface of DR_0079 using chemical shift perturbation methods with the non-hydrolyzable substrates AMPCPP and GMPPNP did not perturb the 1H-15N HSQC spectrum of DR_0079 since these substrates contained purines and they were triphosphates. It is also likely that the excess phosphate in the NMR buffer, at 100-fold molar excess relative to the protein, did not affect the chemical shift mapping experiments significantly as enzymatic studies showed that 50,000-fold molar excess phosphate did not inhibit catalysis (data not shown). Consequently, chemical shift mapping experiments were performed with TMP-CP (chosen over the CMP analogue because it is commercially available) in the same NMR buffer used in the initial studies. Relative to TDP, the oxygen bridging the two phosphorus atoms is replaced with a methylene group in TMP-CP and this prevents hydrolysis of the phosphodiester bond. Figure 7A shows the chemical shift perturbations to the 1H-15N HSQC spectrum of DR_0079 (black) upon the addition of 4-fold molar excess TMP-CP (red) to the protein. The amide resonances for eight residues disappear completely or their intensity is significantly reduced and there are slight perturbations to six additional residues. Note that at intermediate molar ratios there were no ‘perturbed only’ cross peaks, but, there was a gradual decrease in intensity for the eight ‘disappearing’ cross peaks with increasing TMP-CP concentration.
Fig. 7.
A) Overlay of the 1H–15N HSQC spectra of DR_0079 in the absence (black) and presence (red) of ~4:1 molar ratio of TMP-CP:DR_0079. The chemical shifts of backbone amide resonances that disappear or shift (underlined) upon the addition of TMP-CP are labeled. Spectra were collected at a 1H resonance frequency of 600 MHz, 25°C, in buffer containing 100 mM KCl, 100 mM MgCl2, 20 mM potassium phosphate, 2 mM DTT, pH 7.1. B + C) Residues of backbone amides that disappear (dark blue) or shift slightly (light blue) upon the addition of TMP-CP to DR_0079 at a substrate:protein molar ratio of ~4.0:1.0 are mapped unto the structure of DR_0079 and shown in two orientations that differ by ~180°. Also highlighted are E85 and E89 (orange), the residues that most likely bind the divalent cation, and G70 (magenta), the residue the cation may sit over.
The data in Figure 7A is mapped onto the surface of DR_0079 in Figures 7B and 7C. The positions of the 1HN residues that disappear, or intensity decreases, at a 4:1 molar ratio of TMP-CP:DR_0079 are colored dark blue while the position of 1HN residues that are perturbed are colored light blue. As a reference the likely primary metal binding glutamic acid residues, E85 and E89, are colored orange and the residue the metal likely sits over, G70, is colored magenta. In general, the residues perturbed by TMP-CP binding are near the metal binding site and the cleft adjacent to this site. Indeed, A71 is right next to the metal-binding site. Three residues that disappear, E32, S113 and, D156, form a contiguous surface on the far side of the cleft as shown in Figure 7B. Residues in regions that immediately surround the metal-binding core, r3, r1, and the N-terminal β-sheet, as illustrated in Figure 3B and 3C, are not significantly perturbed following the addition of TMP-CP. As illustrated in Figure 4A, TDP isn’t the optimal substrate for DR_0079 and perhaps the binding isn’t very tight. Alternatively, pyrimidine nucleoside diphosphates are not very large molecules and it may be that the binding of (d)NDPs to DR_0079 does not effect significant perturbations to the protein.
