Abstract
Kinetochore attachment to spindle microtubule plus-ends is necessary for accurate chromosome segregation during cell division in all eukaryotes. The centromeric DNA of each chromosome is linked to microtubule plus-ends by eight structural-protein complexes1–9. Knowing the copy number of each of these complexes at one kinetochore–microtubule attachment site is necessary to understand the molecular architecture of the complex, and to elucidate the mechanisms underlying kinetochore function. We have counted, with molecular accuracy, the number of structural protein complexes in a single kinetochore–microtubule attachment using quantitative fluorescence microscopy of GFP-tagged kinetochore proteins in the budding yeast Saccharomyces cerevisiae. We find that relative to the two Cse4p molecules in the centromeric histone1, the copy number ranges from one or two for inner kinetochore proteins such as Mif2p2, to 16 for the DAM–DASH complex8,9 at the kinetochore–microtubule interface. These counts allow us to visualize the overall arrangement of a kinetochore–microtubule attachment. As most of the budding yeast kinetochore proteins have homologues in higher eukaryotes, including humans, this molecular arrangement is likely to be replicated in more complex kinetochores that have multiple microtubule attachments.
Accurate segregation of sister chromosomes during mitosis depends on the assembly of structural proteins at the kinetochore that link spindle microtubule plus-ends to centromeric DNA (CEN DNA). The structural arrangement of these proteins within the kinetochore underlies its function in force generation. It may also influence how the spindle assembly checkpoint senses kinetochore–microtubule attachment, and how errors in attachment are corrected to prevent chromosome mis-segregation. Although serial-section transmission electron microscopy has revealed the overall three-dimensional architecture of vertebrate kinetochores, the structure of individual kinetochore–microtubule attachment remains poorly characterized. Consequently, a mechanistic model of kinetochore function that integrates the details of its structure cannot currently be constructed. To understand the molecular architecture of a kinetochore–microtubule attachment site, we focused on counting the copy number for the core structural kinetochore proteins and protein complexes that are necessary for stable kinetochore–microtubule attachment.
Vertebrate kinetochores have a complex and dynamic structure with multiple attachment sites for kinetochore microtubules (about 20–25 in humans). In contrast, each kinetochore in budding yeast makes only one stable microtubule attachment during metaphase, suggesting that its molecular composition may be stable and making it an ideal organism for investigating kinetochore structure. There are more than 60 known kinetochore proteins in budding yeast and homologues for most of these proteins have been identified in vertebrate systems10,11. Moreover, kinetochore function in metaphase chromosome alignment and segregation in anaphase is similar in budding yeast and higher eukaryotes12. Therefore, insight into the molecular architecture of the budding yeast kinetochore will be valuable for understanding the kinetochore–microtubule interaction in higher eukaryotes, including humans.
The DNA and protein composition of the budding-yeast kinetochore is well-understood. The kinetochore is built on a 125 base-pair stretch of CEN DNA that is composed of three distinct regions, CDE I, II and III. It is wrapped around a single nucleosome containing a centromere-specific histone H3 variant1,13 named Cse4p (the human homologue is hsCENP-A). Proximal to this nucleosome is the CBF3 complex3 of four proteins that includes Ndc10p and Cep3p. The CBF3 complex specifically binds to the CDE III region of the CEN DNA. Another protein, Mif2p (hsCENP-C), binds to the CDE II region of CEN DNA. Next to this is the COMA complex of four proteins4 including Okp1p (hsCENP-F) and Ctf19p. Recently, Spc105p was confirmed as a kinetochore protein5. The homologue of Spc105p in Caenorhabditis elegans, KNL-1 (the uncharacterized human homologue is AF15q14), is necessary for microtubule attachment. Spc105p associates with the MIND complex14, which contains Mtw1p (hsMis12). The COMA complex also recruits the non-essential protein Ctf3p (hsCENP-I). In vertebrates, both CENP-I and Mis12 are required to recruit the outer kinetochore complex NDC80 (ref. 10). The NDC80 complex is a rod-like molecule that is approximately 50 nm long, and has globular ends7. It contains four proteins including Ndc80p (hsHec1) and Nuf2p (hsNuf2). Localization of antibodies to Ndc80 in vertebrate cells suggests that the Ndc80p–Nuf2p end of the NDC80 complex localizes proximal to the microtubule attachment site, whereas the other end localizes proximal to the inner centromere7,15. In budding yeast, the NDC80 complex and the microtubule associated protein complex, DAM–DASH, are both necessary for microtubule attachment10,11. The DAM–DASH complex is a heterodecamer and contains the protein Ask1p. Purified DAM–DASH complexes assemble into rings around microtubules in vitro8,9, which can slide passively along the microtubule lattice8.
