Abstract
Surface patterns of single-stranded DNA (ssDNA) consisting of nanoscale lines as thin as 40 nm were fabricated on polymer substrates for nanotechnology and bioaffinity sensing applications. Large scale arrays (with areas up to 4 cm2) of ssDNA “nanolines” were created on streptavidin-coated polymer (PDMS) surfaces by transferring biotinylated ssDNA from a master pattern of gold nanowires attached to a glass substrate. The gold nanowires were first formed on the glass substrate by the process of lithographically patterned nanowire electrodeposition (LPNE), and then “inked” with biotinylated ssDNA by hybridization adsorption to a thiol-modified ssDNA monolayer attached to the gold nanowires. The transferred ssDNA nanolines were capable of hybridizing with ssDNA from solution to form double-stranded DNA (dsDNA) patterns; a combination of fluorescence and AFM measurements were used to characterize the dsDNA nanoline arrays. To demonstrate the utility of these surfaces for biosensing, optical diffraction measurements of the hybridization adsorption of DNA-coated gold nanoparticles onto the ssDNA nanoline arrays were used to detect a specific target sequence of unlabeled ssDNA in solution.
Introduction
There are a multitude of very good reasons for fabricating patterned DNA monolayers on surfaces. For example, patterned DNA monolayers can be used to: (i) create single-stranded DNA (ssDNA) microarrays for DNA diagnostics and detecting gene expression,1 (ii) generate ssDNA patterns for the adsorption of multiple ssDNA-linked antibodies for the detection of protein biomarkers,2 (iii) position DNA molecules at specific distances and orientations for the formation of DNA nanomachines,3, 4 and (iv) deposit ordered DNA surface patterns for the sequence-specific surface arrangement of DNA tiles for DNA computing applications.5, 6 A subset of these applications requires ssDNA monolayer patterns that simultaneously contain nanometer scale features and also span overall length scales of up to centimeters. There are currently only a few serial deposition strategies such as e-beam deposition methods7, 8 or dip-pen nanolithography9 that can make large scale nanopatterns, but the time required for any serial deposition/fabrication process will naturally scale with the overall size of the surface. One potential methodology that does not have this limitation is the “mechanical transfer process” developed by R. M. Crooks et al. to create replicas of DNA microarrays on polymer (PDMS) surfaces from a master pattern of ssDNA attached to a glass substrate. 10–14 A similar transfer process for two gold surfaces was developed by Stellacci et al.15, 16 This method is a very fast, reproducible, and inexpensive process for making multiple copies of large scale ssDNA patterns with features on length scales down to 10 microns. In this paper, we use a modified version of this master-replica transfer methodology of Crooks et al. to create DNA patterns with nanometer-scale features on PDMS substrates by using master surfaces that consist of patterns of gold nanowires on glass substrates. Nanolines of ssDNA as thin as 40 nm can be created on PDMS substrates with total areas as large as 4 cm2. A combination of SEM, fluorescence, and AFM measurements is first used to characterize the DNA nanoline arrays. We then demonstrate that these nanoscale DNA patterns can be used for surface bioaffinity sensing experiments by detecting the simultaneous specific adsorption of ssDNA target molecules and DNA-coated nanoparticles from solution with optical diffraction measurements.
Experimental Section
Materials
3-aminopropyltriethoxysilane (APTES, Aldrich), streptavidin (SAV, Thermo Scientific), amine-reactive biotin labeling reagents with polyethylene oxide spacer (NHS-PEO4-Biotin, Thermo Scientific), N-hydroxysuccinimidyl ester of methoxypoly (ethylene glycol) propionic acid MW 10,000 (mPEG-NHS, Creative PEGWorks), polydimethylsiloxane (PDMS) curing agent and prepolymer (Sylgard 184, Dow Corning), were used as received. 20xSSC (USB corporation) was diluted to 2x. Five HPLC purified ssDNA sequences (Table I) were purchased from Integrated DNA Technologies. A PBS buffer (100 mM Na2HPO4, 0.3 M NaCl, 5 mM MgCl2, 1 mM EDTA, adjusted to pH 7.4) was used for all DNA solution. Millipore water was used throughout.
Table I.