Speculative substrate binding events
Using the available chemical shift data, the crystal structure for DR_0079, and the knowledge of how other Nudix substrates bind to protein it is possible to speculate which regions and residues in DR_0079 might play a role in binding (d)NDPs. Foremost, DR_0079 has a well-defined pocket at the mouth of the Nudix box, illustrated best in Figure 6B. Differences in the features of such pockets are responsible for the substrate specificity of different Nudix proteins (21, 27). Next, the Co2+ chemical shift mapping data presented here and the data previously reported for Mg2+ (27) suggest that at least one divalent metal cation sits over G70 and it could be held in position by the side chains of E85 and/or E95. As observed in other Nudix protein-substrate complexes, this metal probably coordinates with the phosphodiester group of the substrate, either directly or via a second sphere of water (13). Surrounding the hydrophobic part of the substrate binding cleft in Nudix proteins of known structure are polar residues that form hydrogen bonds with the nucleoside and/or interact with the sugar and phosphate groups of the substrate (21). While four polar residues, R24, R53, K57, and K156, surround the metal binding site (Figure 6B), no perturbations of these residues were observed upon binding to TMP-CP (Figures 7), and therefore, there is no evidence that they participate in the catalysis. The substrate binding cleft in Nudix proteins of known structure also often contain a hydrophobic surface that interacts with the hydrophobic parts of the substrate, especially with regards to ‘base-stacking’-like interactions with the nucleoside base (16). The side chain of W31 sits on the ‘floor’ of the substrate-binding cleft (Figure 6B) next to the ring of polar residues in DR_0079. While no chemical shift perturbations were directly observed for the backbone amide of W31 upon the addition of TMP-CP, the intensity of backbone amides of residues around this region (E32, S113, and D156) were effected (Figure 7B) suggesting that some type of interactions with the substrate may be occurring on this face of the protein.
Biological function
While the substrates tested here show that DR_0079 has a marked preference for (d)CDP and (d)CTP nucleotides, such in vitro testing for biological function is limited to the number of potential substrates available for study (22). For example, DR_0079 may have yet greater specificity for an untested, oxidized (d)CDP or (d)CTP. This problem is compounded because Nudix hydrolases generally show overlapping substrate specificity (1, 34). Indeed, such overlapping specificity will make it difficult to test biological functionality in vivo by systematic gene disruption in D. radiodurans because when a specific Nudix hydrolase is knocked out another one (or more) of the 20 Nudix hydrolases may substitute (22).
The structure of DR_0079 represents the first Nudix hydrolyase to show a preference for cytidine (deoxy)nucleoside 5’-diphosphates. To identify other proteins with similar substrate specificity, the Protein Data Bank was searched with Dali (48) for structures with similarities to DR_0079. Thirty-one proteins with Z-scores greater than four were identified. All of the proteins, except two, are suspected Nudix hydrolases or known Nudix hydrolases with substrate specificities that vary across the known Nudix substrate range. The structure with the highest Z-score, 19.8, is for the crystal structure of the putative Nudix hydrolase yfcD from E. coli strain K12 (PDB ID = 2FKB). The function of this protein is unknown (25) but it contains a N-terminal β-sheet and C-terminal helix-turn-helix motif (in this case it is a helix-helix-helix motif) that is very similar to DR_0079. Perhaps the similarities in the structure of DR_0079 and yfcD will reflect a similar preference for nucleoside diphosphates. On the other hand, it may not, as the next two highest Z-scores, 18.1 and 17.1, were for type I isopentenyl-diphosphate δ-isomerase (IDI) from E. coli (49). This enzyme is an important component of the ubiquitous sterol/isoprenoid biosynthetic pathway, catalyzing the interconversion of isopentenyl diphosphate and dimethylallyl diphosphate. However, these compounds were ruled out as substrates for DR_0079 for a number of compelling reasons, most important of which was a negative result when testing for IPP isomerase activity (27). Consequently, the large number of DALI “hits” with DR_0079 may simply reflect similarities in the basic ‘Nudix fold’ that concomitantly may result in overlap of substrate specificities.