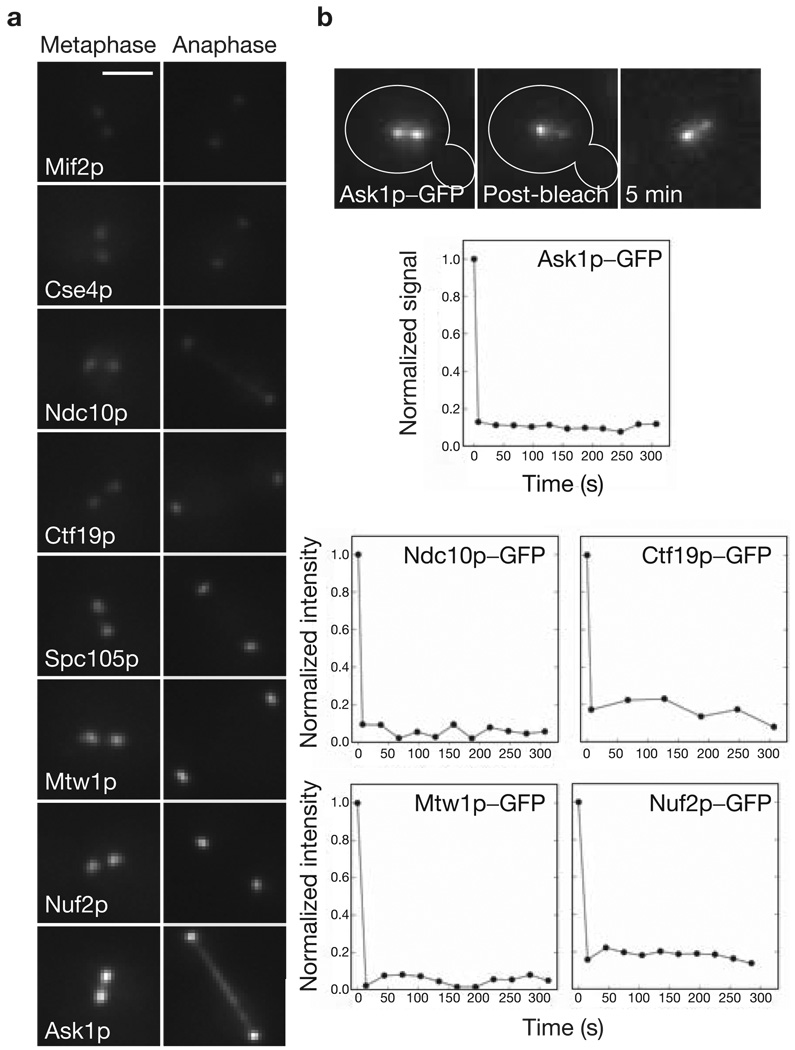
There are important practical advantages in using budding yeast to obtain accurate protein counts using fluorescence microscopy. A protein of interest can be tagged at the carboxyl (C)-terminus with GFP at its chromosomal locus. The GFP-fluorescence signal is then a direct readout of the copy number for that protein16. The geometry of the budding-yeast spindle simplifies the quantification of the fluorescence signal from a GFP-tagged kinetochore protein. In metaphase, the sister kinetochores on the 16 chromosomes become arranged into two distinct clusters of sub-resolution size on either side of the spindle equator along the spindle axis (Fig. 1a). During anaphase, the kinetochore clusters move close to the spindle poles (anaphase A) as the spindle elongates (anaphase B) to push the poles apart (Fig. 1a). The core structural proteins are concentrated exclusively at kinetochores in metaphase and anaphase, with the exception of the CBF3 and DAM–DASH complexes. In anaphase, components of the latter two complexes partially dissociate from the kinetochore, and also associate with the spindle microtubules (Fig. 1a). Quantification of the fluorescence signal for a GFP-tagged protein in each kinetochore cluster provides the cumulative signal for 16 kinetochores, which can be used to measure the average number of molecules of that protein per kinetochore.
Figure 1.
Localization and turnover of kinetochore proteins in metaphase and in anaphase–telophase. (a) Localization of representative GFP-tagged kinetochore proteins during metaphase and anaphase–telophase. At metaphase, the sister kinetochores become aligned on either side of the spindle equator into two distinct clusters each containing 16 kinetochores. Note that both CBF3 and the DAM–DASH complex also localize to the spindle in anaphase. The scale bar represents 2 µm. (b) Pre- and post-photobleaching images of a metaphase cell expressing Ask1p–GFP. Signal recovery (shown in the relative intensity versus time graphs) is undetectable for at least 300 s. Recovery is similarly low for representative proteins from four other complexes.
The centromeric histone Cse4p is a core component of the kinetochore. Cse4p shows virtually no turnover within a kinetochore cluster in metaphase, either through dissociation or through kinetochore movement from one spindle half to the other17. To test whether other structural kinetochore complexes are also similarly stable, fluorescence recovery after photobleaching (FRAP) was measured for representative GFP-tagged proteins from the CBF3, COMA, MIND, NDC80 and DAM–DASH complexes. One of the two sister kinetochore clusters was photobleached in both metaphase and anaphase cells (Fig. 1b, see Supplementary Information, Note 1 and Table S1). The recovery was found to be undetectably low (< 5%) in all cases. This low recovery is indicative of high protein stability at the kinetochore. FRAP measurements for CENP-I, CENP-H and Nuf2 in vertebrate cells show that these proteins are similarly stable18,19. These observations demonstrate that the core protein linkage between the CEN DNA and microtubule plus-ends is stable in metaphase and anaphase.
