Abstract
Silica nanoparticle supported cationic lipids can effectively bind plasmid DNAs and transfect mammalian cells with an efficiency that depends on both the particle size and lipid composition; here the gene delivery and expression process has been confirmed by confocal fluorescence microscopy.
Gene delivery to mammalian cells has gained significant attention due to its importance in gene therapy.1–3 Genes embedded in plasmid DNAs provide a stable source for therapeutic proteins and RNAs. Naked DNAs by themselves cannot cross the cell membrane barrier and are easily degraded by nucleases in biological fluids.4 As a result, delivery vehicles are needed for efficient transfection.5 Due to the intrinsic toxicity and immunogenicity of viral vectors, current research focus has shifted to the development of nonviral carriers.6–12 Cationic lipids and liposomes are quite effective in gene delivery.13, 14 However, highly negatively charged DNAs can induce fusion of such liposomes to generate large particles, which may reduce the transfection efficiency and increase toxicity.15 To minimize this problem, crosslinked or PEGylated liposomes have been tested.15, 16 Crosslinked liposomes, however, may have biodegradation problems in vivo. We have recently explored the use of silica nanoparticle (NP) supported lipid bilayers for drug delivery applications.17, 18 Such supported bilayers have higher stability compared to empty liposomes and the lipid layers are unlikely to fuse with each other due to the presence of a solid core. On the other hand, the lipids are not covalently linked and can still exchange and fuse with cellular lipids and be metabolized. Herein we report the use of supported lipid bilayers for gene delivery to Chinese hamster ovary cells (CHO). One of the advantages of the supported bilayer system is that both the silica core size and lipid composition of the shell can be systematically varied, which provides us a useful system to tune and understand the gene delivery process.
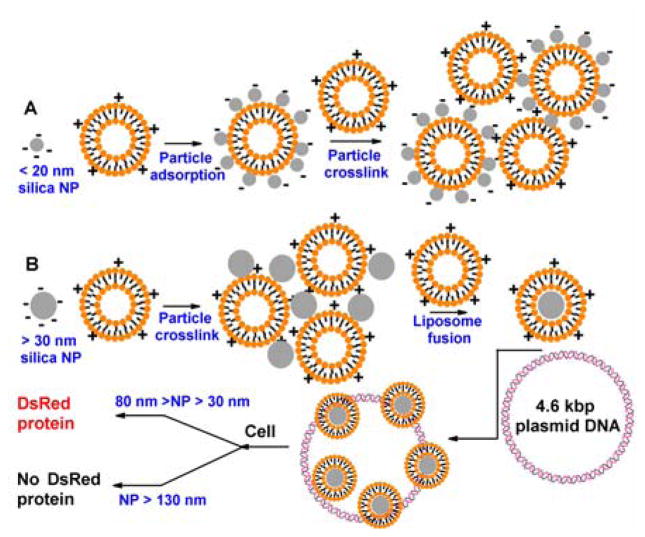
Pure silica is negatively charged at pH 7 and thus requires charge reversal to bind DNA. We employed silica NPs with sizes ranging from 8 to 130 nm, and tested their interactions with cationic liposomes and DNA. Silica NPs with a diameter of 8 nm appear transparent in solution. Addition of a small amount of cationic DOTAP liposomes resulted in a white suspension, suggesting the formation of larger NP assemblies that scattered light strongly (Fig 1A). These larger NPs showed a negative surface charge by zeta-potential measurement (Fig 2A). This is consistent with the formation of liposome-supported, sub-20-nm diameter NPs as reported by Zhang and Granick.19 Further addition of DOTAP led to the formation of gel-like large aggregates, suggesting the crosslinking of supported particles by liposomes (Fig 1A). Since these very small particles gave either a negative surface charge or very large aggregates when mixed with cationic lipids, they are not suitable for binding negatively charged DNA and are therefore abandoned for further studies.
Fig. 1.