Comparision to the structure of other D. radiodurans Nudix proteins
The genome of the bacterium D. radiodurans codes for 21 Nudix hydrolases containing a fully conserved Nudix box (50). Crystal structures have been determined for three of these gene products: DR1184 (20), DR1025 (38) and now DR_0079. There is convincing evidence that the native substrate for DR1184 is coenzyme A (20), the native substrate for DR1025 is uncertain but it has been co-crystallized with both a GTP analog and Ap4A (38), and DR_0079 shows a perference for (d)CDP and (d)CTP. Given different preferred substrates for each D. radiodurans hydrolase it is not surprising that while the same basic Nudix fold, the mixed ‘four’ strand β-sheet sandwiched between orthogonal α-helices, is similar in all three proteins, the length and nature of the folds and appendages connecting to the ‘four’ strand β-sheet differ. In DR_1184 the appendages are an N-terminal α-helix followed by a β-strand, an extra three strand β-sheet attached to the ‘four’ strand β-sheet, and an α-helix and three strand anti-parallel β-sheet inserted near the N-terminal. In DR_1025 these appendages are a short N-terminal helix, a small three strand β-sheet attached to the ‘four’ strand β-sheet, and extra α-helix near the C-terminal. In DR_0079 these appendages are an N-terminal anti-parallel β-sheet (β1 and β2), an extra three strand β-sheet (β4, β4*, and β8) attached to the ‘four’ strand β-sheet, and an α-helix (α2) inserted near the C-terminus. The structural and functional diversity of these three proteins, despite their sequence similarities, highlight the need to explore the diversity in protein families with the same basic bold, but, differing sequence and function (38).
The Nudix hydrolysis DR_0975 has a marked specificity for (d)NDPs and is nearly the same length as DR_0079 at 169 versus 171 residues. Could the overall structure of both proteins be similar, especially with regards to the substrate-binding site? While some details of the substrate-binding cleft may be similar between the proteins, the overall structure will likely differ because the first residue of the Nudix box in DR_0079 is G70 while the first residue in DR_0975 is G40. Consequently, the region N-terminal to the Nudix box will be 30 residues shorter in DR_0975 while the region C-terminal to the Nudix box will be ~30 residues longer.
Concluding remarks
There had been a lot of interest in the bacterium Deinococcus radiodurans because of its unique capacity to withstand relatively high doses of ionizing radiation and the lethal and mutagenic effects of ultraviolet radiation and other physical and chemical DNA-damaging agents (51, 52). Such properties likely are a consequence of the organism’s ability to tolerate dessication (53). Another unique property of D. radiodurans is that its genome contains an uncommonly high number of genes with potential DNA repair activities (50). Consequently, it has been suggested that D. radiodurans dessication/radiation tolerance is related to the redundancy in DNA repair genes that include the 21 genes with sequence homology to the Nudix family of polyphosphate pyrophosphohydrolases (3). However, emerging evidence suggests that DNA repair is not the primary function of the Nudix hydrolase suite of proteins in D. radiodurans. A transcriptomic study indicated that only five of the 21 D. radiodurans genes were induced following exposure to γ-irradiation (54). Global analysis of the D. radiodurans proteome using high-resolution mass spectrometry methods showed that DR_0079 and the other 20 hypothetical Nudix proteins were predominately expressed in unstressed cells (55). Induction assays showed that neither peroxide nor superoxide induced the expression of the Nudix hydrolase DR_0975 in D. radiodurans (22). Instead of repairing oxidized DNA, the primary function of the Nudix hydrolases may be to maintain the physiological balance in the cell during the hours of chromosomal repair immediately following exposure to ionizing radiation or stress by removing or inactivating potentially toxic endogenous metabolites that contain at least one diphosphate linkage (9, 34). The promiscuous nature of many of these hydrolases, especially towards oxidized nucleosides, may enhance their housekeeping role. Towards ‘keeping house’, it has been observed that D. radiodurans cells in stationary phase are more resistant to radiation that cells in logarithmic phase (56). Because induction assays show that DR_0975 expression is induced only upon entry into stationary phase, the housekeeping duties that DR_0975 performs to prepare the cell for stationary phase may indirectly contribute to the organism’s radiation resistant properties (22). Further biochemical studies are necessary to determine if DR_0079 specificity for cytosine ribonucleoside 5’-diphosphate (CDP) and cytosine ribonucleoside 5’-triphosphate (CTP) is part of its in vivo housekeeping duties.