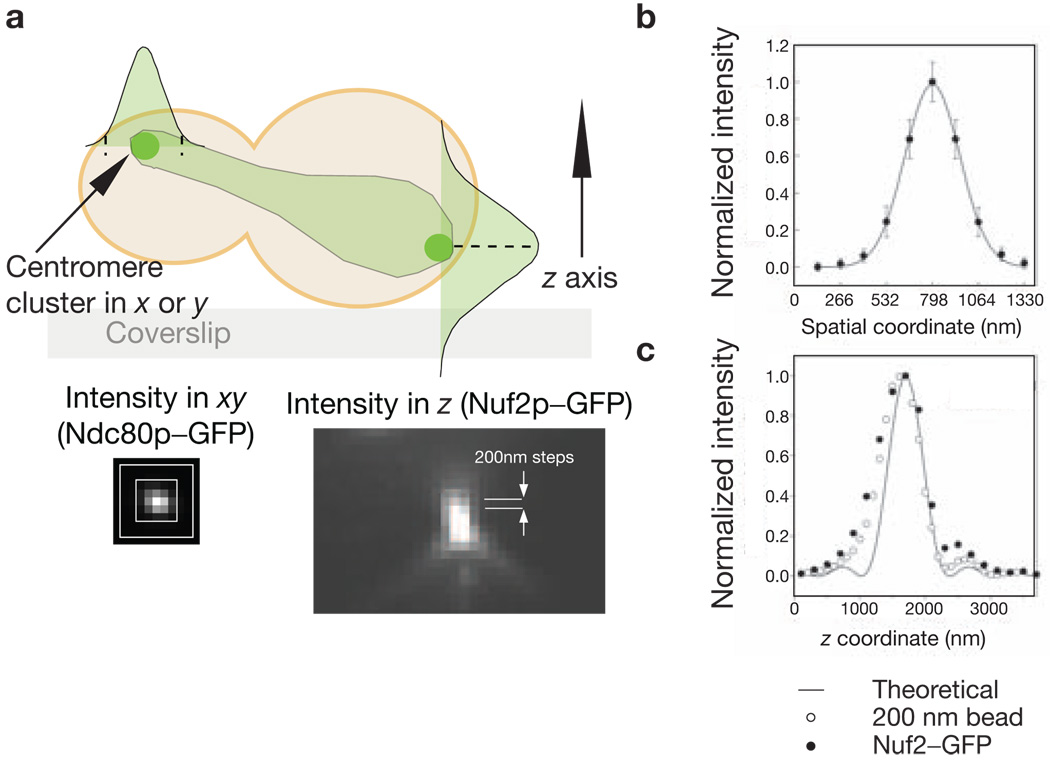
A comparative approach was used to count the number of molecules of a specific protein at the kinetochore. There are two Cse4p molecules in the centromeric nucleosome1,13 (see Supplementary Information, Note 2). The ratio of the average fluorescence signal for a kinetochore protein to the signal for Cse4p–GFP was multiplied by two to yield the average copy number per kinetochore for that protein. In each experiment, cells of two different strains — one expressing a GFP-tagged protein of interest and the other expressing Cse4p–GFP — were mixed together and immobilized on a coverslip for imaging (see Methods). An image stack, with a 200 nm separation along the z axis between consecutive images, was then obtained for each cell (Fig. 2a). The maxima of the intensity distribution of each kinetochore cluster along the z axis was determined with an average underestimation of 4% (Fig. 2c, see Supplementary Information, Note 3). At this maxima, the fluorescence was integrated over a 5 × 5 pixel (for anaphase–telophase cells) or 6 × 6 pixel (for metaphase cells) region. The dimensions of the signal and the background region are dictated by the spread of the fluorescence intensity for a kinetochore cluster, which can be characterized by fitting the spatial intensity distribution with a Gaussian curve (Fig. 2b, see Supplementary Information, Note 4). The total signal was then obtained by integration over the intensity distribution using the 2σ limit (where σ = s.d. for the Gaussian curve), after subtracting the background signal. Obtaining the ratio of the average signal values for the GFP-tagged protein of interest and Cse4p–GFP in each experiment avoids a direct evaluation of the in vivo fluorescence signal for one GFP molecule. This method also minimizes measurement errors.
Figure 2.
Characterization of the intensity distribution of a kinetochore cluster. (a) A schematic representation of a budding yeast cell expressing a GFP-tagged kinetochore protein in anaphase–telophase is shown. Signal was measured by integrating the signal intensity in the xy direction (shown for Ndc80p–GFP) in the plane that contains the maximum intensity pixel along the z axis (shown for Nuf2p–GFP). (b) Fitting a Gaussian function to the anaphase–telophase intensity distribution in the xy plane for Nuf2p–GFP yields σ = 159 nm (s.d. for the Gaussian curve). Similar measurement for metaphase xy intensity distribution yields σ = 189 nm. (c) The intensity distribution along the z axis for a Nuf2p-GFP cluster and a 200 nm green fluorescent bead. The solid line represents the theoretical intensity distribution along the z axis.