(A) Small silica NPs are adsorbed onto positively charged DOTAP liposomes, resulting a negatively charged surface. With excess DOTAP, the system crosslinks to form large aggregates. (B) Larger silica NPs first formed aggregates by a small amount of DOTAP. Excess DOTAP leads to liposome fusion and formation of supported bilayers with a positively charged surface useful for DNA association and delivery.
Fig. 2.
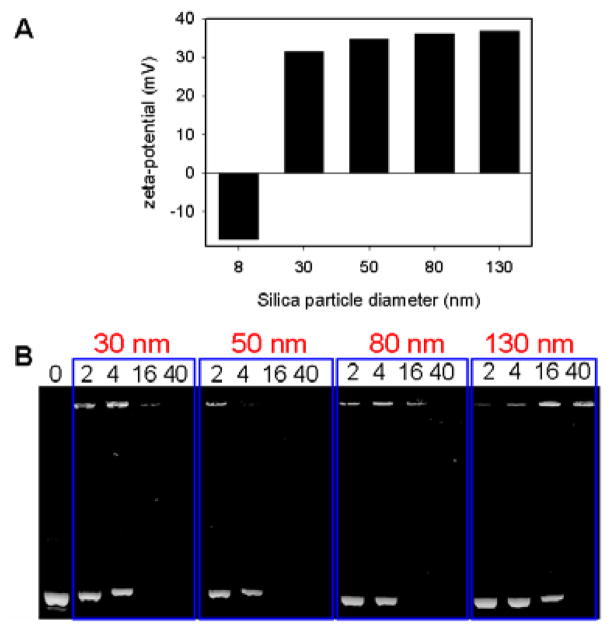
(A) Zeta-potential of complexes formed by silica NPs of different sizes and DOTAP liposomes. For the 8 nm silica NPs, excess silica was used; while for the other particles, excess liposomes were used (both purified by centrifugation). (B) Association of supported bilayers with the plasmid DNA studied by agarose gel electrophoresis. The retained DNAs are on the top and free DNAs migrated to the bottom. The numbers on the top of each lane indicate the mass of silica NPs relative to DNA.
Next, larger silica NPs (diameters 30, 50, 80, 130 nm) were tested. For all these particles, large gel-like aggregates were observed in the presence of a low concentration of DOTAP liposomes, which disappeared with increasing amount of lipids, suggesting the formation of NP supported bilayers.17 A schematic of this process is shown in Fig 1B. Zeta-potential measurements showed that the supported bilayers all had positively charged surfaces (Fig 2A) and therefore, should bind DNA. Indeed, after mixing these NP-supported bilayers with a plasmid DNA encoding for the DsRed fluorescent protein, particle size-dependent DNA association was observed from agarose gel electrophoresis studies (Fig 2B). For each lane in the gel, 0.25 μg of plasmid DNA was used and the mass of silica NPs in supported bilayers was varied from 2x to 40x that of the DNA (lanes labeled 2–40). The lower bands are the free plasmid DNA and the upper bands are DNA trapped in the gel loading wells due to the complex formation with supported lipid bilayers. For NPs of 30 to 80 nm, complete DNA binding was observed for silica contents corresponding to 4x or 16x the DNA mass; while for the 130 nm NPs, a silica content of ~40x the DNA mass was needed, possibly due to decreased surface area with increase of particle size when the mass is fixed.