ACKNOWLEDGMENT
We thank Dr. Stephen R. Holbrook at Lawrence Berkeley National Laboratory for first getting us involved in studies on D. radiodurans Nudix hydrolases.
Abbrevations
- (CDP)
cytosine ribonucleoside 5’-diphosphate
- (CTP)
cytosine ribonucleoside 5’-triphosphate
- (IDP)
inosine ribonucleoside 5’-diphosphate
- (TMP-CP)
thymidine-5’-O-(α,β-methylenediphosphate)
- (8-oxo-dGTP)
7,8-dihydro-8-oxoguanosine triphosphate
- (8-oxo-dGDP)
7,8-dihydro-8-oxoguanosine diphosphate
- (IDI)
isopentenyl-diphosphate δ-isomerase
- ((d)NMP)
ribonucleoside and deoxyribonucleoside 5’-monophosphate
- ((d)NDP)
ribonucleoside and deoxyribonucleoside 5’-diphosphate
- ((d)NTP)
ribonucleoside and deoxyribonucleoside 5’-triphosphate
- (PPi)
pyrophosphate
- (Pi)
orthophosphate
Footnotes
The research was performed in the Environmental Molecular Sciences Laboratory (a national scientific user facility sponsored by the DOE Biological and Environmental Research) located at Pacific Northwest National Laboratory and operated for DOE by Battelle, as well as in the Ontario Centre for Structural and Functional Proteomics at the University of Toronto. Support for beamline X29A at the National Synchrotron Light Source comes principally from the Offices of Biological and Environmental Research and of Basic Energy Sciences of the US Department of Energy, and from the National Center for Research Resources of the National Institutes of Health. This work was funded by grants from the U.S. Department of Energy, Office of Biological Energy Research, Contract No. (DE-AC03-76SF00098), Genome Canada (through the Ontario Genomics Institute), the Ontario Research and Development Challenge Fund, and the Protein Structure Initiative of the National Institutes of Health (Midwest Center for Structural Genomics, NIH grant GM 62414).
REFERENCES
- 1.McLennan AG. The Nudix hydrolase superfamily. Cell. Mol. Life Sci. 2006;63:123–143. doi: 10.1007/s00018-005-5386-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abeygunawardana C, Weber DJ, Gittis AG, Frick DN, Lin J, Miller AF, Bessman MJ, Mildvan AS. Solution structure of the MutT enzyme, a nucleoside triphosphate pyrophosphohydrolase. Biochemistry. 1995;34:14997–15005. doi: 10.1021/bi00046a006. [DOI] [PubMed] [Google Scholar]
- 3.Bessman MJ, Frick N, O’Handley SF. The MutT proteins or "Nudix" hydrolases, a family of versatile, widely distributed, "housecleaning" enzymes. J. Biol. Chem. 1996;271:25059–25062. doi: 10.1074/jbc.271.41.25059. [DOI] [PubMed] [Google Scholar]
- 4.Wang Z, Jiao X, Carr-Schmid A, Kiledjian M. The hDcp2 protein is a mammalian mRNA decapping enzyme. Proc. Natl. Acad. Sci. USA. 2002;99:12663–12668. doi: 10.1073/pnas.192445599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunn CA, O’Handley SF, Frick DN, Bessman MJ. Studies on the ADP-ribose pyrophosphatase subfamily of the nudix hydrolases and tentative identification of trgB, a gene associated with tellurite resistance. J. Biol. Chem. 1999;274:32318–32324. doi: 10.1074/jbc.274.45.32318. [DOI] [PubMed] [Google Scholar]
- 6.Lawhorn BG, Gerdes SY, Begley TP. A genetic screen for the identification of thiamin metabolic genes. J. Biol. Chem. 2004;279:43555–43559. doi: 10.1074/jbc.M404284200. [DOI] [PubMed] [Google Scholar]