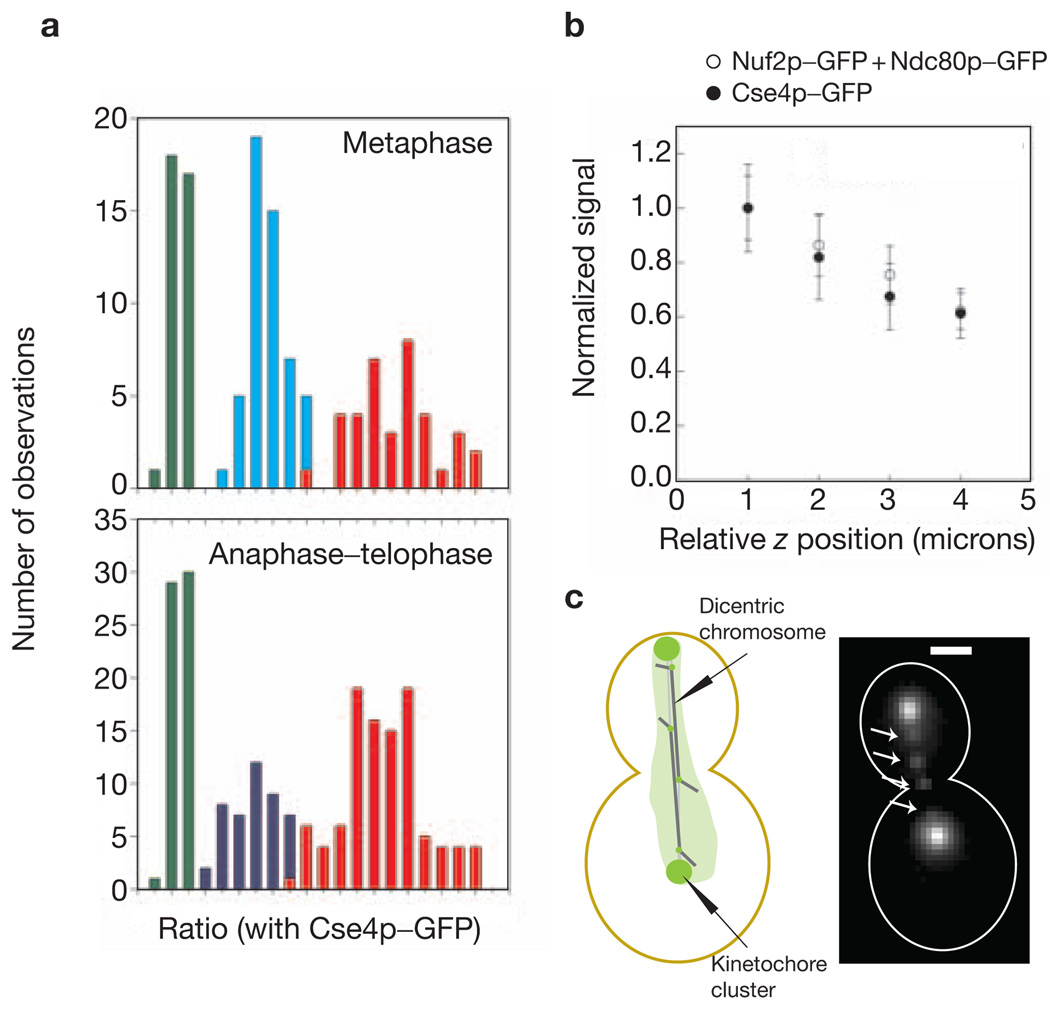
To test system linearity and accuracy, the metaphase and anaphase–telophase signals were evaluated for three strains expressing Nuf2p–GFP, Ndc80p–GFP or Nuf2p–GFP + Ndc80p–GFP. These measurements confirmed the 1:1 stoichiometry of Ndc80p and Nuf2p in the NDC80 complex7. As expected, the Nuf2–GFP + Ndc80p–GFP signal was twice the signal for either Nuf2p–GFP or Ndc80p–GFP alone (Fig. 3a). The variation in protein number per kinetochore is given by the s.d. of the average signal when the contribution of experimental errors is minimal. As the signal and the background can both be measured accurately in anaphase–telophase cells, the anaphase–telophase data was used for this analysis. It was found that the measured s.d. in anaphase–telophase cells was dominated by the signal loss due to spherical aberrations with increasing depth of the kinetochore cluster from the coverslip surface (Fig. 3b). Therefore, the difference in measured signal was evaluated for two kinetochore clusters in the same cell that were separated by 600 nm or less along the z axis. The average value for this difference was approximately 10% of the average signal, suggesting that the variation in the protein number is less than one molecule per kinetochore (see Supplementary Information, Note 5). These results also apply to metaphase cells, as the protein complexes are stably anchored at the kinetochore in metaphase and anaphase.
Figure 3.
Linearity and sensitivity of the measurement technique. (a) A frequency histogram for signal measurements in metaphase and anaphase–telophase for Cse4p–GFP (green), Nuf2p–GFP (cyan), Ndc80p–GFP (blue) and Nuf2p–GFP + Ndc80p–GFP (red). The proportional increase in the fluorescence intensity for Nuf2p–GFP + Ndc80p–GFP also demonstrates that the proximity of fluorophores does not detectably affect their fluorescence. (b) Normalized signal plotted as a function of the relative z coordinate of the kinetochore clusters of Cse4p–GFP and Nuf2p–GFP + Ndc80p–GFP. The error bars represent the s.d. of the mean signal value in each bin. (c) Four lagging kinetochores (arrows, Nuf2p–GFP) on the two dicentric chromosomes in a mid-anaphase cell. The chromatin between the two centromeres is 40 kb long. The scale bar represents 2 µm.