DNA Sequences
| Symbol | Sequence |
|---|---|
| D1-S | 5′-HS-(CH2)6-TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT -3′ |
| D2-B | 5′-Biotin -(CH2)6 - AAA AAA AAA AAA AAA AAA AAA AAA AAA AAA -3′ |
| D1-F | 5′-Cy3 -(CH2)6 - TTT TTT TTT TTT TTT TTT TT-3′ |
| D3-F | 5′-Cy3 -(CH2)6 – ACA CAT AAG CAC GAA CCA CT - 3′ |
| DT | 5′-TGT GTC TGG ATT TCG TTA TGT GTC TGG ATT TTT TTT TTT TTT TTT TTT TT -3′ |
| D4-NP | 5′ - AAT CCA GAC ACA TAA CGA AAT CCA GAC ACA TAA GCA CGA ACC GAA - (CH2)3- SH - 3′ |
Master-Replica Transfer Procedure
A few modifications were made to the DNA microarray replication technique developed by Crooks et al.10–14, 17 The first step is to create a streptavidin coated PDMS surface. The PDMS substrate was cast on polished Si wafers, cured, and then thoroughly cleaned by sonication in ethanol and water. The PDMS surface was then exposed to an O2 plasma at 200–250 mTorr for 30 seconds (from our experience, the PDMS surface is very sensitive to the plasma conditions). The PDMS surface was exposed to HCl vapor by being placed face down above a 12M solution for 1 minute. Immediately following, the PDMS was exposed to APTES vapor for 1 hour in the same manner, however, in a vacuum chamber. The amine-modified PDMS was then treated with a 2 mg/mL solution of NHS-PEO4-Biotin in PBS buffer for 2 hours, and then blocked with a 1 mg/mL mPEG-NHS in PBS buffer for 1 hour. The biotinylated PDMS surface was finally exposed to a 0.1 mg/mL streptavidin solution in 0.1M NaHCO3 buffer (pH8.0) for 30 minutes.
The second step in the master-replica transfer process was to “ink” the gold nanowire array master with biotinylated ssDNA. The gold nanowire master was created by a novel electrodeposition process called “lithographically patterned nanowire electrodeposition” or “LPNE” as described elsewhere.18, 19 The gold nanowire master was first exposed to intense UV light from a mercury arc lamp for 15 minutes then rinsed with ethanol and water to remove any residual photoresist from the LPNE process. The gold nanowire master was first exposed to a 250 μM solution of 5′ thiol-modified T45 oligonucleotide denoted as D1-S (see Table I) for 4 hours, rinsed, gently blown dry with N2 then exposed to a 10 μM 5′-biotinylated A30 oligonucleotide denoted as D2-B for at least 2 hours.
The third step in the master-replica transfer process is to transfer the biotinylated oligonucleotide, D2-B, from the inked gold nanowire master to the streptavidin-coated PDMS substrate. The streptavidin-coated PDMS was thoroughly rinsed with water and gently dried with nitrogen. The inked nanowire master was immersed in 2xSSC for one minute, briefly rinsed with water and gently dried with nitrogen. The streptavidin-coated PDMS surface was placed in contact with the inked gold nanowire master by applying a minimum amount of pressure ~1.8 × 10−3 N cm−2, corresponding to the weight of the PDMS alone. After 15 minutes of contact, the PDMS replica was gently peeled off from the master surface. The resultant ssDNA nanoline array on the PDMS surface was thoroughly rinsed with water.
Fluorescence Measurements
The ssDNA (D2-B) nanoline array on the PDMS surface was exposed to 1 μM solution of complimentary fluorescently tagged ssDNA, D1-F, for 15 minutes, then immersed in 2xSSC for 1 minute and briefly rinse with water. For control experiments, a non-complimentary ssDNA, D3-F was used. Fluorescence images were taken on an inverted fluorescence microscope Olympus IX71 and an Eclipse E800 with an oil emersion lens (60XA/1.40 oil).
SEM and AFM measurements
Scanning electron microscopy (SEM) images were carried out using a Zeiss Ultra 55, operated at 10 keV. All gold nanowire samples were sputter coated before imaging to prevent charging. Atomic force microscopy (AFM) imaging was performed in AC mode in air at ambient pressure and humidity using a MFP-3D AFM (Asylum Research). The AFM tips were silicon with an aluminum coating (AC160TS, Olympus).
Diffraction Measurements
The ssDNA nanoline array on the PDMS surface was exposed to 1 μM solution of ssDNA denoted as DT, and then to a 1 nM solution of ssDNA-coated gold nanoparticles denoted as D4-NP. The detailed procedure of the three sequence hybridization adsorption reaction was as described elsewhere.17 Diffraction measurements were obtained using an optical setup of a HeNe laser, a λ/2 plate, a polarizer, and a monochrome CCD camera as described elsewhere. 20 In TIR geometry, a PDMS prism was coupled to the DNA nanowires PDMS substrate with a refractive index of 1.43.