- 7.Safrany ST, Caffrey JJ, Yang XN, Bembenek ME, Moyer MB, Burkhart WA, Shears SB. A novel context for the 'MutT' module, a guardian of cell integrity, in a diphosphoinositol polyphosphate phosphohydrolase. EMBO J. 1998;17:6599–6607. doi: 10.1093/emboj/17.22.6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu W, Jones CR, Dunn CA, Bessman MJ. Gene ytkD of Bacillus subtilis encodes an atypical nucleoside triphosphate member of the Nudix hydrolase superfamily. J. Bacteriol. 2004;186:8380–8384. doi: 10.1128/JB.186.24.8380-8384.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galperin MY, Moroz OV, Wilson KS, Murzin AG. House cleaning, a part of good housekeeping. Mol. Microbiol. 2006;59:5–19. doi: 10.1111/j.1365-2958.2005.04950.x. [DOI] [PubMed] [Google Scholar]
- 10.Grollman AP, Moriya M. Mutagenesis by 8-oxoguanine: an enemy within. Trends Genet. 1993;9:246–249. doi: 10.1016/0168-9525(93)90089-z. [DOI] [PubMed] [Google Scholar]
- 11.Maki H, Sekiguchi M. MutT protein specifically hydrolyzes a potent mutagenic substrate for DNA synthesis. Nature. 1992;355:273–275. doi: 10.1038/355273a0. [DOI] [PubMed] [Google Scholar]
- 12.Weber DJ, Bhatnagar SK, Bullions LC, Bessman MJ, Mildvan AS. NMR and isotopic exchange studies of the site of bond cleavage in the MutT reaction. J. Biol. Chem. 1992;267:16939–16942. [PubMed] [Google Scholar]
- 13.Lin J, Abeygunawardana C, Frick DN, Bessman MJ, Mildvan AS. Solution structure of the quaternary MutT-M2+-AMPCPP-M2+ complex and mechanism of its pyrophosphohydrolase action. Biochemistry. 1997;36:1199–1211. doi: 10.1021/bi962619c. [DOI] [PubMed] [Google Scholar]
- 14.Bailey S, Sedelnikova SE, Blackburn GM, Abdelghany HM, Baker PJ, McLennan AG, Rafferty JB. The crystal structure of diadenosine tetraphosphate hydrolase from caenorhabditis elegans in free and binary complex forms. Structure. 2002;10:589–600. doi: 10.1016/s0969-2126(02)00746-3. [DOI] [PubMed] [Google Scholar]
- 15.Fletcher JI, Swarbrick JD, Maksel D, Gayler KR, Gooley PR. The structure of Ap(4)A hydrolase complexed with ATP-MgF(x) reveals the basis of substrate binding. Structure. 2002;10:205–213. doi: 10.1016/s0969-2126(02)00696-2. [DOI] [PubMed] [Google Scholar]
- 16.Gabelli SB, Bianchet MA, Bessman MJ, Amzel LM. The structure of ADP-ribose pyrophosphatase reveals the structural basis for the versatility of the Nudix family. Nat. Struct. Biol. 2001;8:467–472. doi: 10.1038/87647. [DOI] [PubMed] [Google Scholar]
- 17.Kang L-W, Gabelli SB, Cunningham JE, O’Handley SF, Amzel LM. Structure and mechanism of MT-ADPRase, a Nudix hydrolase from Mycobacterium tuberculosis. Structure. 2003;11:1015–1023. doi: 10.1016/s0969-2126(03)00154-0. [DOI] [PubMed] [Google Scholar]
- 18.Zha M, Zhong C, Peng Y, Hu H, Ding J. Crystal structure of human NUDT5 reveals insights into the structural basis of the substrate specificity. J. Mol. Biol. 2006;364:1021–1033. doi: 10.1016/j.jmb.2006.09.078. [DOI] [PubMed] [Google Scholar]
- 19.Gabelli SB, Bianchet MA, Xu W, Dunn CA, Niu ZD, Amzel LM, Bessman MJ. Structure and function of the Escherichia coli dihydroneopterin triphosphate pyrophosphatase: a Nudix enzyme involved in folate biosynthesis. Structure. 2007;15:1014–1022. doi: 10.1016/j.str.2007.06.018. [DOI] [PubMed] [Google Scholar]