To image single kinetochores, we used cells carrying a conditional dicentric chromosome (containing one conditional centromere in addition to the wild-type centromere20) and expressing Nuf2p–GFP. The induction of the conditional centromere produces one or two lagging chromatids in mid-anaphase, with their kinetochores visible as separated fluorescence foci along the spindle axis (Fig. 3c). The ratio of signal for the nearest kinetochore cluster to the signal from these foci was found to be 16:1 ± 2, thus verifying that these foci are single kinetochores. These measurements demonstrate that accurate measurement of as few as seven closely clustered GFP molecules is possible in vivo (seven being the anaphase number of Nuf2p molecules per kinetochore; see Table 1). More importantly, these measurements validate our method of calculating the average number of proteins per kinetochore based on the cumulative fluorescence of 16 kinetochores in a cluster.
Table 1.
Metaphase and anaphase–telophase ratio measurements
| Complex | Protein | Vertebrate homologue | Metaphase ratio | Anaphase ratio | Metaphase number | Anaphase number |
|---|---|---|---|---|---|---|
| Nucleosome | Cse4p | hsCENP-A | 1 | 1 | 2 | 2 |
| CBF3 | Ndc10p | – | 1.9 ± 0.2 | 1.3 ± 0.01 | 4 | 2–3 |
| CBF3 | Cep3p | – | 0.9 ± 0.2 | 0.6 ± 0.01 | 2 | 1–2 |
| – | Mif2p | hsCENP-C | 5.4 ± 0.4* | 5.5 ± 0.10* | 1–2 | 1–2 |
| COMA | Ctf19p | hsCENP-F | 3.4 ± 0.3* | 3.4 ± 0.20* | 3 | 2 |
| – | Spc105 | CeKNL-1 | 2.4 ± 0.01 | 2.4 ± 0.01 | 5 | 5 |
| MIND | Mtw1p | hsMis12 | 3.3 ± 0.2 | 2.4 ± 0.10 | 6–7 | 4–5 |
| NDC80 | Nuf2p | hsNuf2 | 4.0 ± 0.2 | 3.6 ± 0.20 | 8 | 7 |
| DAM–DASH | Ask1p | – | 9.0 ± 1 | 5.3 ± 0.30 | 16–20 | 10–11 |
| CTF3 | Ctf3p | hsCENP-I | – | 0.5 ± 0.01 | – | 1 |
| CHL4–IML3 | Chl4p | – | – | 0.26 ± 0.01 | – | <1 |
| NKP1–NKP2 | Nkp2p | – | – | 6.1 ± 0.05* | – | 1 |
The ratios shown are the average ratios obtained from three experiments with at least 20 measurements for metaphase cells, and at least two experiments with up to 80 measurements for late anaphase–telophase cells. The coefficient of variation (s.d. / mean) was better than 0.26 in all the measurements with the exception of Cep3p, for which the coefficient is 0.5. The asterisks indicate that the reported ratio is (Nuf2p – signal) : (protein signal). Mif2p–GFP, Ctf19p–GFP and Nkp2–GFP measurements were carried out with Nuf2p–GFP as the reference signal (see Methods).
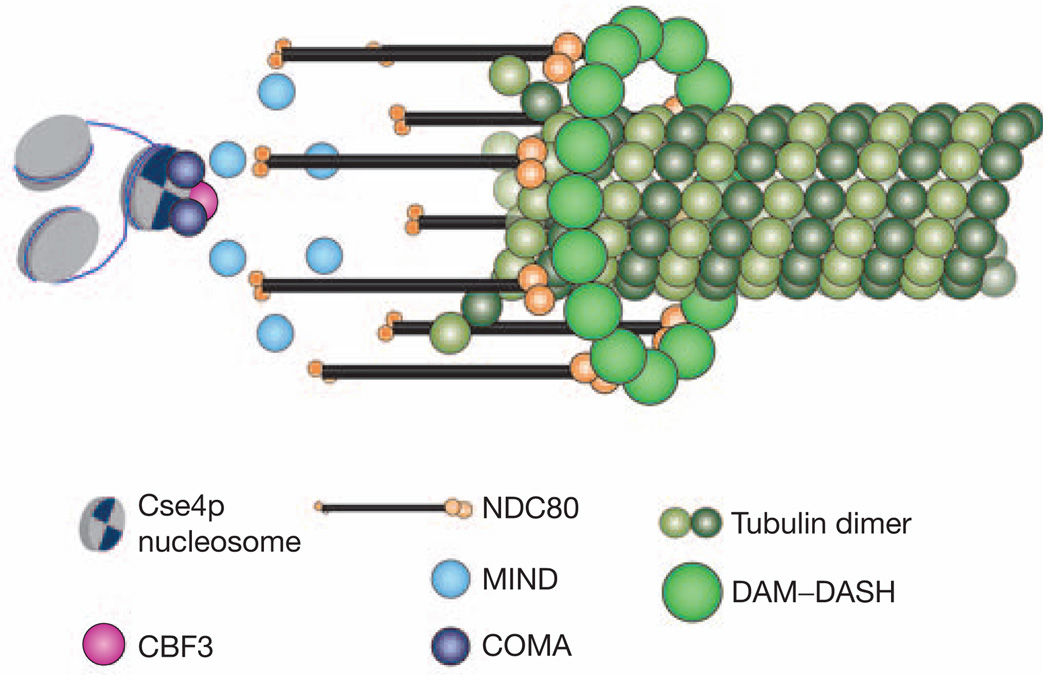
Table 1 lists the observed counts for the average protein number per kinetochore for representative proteins from each protein complex. A possible arrangement of the essential structural protein complexes at the budding yeast kinetochore in metaphase, based on these protein numbers, and the structures of the DAM–DASH complex and the NDC80 complex7–9,21, is shown in Fig. 4. The protein linkage between the CEN DNA and a microtubule plus-end begins with the centromeric nucleosome incorporating two Cse4p molecules. Next is the CBF3 complex that incorporates one dimer each of Ndc10p and Cep3p. The counts for Ndc10p and Cep3p (four and two, respectively; see Supplementary Information, Note 6) together show that there is only one CBF3 complex per kinetochore22. It is likely that the extra Ndc10p dimer binds to the CDE II region of CEN DNA, independent of its inclusion in the CBF3 complex, as suggested by in vitro experiements23. The count for Cep3p also supports the inclusion of only two Cse4p molecules per kinetochore. The average fluorescence signal for Mif2p shows that each kinetochore has at least one Mif2p molecule, although some kinetochores carry two. Members of the COMA complex show interactions with Cse4p in two-hybrid assays4,24, along with a genetic interaction between Ctf19p and the amino (N) terminus of Cse4p24. The low copy number (1–2) for these protein complexes supports the presence of a network of inner kinetochore proteins4, suggested by genetic and biochemical interactions between members of these protein complexes.