Results and Discussion
Master-replica transfer process
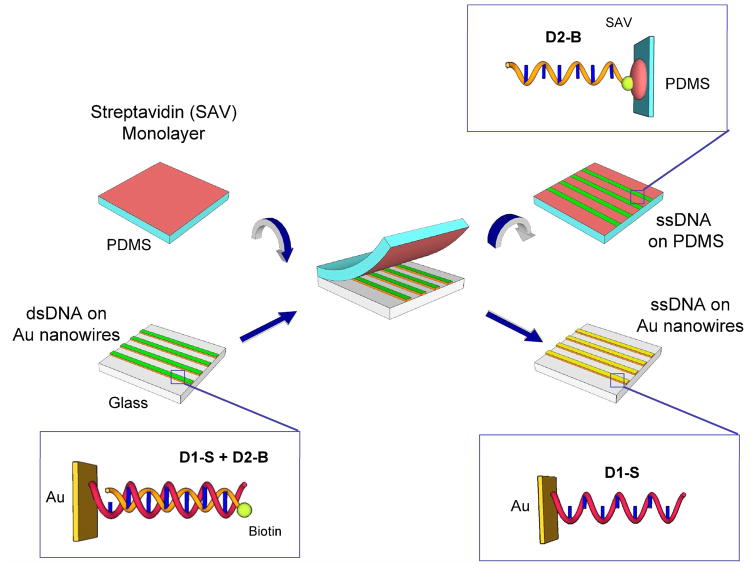
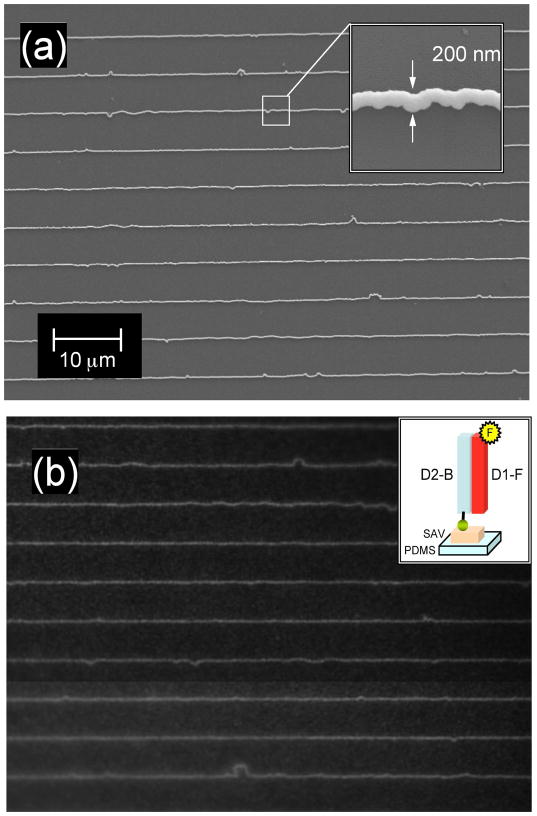
The master-replica transfer process for creating nanoscale ssDNA patterns on PDMS polymer substrates is shown schematically in Figure 1. The first step is to create a large scale master pattern of gold nanowires on a glass substrate. In a series of recent papers,18, 19, 21 we have described a novel electrodeposition process for creating patterns of metallic nanowires that we call “lithographically patterned nanowire electrodeposition” or “LPNE”; the nanowires created by LPNE can range in width from 500 nm down to 11 nm, and the overall LPNE surface pattern can cover areas of up to wafer length scale.21 For example, we have applied the LPNE methodology to create arrays of Au and Pd nanowires with unique optical diffraction properties for biosensing applications.20, 22 LPNE is used in this paper to create an array of gold nanowires that are spaced by approximately 5 microns; the LPNE process actually creates nanowire arrays with a slightly asymmetric alternate spacing that leads to a unique signature in the diffraction experiments.20 Figure 2a shows a representative SEM image of a portion of a 200 nm gold nanowire array (the total area was 1 cm2). The inset in Figure 2a shows a higher magnification SEM image of a gold nanowire in the array; the grain structure of the electrodeposited nanowires has been discussed elsewhere.21 The average nanowire width of the arrays used in this paper was varied from sample to sample in the range of 40 to 250 nm.
Figure 1.
Schematic diagram of the master-replica transfer process for creating nanoscale ssDNA patterns on PDMS polymer substrates. The gold nanowire array master was “inked” by first exposing it to thiol-modified ssDNA, D1-S, then hybridizing with complimentary biotinylated ssDNA, D2-S, to create a dsDNA monolayer on the nanowire surface (bottom left). A streptavidin-modified PDMS substrate (top left) was placed in contact with the inked gold nanowire array with minimum pressure. When peeled apart, D2-B was bound to PDMS replica by the strong biotin-streptavidin interaction (top right), leaving D1-S on the nanowire master (bottom right).