- 20.Kang L-W, Gabelli SB, Bianchet MA, Xu WL, Bessman MJ, Amzel LM. Structure of a coenzyme A pyrophosphatase from Deinococcus radiodurans: a member of the Nudix family. J. Bacteriol. 2003;185:4110–4118. doi: 10.1128/JB.185.14.4110-4118.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mildvan AS, Xia Z, Azurmendi HF, Saraswat V, Legler PM, Massiah MA, Gabelli SB, Bianchet MA, Kang L-W, Amzel LM. Structures and mechanisms of Nudix hydrolases. Arch. Biochem. Biophys. 2005;433:129–143. doi: 10.1016/j.abb.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 22.Fisher DJ, Cartwright JL, Harashima H, Kamiya H, McLennan AG. Characterization of a Nudix hydrolase from Deinococcus radiodurans with a marked specificity for deoxyribonucleoside 5’-diphosphates. BMC Biochem. 2004;5:7–14. doi: 10.1186/1471-2091-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishibashi T, Hayakawa H, Sekiguchi M. A novel mechanism for preventing mutations caused by oxidation of guanine nucleotides. EMBO Rep. 2003. 2003;4:479–483. doi: 10.1038/sj.embor.embor838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hori M, Fujikawa K, Kasai H, Harashima H, Kamiya H. Dual hydrolysis of diphosphate and triphosphate derivatives of oxidized deoxyadenosine by Orf17 (NtpA), a MutT-type enzyme. DNA Repair. 2005;4:33–39. doi: 10.1016/j.dnarep.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 25.Xu W, Dunn CA, O’Handley SF, Smith DL, Bessman MJ. Three new Nudix hydrolases from Escherichia coli. J. Biol. Chem. 2006;281:22794–22798. doi: 10.1074/jbc.M603407200. [DOI] [PubMed] [Google Scholar]
- 26.Ito R, Hayakawa H, Sekiguchi M, Ishibashi T. Multiiple enzyme activities of Escherichia coli MutT protein for sanitization of DNA and RNA precursor pools. Biochemistry. 2005;44:6670–6674. doi: 10.1021/bi047550k. [DOI] [PubMed] [Google Scholar]
- 27.Buchko GW, Ni S, Holbrook SR, Kennedy MA. Solution structure of hypothetical Nudix hydrolase DR0079 from extremely radiation-resistant Deinococcus radiodurans bacterium. Proteins. 2004;56:28–39. doi: 10.1002/prot.20082. [DOI] [PubMed] [Google Scholar]
- 28.Buchko GW, Ni S, Holbrook SR, Kennedy MA. 1H, 13C, and 15N NMR assignments of the hypothetical Nudix protein DR0079 from the extremely radiation-resistant bacterium Deinococcus radiodurans. J. Biomol. NMR. 2003;25:169–170. doi: 10.1023/a:1022243724501. [DOI] [PubMed] [Google Scholar]
- 29.Zuiderweg ERP. Mapping protein-protein interactions in solution by NMR spectroscopy. Biochemistry. 2002;41:1–7. doi: 10.1021/bi011870b. [DOI] [PubMed] [Google Scholar]
- 30.Buchko GW, Daughdrill GW, de Lorimier R, Rao BK, Isern NG, Lingbeck JM, Taylor JS, Wold MS, Gochin M, Spicer LD, Lowry DF, Kennedy MA. Interactions of human nucleotide excision repair protein XPA with DNA and RPA70ΔC327: Chemical shift mapping and 15N NMR relaxation studies. Biochemistry. 1999;38:15116–15128. doi: 10.1021/bi991755p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holbrook EL, Schulze-Gahmen U, Buchko GW, Ni S, Kennedy MA, Holbrook SR. Purification, crystallization and preliminary X-ray analysis of two Nudix hydrolases from Deinococcus radiodurans. Acta Crystallogr. 2003;D59:737–740. doi: 10.1107/s0907444903002671. [DOI] [PubMed] [Google Scholar]