We next studied the effect of silica particle size on the transfection efficiency. In all the experiments, the silica/DNA mass ratio was maintained at the intermediate level of 10 (1 μg DNA and 10 μg silica-NPs). The lipid contained an equal mass mixture of DOTAP and cholesterol. The supported bilayers and DNA were first suspended in a serum free F-12K media and then incubated with CHO cells. After 4 hr incubation at 37 °C, the old media containing the transfection agent was removed and replaced with fresh serum containing media. The transfection efficiency was determined by flow cytometry after 24 hrs. As shown in Fig 3A, the transfection efficiency decreased dramatically with increasing particle size. With a 130 nm silica core, the efficiency is close to zero. Increasing the mass of 130-nm silica NPs from 10 to 20 or 40 μg did not improve transfection (data not shown), suggesting that low transfection was not due to insufficient NPs. This size-dependent trend is quite different from that observed by Saltzman et al, who first mixed plasmid DNAs with commercial transfection agents (cationic dendrimers or lipids) and then added silica NPs before transfection. In that case, transfection efficiency increased significantly with increasing silica NP size, which Saltzman attributed to a simple increase of physical settling of transfection agents due to gravity.20 The difference in the size-dependent transfection efficiency we observe suggests a different mechanism of DNA condensation/delivery. In our case, silica is part of the transfection agent and its size directly affects the condensation of DNA. Interestingly, compared to the empty liposomes of the same composition, NP-supported bilayers showed about 3-fold higher transfection efficiency. Further, the silica core may also help to settle the delivery system and thus facilitate endocytosis of the plasmid DNA.20 Finally, for smaller particles, the release of DNA from the supported bilayers inside cells may be facilitated, therefore resulting in higher transfection efficiency.
Fig. 3.
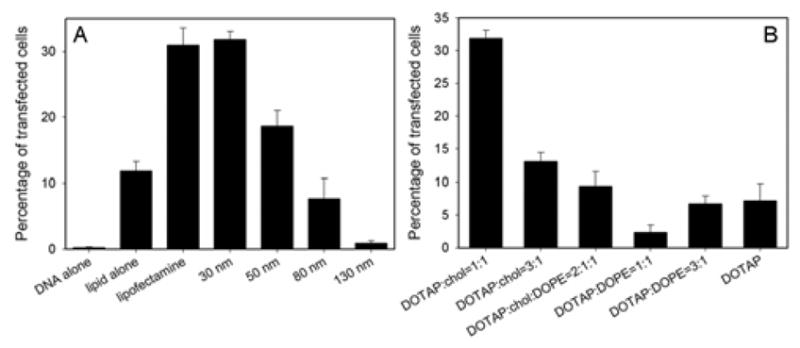
Transfection efficiency as a function of silica NP core size (A) and lipid shell composition (B).
To study the effect of lipid composition, we have tested lipid mixtures made of DOTAP, cholesterol, and DOPE. DOPE is known to facilitate liposome fusion and transfection. It has been recently demonstrated that cholesterol can enhance transfection efficiency in liposome-based formulations when mixed with cationic lipids.21–23 As presented in Fig 3B, the 1:1 DOTAP:cholesterol lipid has the highest transfection efficiency, which is similar to the commercially available lipofectamine. Decreasing cholesterol amount or introducing DOPE resulted in decreased efficiency. Under all conditions, cell viability was greater than 90% based on staining cells with ethidium homodimer and calcein-AM.