Figure 4.
A schematic representation of the possible architecture of a kinetochore–microtubule attachment site in metaphase. This structure is based on the protein counts reported here, and on the in vitro structures of the DAM–DASH and the NDC80 complex. It depicts only five essential kinetochore protein complexes.
Five molecules of Spc105p were found. This protein immunoprecipitates with members of the MIND complex5, of which there are six or seven copies. The number of Mtw1p molecules is also close to the number of NDC80 complex molecules (eight). In vertebrate cells, the NDC80 complex is arranged with the Ndc80 N-terminal domain within the outer kinetochore, where microtubule plus ends are located15. Although a link between the NDC80 and the DAM–DASH complexes has not been directly established, there is indirect evidence for interactions between these complexes in the form of in vitro binding between Ndc80p and Dam1p25, and two-hybrid interactions between Ndc80p and DAM–DASH complex members (Dam1p and Spc19p)26. Moreover, both of these complexes are necessary for end-on microtubule attachments. The two-headed N-termini of the NDC80 molecules are therefore positioned symmetrically around an oligomeric DAM–DASH complex ring in Fig. 4.
The metaphase count for the DAM–DASH complex (16–20 copies) reported here agrees well with the number of molecules required to form one DAM–DASH ring around a microtubule in vitro8. Biochemical estimation of the number of DAM–DASH complexes in a cell also suggests that there are not many more molecules than those needed to form one ring per kinetochore microtubule9. Because DAM–DASH is a microtubule associated protein complex, the DAM–DASH complex may also localize along the microtubule outside of the kinetochore. However, the high stability of this complex within a kinetochore cluster, in comparison with the rapid tubulin turnover within spindle microtubules (half life = 60 s)27, suggests that most of the DAM–DASH complex molecules must bind stably at the kinetochores (see Supplementary Information, Note 7). The metaphase and late anaphase–telophase counts for all the kinetochore complexes are similar, with the exception of DAM–DASH and Ndc10p, both of which also localize to the spindle in anaphase (Fig. 1a). Interestingly, the copy numbers for the non-essential proteins11 (Ctf3p, Chl4p and Nkp2p) show that these complexes exist in minimal copy number (one per kinetochore). Preliminary measurements show that the metaphase signal for these proteins is similar to their respective anaphase–telophase signal.
This study demonstrates that the kinetochore–microtubule attachment site is built from a low and standardized number of copies of structural protein complexes. Previous studies of the structure of vertebrate kinetochores with multiple microtubule attachment sites indicate that it is constructed from a number of identical units spread along the centromere28. This repeat-subunit structure of the kinetochore may be attained by replicating individual microtubule attachment sites, similar to the one at the budding yeast kinetochore. The molecular counts allow us to visualize possible arrangements of the structural proteins within the kinetochore. Kinetochore structure influences its function in microtubule polymerization–depolymerization-coupled force generation, in regulation of attachment stability based on mechanical cues such as tension, and in communicating attachment status to the spindle-assembly checkpoint. The combination of previous biochemical and genetic analyses with the molecular counts for structural proteins reported here provides the foundation for building a mechanistic model of these kinetochore functions.