- 32.Doublie S. Preparation of selenomethionyl proteins for phase determination. Methods Enzymol. 1997;276:523–530. [PubMed] [Google Scholar]
- 33.Baykov AA, Evtushenko OA, Avaeva SM. A malachite green procedure for orthophosphate determination and its use in alkaline phosphatase-based enzyme immunoassay. Anal. Biochem. 1988;171:266–270. doi: 10.1016/0003-2697(88)90484-8. [DOI] [PubMed] [Google Scholar]
- 34.Xu W, Shen J, Dunn CA, Desai S, Bessman MJ. The Nudix hydrolases of Deinococcus radiodurans. Mol. Microbiol. 2001;39:286–290. doi: 10.1046/j.1365-2958.2001.02267.x. [DOI] [PubMed] [Google Scholar]
- 35.Maiti R, van Domselaar GH, Zhang H, Wishart DS. SuperPose: a simple server for sophisticated structural superposition. Nucleic Acids Res. 2004;32:W590–W594. doi: 10.1093/nar/gkh477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koonin EV. A highly conserved sequence motif defining the family of MutT-related proteins from eubacteria, eukaryotes and viruses. Nucleic Acids Res. 1993;21:4847. doi: 10.1093/nar/21.20.4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang S, Mura C, Sawaya MR, Cascio D, Eisenberg D. Structure of a Nudix protein from Pyrobaculum aerophilum reveals a dimer with two intersubunit beta-sheets. Acta Crystallogr. 2002;D58:571–578. doi: 10.1107/s0907444902001191. [DOI] [PubMed] [Google Scholar]
- 38.Ranatunga W, Hill EE, Mooster JL, Holbrook EL, Schulze-Gahmen U, Xu W, Bessman MJ, Brenner SE, Holbrook SR. Structural studies of the Nudix hydrolase DR1025 from Deinococcus radiodurans and its ligand complexes. J. Mol. Biol. 2004;339:103–116. doi: 10.1016/j.jmb.2004.01.065. [DOI] [PubMed] [Google Scholar]
- 39.Massiah MA, Saraswat V, Azurmendi HF, Mildvan AS. Solution structure and NH exchange studies of the MutT pyrophosphohydrolase complexed with Mg2+ and 8-oxo-dGMP, a tightly bound product. Biochemistry. 2003;42:10140–10154. doi: 10.1021/bi030105p. [DOI] [PubMed] [Google Scholar]
- 40.Stogios PJ, Chen L, Prive GG. Crystal structure of the BTB domain from the LRF/ZBTB7 transcriptional regulator. Protein Sci. 2007;16:336–342. doi: 10.1110/ps.062660907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dunker AK, Brown CJ, Lawsor JD, Iahoucheva LM, Obradovic Z. Intrinsic disorder and protein function. Biochemistry. 2002;41:6573–6582. doi: 10.1021/bi012159+. [DOI] [PubMed] [Google Scholar]
- 42.Cohn M, Hu A. Isotopic (18O) shift in 31P nuclear magentic resonance applied to a study of enzyme-catalyzed phosphate-phosphate exchange and phosphate (oxygen)-water exchange reactions. Proc. Natl. Acad. Sci. USA. 1978;75:200–203. doi: 10.1073/pnas.75.1.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cohn M, Hughes TR. Phosphorus magnetic resonance spectra of adenosine di-and triphosphate. J. Biol. Chem. 1960;235:3250–3253. [PubMed] [Google Scholar]
- 44.Tran-Dinh S, Neumann JM. A 31P-NMR study of the interaction of Mg2+ with nucleoside diphosphates. Nucleic Acids Res. 1977;4:397–403. doi: 10.1093/nar/4.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shukar SB, Hajduk PJ, Meadows RP, Fesik SW. Discovering high-affinity ligands for proteins. Science. 1996;274:1531–1534. doi: 10.1126/science.274.5292.1531. [DOI] [PubMed] [Google Scholar]