To test if the supported bilayers can protect DNA from enzymatic degradation, DNase I was added to the plasmid DNA before mixing with supported bilayers and essentially no transfected cells can be found; while if the DNA and supported bilayers were mixed first before adding the DNase, high transfection can still be obtained (see Supporting Information). Therefore, the supported bilayers can effectively protect DNA from nuclease degradation. Similar protection effects were also reported with cationic silica NPs or dendrimer-functionalized mesoporous silica NPs.24, 25
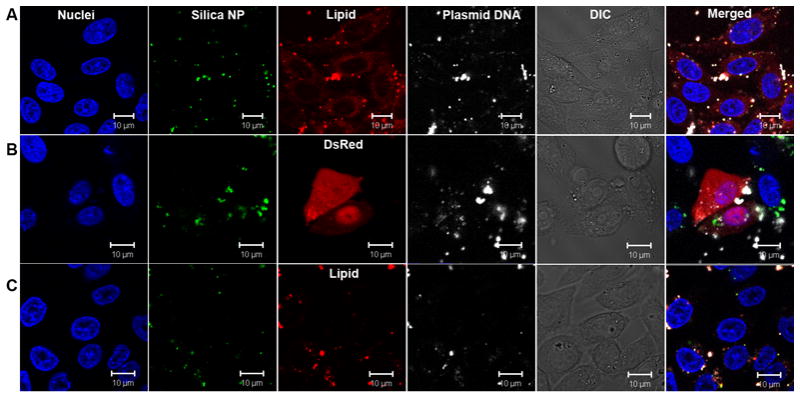
Finally, we studied the DNA uptake and transfection process by confocal fluorescence microscopy. Silica NPs (50 nm diameter) with green emission fluorophore labels were mixed with Texas Red-DHPE-labeled DOTAP/cholesterol liposomes to form supported bilayers. The plasmid DNA was covalently labeled with Cy-5. After incubating with CHO cells for 2 hrs and 12 hrs at 37 °C, the cells were washed by PBS, stained with Hoechst 33342, fixed with formaldehyde, and mounted with an anti-fade agent for microscopy. After 2 hr of mixing, the colocalization of green, red, and far red emissions was observed (Fig 4A), suggesting the silica NPs, lipids, and DNA entered the cell together. The cell surface also appeared to be covered with red dyes, suggesting some lipid on supported bilayers exchanged/fused with the cell membrane. After 12 hrs, some cells were transfected and the red fluoscence in Fig 4B is due to the expressed DsRed protein (lipid unlabeled). Cy-5 labeled plasmid DNA can be observed inside cell nuclei, while most green fluorescence (silica NPs) stayed outside cell nuclei, suggesting that DNA needs to be relased from the NPs to induce transfection. For larger NPs (200 nm diameter, Fig 4C), although there is a significant uptake of the NP/DNA complex, the complex appeared to be trapped in the endosomal compartment and no labeled plasmid DNA was found in cell nuclei, which may explain the inhibited transfection with large silica NPs.
Fig. 4.
DNA delivery studied by confocal fluorescence microscopy. Cell nuclei were stained with Hoechst 33342 (blue), silica NPs were labelled with a green emission fluorophore and the plasmid DNA was labelled with Cy-5 (emission colored white). (A) 50 nm silica NP supported bilayer (lipid labelled with Texas Red) incubated with CHO for 2 hrs. (B) 50 nm NP supported bilayer (lipid unlabeled) incubated with CHO for 12 hrs and some cells expressed the DsRed protein (red). (C) 200 nm NP supported bilayers (lipid labelled with Texas Red) incubated with CHO for 12 hrs.
In summary, we have demonstrated that supported bilayers are useful transfection agents, and the transfection efficiency depends on particle size and lipid composition. Compared to lipofectamine, the supported bilayer complex we describe is more cost-effective with similar performance. If instead of a solid core, a mesoporous silica core is used, additional drugs, siRNA, or imaging agents can be loaded into the pores as we recently described and co-delivered with plasmid DNAs.17, 18, 25, 26 Therefore, our system can potentially be engineered as a multi-functional drug delivery platform.
Supplementary Material
Acknowledgments
We acknowledge funding from the USA NIH through the NIH Roadmap for Medical Research, DOE Office of Science and Air Force Office of Scientific Research.