METHODS
Yeast strains and growth conditions
All strains were constructed in the YEF473A background unless otherwise noted (see Supplementary Information, Table S5). GFP fusions were made by PCR amplification of a GFP–KANr cassette (from pFA6a–GFP(S65T) KANr MX6) flanked with 60 base pairs of homology to the site of integration at the 3′ end of the gene29. Cells were grown in YPD at 25 °C to mid-log phase before imaging. For induction of the conditional centromere on the dicentric chromosome, a mid-log phase culture grown on galactose media was shifted to glucose media for 2 h before imaging. For imaging, cells were suspended in filter sterile synthetic dextrose media, and immobilized on standard glass coverslips coated with 0.5 mg ml−1 of concanavalin A (cat. no. C7275, Sigma).
Image acquisition and data analysis
Image acquisition was carried out on an Eclipse TE2000-U (Nikon, Melville, NY) microscope with a 1.4 NA, 100× DIC oil immersion lens and the standard yEGFP filter set from Chroma (Rockingham, VT). Images were acquired with an ORCA ER cooled CCD camera (Hamamatsu Photonics, Bridgewater, NY) with 2 × 2 binning and using MetaMorph (Molecular Devices, Downingtown, PA) as the image acquisition software. A 300 × 300 pixel-wide region in the middle of the field of view of the microscope was acquired to minimize the non-uniformities in the illumination field. Twenty one z sections were acquired through each cell by stepping the stage in 200 nm steps with respect to the objective. A 400 ms integration time was used for all the measurements. Cells belonging to the two strains (the protein of interest and Cse4p–GFP) were differentiated based on significant differences in the fluorescence of their kinetochore clusters. This was not possible in the case of Mif2p–GFP and Ctf19p–GFP when using Cse4p–GFP in the reference strain. Therefore, a reference strain containing Nuf2p–GFP was used for these two proteins.
Data analysis was carried out in MatLAB (Mathworks Inc., Natick, MA) using a custom written graphical user interface. The plane with the maximum intensity pixel was found in a 10 × 10 pixel user-selected region in the 21 z-plane stack. In this plane, the signal was computed by integrating the signal intensity in a 5 × 5 pixel square centred on the maximum intensity pixel for telophase measurements. A 6 × 6-pixel square was used for metaphase measurements because of the larger size of the signal. For anaphase–telophase cells, a larger concentric box of the appropriate dimension was used to calculate the background. The proximity of the two kinetochore clusters in metaphase cells prevented the use of this method. Therefore, the background region was chosen manually by drawing a 6 × 6-pixel box in the vicinity of the spindle inside the cell.
FRAP measurements
FRAP measurements were carried out on metaphase and anaphase cells expressing Ndc10p–GFP, Ctf19p–GFP, Mtw1p–GFP, Nuf2p–GFP and Ask1–GFP as previously described 30.
Supplementary Material
ACKNOWLEDGEMENTS
We thank A. Hunt, D. Odde, S. Inoué, and members of the Salmon and Bloom laboratory for helpful comments on the manuscript. This work was supported by National Institutes of Health (NIH) grants to K.S.B. (GM32238), and to E.D.S. (GM24364 and GM60678).
Footnotes
Note: Supplementary Information is available on the Nature Cell Biology website.
COMPETING FINANCIAL INTERESTS
The authors declare that they have no competing financial interests.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/
References
- 1.Meluh PB, Yang P, Glowczewski L, Koshland D, Smith MM. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell. 1998;94:607–613. doi: 10.1016/s0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- 2.Meluh PB, Koshland D. Evidence that the MIF2 gene of Saccharomyces cerevisiae encodes a centromere protein with homology to the mammalian centromere protein CENP-C. Mol. Biol. Cell. 1995;6:793–807. doi: 10.1091/mbc.6.7.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Espelin CW, Kaplan KB, Sorger PK. Probing the architecture of a simple kinetochore using DNA–protein crosslinking. J. Cell Biol. 1997;139:1383–1396. doi: 10.1083/jcb.139.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ortiz J, Stemmann O, Rank S, Lechner J. A putative protein complex consisting of Ctf19, Mcm21, and Okp1 represents a missing link in the budding yeast kinetochore. Genes Dev. 1999;13:1140–1155. doi: 10.1101/gad.13.9.1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nekrasov VS, Smith MA, Peak-Chew S, Kilmartin JV. Interactions between centromere complexes in Saccharomyces cerevisiae. Mol. Biol. Cell. 2003;14:4931–4946. doi: 10.1091/mbc.E03-06-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Euskirchen GM. Nnf1p, Dsn1p, Mtw1p, and Nsl1p: a new group of proteins important for chromosome segregation in Saccharomyces cerevisiae. Eukaryot. Cell. 2002;1:229–240. doi: 10.1128/EC.1.2.229-240.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wei RR, Sorger PK, Harrison SC. Molecular organization of the Ndc80 complex, an essential kinetochore component. Proc. Natl Acad. Sci. USA. 2005;102:5363–5367. doi: 10.1073/pnas.0501168102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Westermann S, et al. The Dam1 kinetochore ring complex moves processively on depolymerizing microtubule ends. Nature. 2006;440:565–569. doi: 10.1038/nature04409. [DOI] [PubMed] [Google Scholar]