- 46.Gochin M, Roder H. Protein structure refinement based on paramagnetic NMR shifts: Applications to wild-type and mutant forms of cytochrome c. Protein Sci. 1995;4:296–305. doi: 10.1002/pro.5560040216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin J, Abeygunawardana C, Frick DN, Bessman MJ, Mildvan AS. The role of glu 57 in the mechanism of the Escherichia coli MutT enzyme by mutagenesis and heteronuclear NMR. Biochemistry. 1996;35:6715–6726. doi: 10.1021/bi953071x. [DOI] [PubMed] [Google Scholar]
- 48.Holm L, Sander C. Touring protein fold space with Dali/FSSP. Nucleic Acids Res. 1998;26:316–319. doi: 10.1093/nar/26.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bonanno JB, Edo C, Eswar N, Pieper U, Romanowski MJ, Ilyin V, Gerchman SE, Kycia H, Studier FW, Sali A, Burley SK. Structural genomics of enzymes involved in sterol/isoprenoid biosynthesis. Proc. Natl. Acad. Sci. USA. 2001;98:12896–12901. doi: 10.1073/pnas.181466998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.White O, Eisen JA, Heidelberg JF, Hickey EK, Peterson JD, Dodson RJ, Haft DH, Gwinn ML, Nelson WC, Richardson DL, Moffat KS, Qin H, Jiang L, Pamphile W, Crosby M, Shen M, Vamathevan JJ, Lam P, McDonald L, Utterback T, Zalewski C, Makarova KS, Aravind L, Daly MJ, Minton KW, Fleischmann RD, Ketchum KA, Nelson KE, Salzberg S, Smith HO, Venter JC, Fraser CM. Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science. 1999;286:1571–1577. doi: 10.1126/science.286.5444.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anderson AW, Nordon HC, Cain RF, Parrish G, Duggan D. Studies on a radio-resistant micrococcus. I. Isolation, morphology, cultural characteristics, and resistance to gamma radiation. Food Technol. 1956;10:575–578. [Google Scholar]
- 52.Battista A. Against all odd: The survival strategies of Deinococcus radiodurans. Ann. Rev. Microbiol. 1997;51:203–224. doi: 10.1146/annurev.micro.51.1.203. [DOI] [PubMed] [Google Scholar]
- 53.Mattimore V, Battista JR. Radioresistance of Deinococcus radiodurans: functions necessary to survive ionizing radiation are also necessary to survive prolonged desiccation. J. Bacteriol. 1996;178:633–637. doi: 10.1128/jb.178.3.633-637.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu Y, Zhou J, Omelchenko MV, Beliaev AS, Venkateswaran A, Stair J, Wu L, Thompson DK, Xu D, Rogozin IB, Gaidamakova EK, Zhai M, Makarova KS, Koonin EV, Daly MJ. Transcriptome dynamics of Deinococcus radiodurans recovering from ionizing radiation. Proc. Natl. Acad. Sci. USA. 2003;100:4193–4196. doi: 10.1073/pnas.0630387100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lipton MS, Pasa-Tolic L, Anderson GA, Anderson DJ, Auberry DL, Battista JR, Daly MJ, Fredrickson J, Hixson KK, Kostandarithes H, Masselon C, Markillie LM, Moore RJ, Romine MF, Shen Y, Stritmatter E, Tolic N, Udseth HR, Venkateswaran A, Wong K-K, Zhao R, Smith RD. Global analysis of the Deinococcus radiodurans proteome by using accurate mass tags. Proc. Natl. Acad. Sci. USA. 2002;99:11049–11054. doi: 10.1073/pnas.172170199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Milton KW. DNA repair in the extremely radioresistant bacterium Deinococcus radiodurans. Mol. Microbiol. 1994;13:9–15. doi: 10.1111/j.1365-2958.1994.tb00397.x. [DOI] [PubMed] [Google Scholar]