Footnotes
Electronic Supplementary Information (ESI) available: [experimental section and DNase assays]. See DOI: 10.1039/b000000x/
Notes and references
- 1.Somia N, Verma IM. Nat Rev Genet. 2000;1:91–99. doi: 10.1038/35038533. [DOI] [PubMed] [Google Scholar]
- 2.Edelstein ML, Abedi MR, Wixon J, Edelstein RM. J Gene Med. 2004;6:597–602. doi: 10.1002/jgm.619. [DOI] [PubMed] [Google Scholar]
- 3.Verma IM, Weitzman MD. Ann Rev Biochem. 2005;74:711–738. doi: 10.1146/annurev.biochem.74.050304.091637. [DOI] [PubMed] [Google Scholar]
- 4.Niven R, Pearlman R, Wedeking T, Mackeigan J, Noker P, Simpson-Herren L, Smith JG. J Pharm Sci. 1998;87:1292–1299. doi: 10.1021/js980087a. [DOI] [PubMed] [Google Scholar]
- 5.Boussif O, Lezoualch F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr JP. Proc Natl Acad Sci USA. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Behr JP. Acc Chem Res. 1993;26:274–278. [Google Scholar]
- 7.Luo D, Saltzman WM. Nat Biotechnol. 2000;18:33–37. doi: 10.1038/71889. [DOI] [PubMed] [Google Scholar]
- 8.Mintzer MA, Simanek EE. Chem Rev. 2009;109:259–302. doi: 10.1021/cr800409e. [DOI] [PubMed] [Google Scholar]
- 9.Verma IM, Somia N. Nature. 1997;389:239–242. doi: 10.1038/38410. [DOI] [PubMed] [Google Scholar]
- 10.Bharali DJ, Klejbor I, Stachowiak EK, Dutta P, Roy I, Kaur N, Bergey EJ, Prasad PN, Stachowiak MK. Proc Natl Acad Sci USA. 2005;102:11539–11544. doi: 10.1073/pnas.0504926102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li PC, Li D, Zhang LX, Li GP, Wang EK. Biomaterials. 2008;29:3617–3624. doi: 10.1016/j.biomaterials.2008.05.020. [DOI] [PubMed] [Google Scholar]
- 12.Rosi NL, Giljohann DA, Thaxton CS, Lytton-Jean AKR, Han MS, Mirkin CA. Science. 2006;312:1027–1030. doi: 10.1126/science.1125559. [DOI] [PubMed] [Google Scholar]
- 13.Gao X, Huang L. Gene Therapy. 1995;2:710–722. [PubMed] [Google Scholar]
- 14.Torchilin VP. Nat Rev Drug Disc. 2005;4:145–160. doi: 10.1038/nrd1632. [DOI] [PubMed] [Google Scholar]
- 15.Matsui K, Sando S, Sera T, Aoyama Y, Sasaki Y, Komatsu T, Terashima T, Kikuchi J. J Am Chem Soc. 2006;128:3114–3115. doi: 10.1021/ja058016i. [DOI] [PubMed] [Google Scholar]
- 16.Zuber G, Zammut-Italiano L, Dauty E, Behr JP. Angew Chem, Int Ed. 2003;42:2666–2669. doi: 10.1002/anie.200250446. [DOI] [PubMed] [Google Scholar]
- 17.Liu J, Stace-Naughton A, Jiang X, Brinker CJ. J Am Chem Soc. 2009;131:1354–1355. doi: 10.1021/ja808018y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu J, Jiang X, Ashley C, Brinker CJ. J Am Chem Soc. 2009;131:7567–7569. doi: 10.1021/ja902039y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang LF, Granick S. Nano Lett. 2006;6:694–698. doi: 10.1021/nl052455y. [DOI] [PubMed] [Google Scholar]
- 20.Luo D, Saltzman WM. Nat Biotechnol. 2000;18:893–895. doi: 10.1038/78523. [DOI] [PubMed] [Google Scholar]
- 21.Templeton NS, Lasic DD, Frederik PM, Strey HH, Roberts DD, Pavlakis GN. Nat Biotechnol. 1997;15:647–652. doi: 10.1038/nbt0797-647. [DOI] [PubMed] [Google Scholar]
- 22.Xu L, Anchordoquy TJ. Biochim Biophys Acta-Biomembranes. 2008;1778:2177–2181. doi: 10.1016/j.bbamem.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 23.Zhu L, Lu Y, Miller DD, Mahato RI. Bioconjugate Chem. 2008;19:2499–2512. doi: 10.1021/bc8004039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He XX, Wang KM, Tan WH, Liu B, Lin X, He CM, Li D, Huang SS, Li J. J Am Chem Soc. 2003;125:7168–7169. doi: 10.1021/ja034450d. [DOI] [PubMed] [Google Scholar]
- 25.Radu DR, Lai CY, Jeftinija K, Rowe EW, Jeftinija S, Lin VSY. J Am Chem Soc. 2004;126:13216–13217. doi: 10.1021/ja046275m. [DOI] [PubMed] [Google Scholar]
- 26.Lu J, Liong M, Zink JI, Tamanoi F. Small. 2007;3:1341–1346. doi: 10.1002/smll.200700005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.