- 9.Miranda JJ, De Wulf P, Sorger PK, Harrison SC. The yeast DASH complex forms closed rings on microtubules. Nature Struct. Mol. Biol. 2005;12:138–143. doi: 10.1038/nsmb896. [DOI] [PubMed] [Google Scholar]
- 10.Chan GK, Liu ST, Yen TJ. Kinetochore structure and function. Trends Cell Biol. 2005;15:589–598. doi: 10.1016/j.tcb.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 11.McAinsh AD, Tytell JD, Sorger PK. Structure, function, and regulation of budding yeast kinetochores. Annu. Rev. Cell Dev. Biol. 2003;19:519–539. doi: 10.1146/annurev.cellbio.19.111301.155607. [DOI] [PubMed] [Google Scholar]
- 12.Pearson CG, Maddox PS, Salmon ED, Bloom K. Budding yeast chromosome structure and dynamics during mitosis. J. Cell Biol. 2001;152:1255–1266. doi: 10.1083/jcb.152.6.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collins KA, Furuyama S, Biggins S. Proteolysis contributes to the exclusive centromere localization of the yeast Cse4/CENP-A histone H3 variant. Curr. Biol. 2004;14:1968–1972. doi: 10.1016/j.cub.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 14.De Wulf P, McAinsh AD, Sorger PK. Hierarchical assembly of the budding yeast kinetochore from multiple subcomplexes. Genes Dev. 2003;17:2902–2921. doi: 10.1101/gad.1144403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeLuca JG, et al. Hec1 and nuf2 are core components of the kinetochore outer plate essential for organizing microtubule attachment sites. Mol. Biol. Cell. 2005;16:519–531. doi: 10.1091/mbc.E04-09-0852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu JQ, Pollard TD. Counting cytokinesis proteins globally and locally in fission yeast. Science. 2005;310:310–314. doi: 10.1126/science.1113230. [DOI] [PubMed] [Google Scholar]
- 17.Pearson CG, et al. Stable kinetochore–microtubule attachment constrains centromere positioning in metaphase. Curr. Biol. 2004;14:1962–1967. doi: 10.1016/j.cub.2004.09.086. [DOI] [PubMed] [Google Scholar]
- 18.Nishihashi A, et al. CENP-I is essential for centromere function in vertebrate cells. Dev. Cell. 2002;2:463–476. doi: 10.1016/s1534-5807(02)00144-2. [DOI] [PubMed] [Google Scholar]
- 19.Hori T, Haraguchi T, Hiraoka Y, Kimura H, Fukagawa T. Dynamic behavior of Nuf2-Hec1 complex that localizes to the centrosome and centromere and is essential for mitotic progression in vertebrate cells. J. Cell Sci. 2003;116:3347–3362. doi: 10.1242/jcs.00645. [DOI] [PubMed] [Google Scholar]
- 20.Yang SS, Yeh E, Salmon ED, Bloom K. Identification of a mid-anaphase checkpoint in budding yeast. J. Cell Biol. 1997;136:345–354. doi: 10.1083/jcb.136.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ciferri C, et al. Architecture of the human ndc80-hec1 complex, a critical constituent of the outer kinetochore. J. Biol. Chem. 2005;280:29088–29095. doi: 10.1074/jbc.M504070200. [DOI] [PubMed] [Google Scholar]
- 22.Russell ID, Grancell AS, Sorger PK. The unstable F-box protein p58–Ctf13 forms the structural core of the CBF3 kinetochore complex. J. Cell Biol. 1999;145:933–950. doi: 10.1083/jcb.145.5.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Espelin CW, Simons KT, Harrison SC, Sorger PK. Binding of the essential Saccharomyces cerevisiae kinetochore protein Ndc10p to CDEII. Mol. Biol. Cell. 2003;14:4557–4568. doi: 10.1091/mbc.E02-08-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Y, et al. The N terminus of the centromere H3-like protein Cse4p performs an essential function distinct from that of the histone fold domain. Mol. Cell Biol. 2000;20:7037–7048. doi: 10.1128/mcb.20.18.7037-7048.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shang C, et al. Kinetochore protein interactions and their regulation by the Aurora kinase Ipl1p. Mol. Biol. Cell. 2003;14:3342–3355. doi: 10.1091/mbc.E02-11-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uetz P, et al. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 27.Maddox PS, Bloom KS, Salmon ED. The polarity and dynamics of microtubule assembly in the budding yeast Saccharomyces cerevisiae. Nature Cell Biol. 2000;2:36–41. doi: 10.1038/71357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zinkowski RP, Meyne J, Brinkley BR. The centromere–kinetochore complex: a repeat subunit model. J. Cell Biol. 1991;113:1091–1110. doi: 10.1083/jcb.113.5.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Longtine MS, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 30.Roumanie O, et al. Rho GTPase regulation of exocytosis in yeast is independent of GTP hydrolysis and polarization of the exocyst complex. J. Cell Biol. 2005;170:583–594. doi: 10.1083/jcb.200504108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.