Figure 2.
SEM image of a gold nanowire array master and fluorescence image of DNA nanolines on PDMS replica. (a) A representative large area scan showing a portion of gold nanowire array with average spacing of 5 μm. The inset is a high-magnification image of a single gold nanowire with an average width of 200 nm. (b) A 60x fluorescence image of the PDMS replica surface obtained after the master-replica transfer process and exposure to D1-F, a fluorescently labeled ssDNA complementary to D2-B. The inset is a schematic diagram of the replica surface.
The second step in the master-replica transfer process is to “ink” the gold nanowire array pattern with biotinylated ssDNA molecules that will eventually be transferred to the PDMS substrate. The biotinylated ssDNA molecules are attached to the gold nanowires by hybridization adsorption to form DNA duplexes with a monolayer of a thiol-modified ssDNA that was first adsorbed onto the gold nanowires (see Figure 1: the thiol-modified ssDNA is denoted as D1-S and the complementary biotinylated ssDNA is denoted as D2-B). The sequence and chemical modfications of all DNA molecules used in this paper are listed in Table I. The adsorption of thiol-modified ssDNA onto gold surfaces has been characterized by several research groups; a typical ssDNA surface density is 4 × 1012 molecules/cm2 and hybridization efficiencies of these ssDNA monolayers are typically 80–100%.1, 23 A 100% hybridization efficiency would correspond to an average of 8,000 D2-B oligonucleotides/micron for a 200 nm gold nanowire.
The third and final step in forming the nanoscale ssDNA pattern is to use a stamping process to transfer biotinylated ssDNA molecules from the nanowire master surface to a streptavidin-coated PDMS surface. Transfer of the D2-B molecules from the nanowire array to the PDMS surface was accomplished through the strong non-covalent binding of the D2-B biotin moiety to a streptavidin monolayer on the PDMS. This biotin-streptavidin interaction is much stronger than the hybridization energy of the D1–D2 duplex. A monolayer of streptavidin was created on the PDMS surface using a standard surface modification procedure of sequential reactions with an aminosilane, NHS-biotin and then streptavidin. The streptavidin-coated PDMS surface and the inked gold nanowire array master surfaces were first both briefly dried with nitrogen, and then placed in contact with each other for 15 minutes. Crooks et al. used pressures of approximately 1.4 N cm−2 for the mechanical transfer process, but we found for our master-replica transfer process that any pressure above 0.5 N cm−2 resulted in the indentation of the PDMS substrate and even some transfer of gold nanowires to the PDMS. The efficient transfer of biotinylated ssDNA molecules (D2-B) without indentation or nanowire transfer was observed for pressures as low as 0.01 N cm−2. After 15 min, the PDMS replica was gently peeled off from the master surface (see Figure 1). Finally, the resultant ssDNA nanoline array on the PDMS surface was rinsed thoroughly with Millipore water to remove any thiol-modified ssDNA molecules (D1-S) that were accidentally transferred in the stamping process along with the D2-B molecules. In order to faithfully replicate the pattern on the nanoscale, there should be no visible liquid between the PDMS and the nanowire interface when brought into contact; If contact is made under wet condition as described by Crooks et al., fluorescence images showed indistinct, blurry nanolines, possibly due to diffusion of D2-B from a partial denaturation of DNA duplex in aqueous solution. For convenience, we show data from poly(T) and poly(A) oligonucleotides in this paper, but we have also successfully used other sequences containing all four bases.
Fluorescence hybridization adsorption measurements
The sequence-specific adsorption of complementary ssDNA molecules onto the ssDNA nanoline patterns created by the master-replica transfer process was characterized with a series of fluorescence microscopy measurements. Nanoline arrays formed by the attachment of biotinylated ssDNA (sequence D2-B) onto streptavidin-coated PDMS surfaces were exposed to phosphate buffer solutions that contained fluorescently labeled ssDNA (either complementary ssDNA D1-F, or non-complementary ssDNA D3-F). After 15 min, the PDMS replica was rinsed with buffer and examined with a fluorescence microscope. Figure 2b shows a typical 60x fluorescence image of the PDMS surface obtained after exposure to complementary ssDNA D1-F; exposure to the noncomplementary ssDNA D3-F resulted in no observable fluorescence in the image. Lines of fluorescence spaced by approximately 5 microns with an overall morphology very similar to the gold nanowires seen in the SEM data (Fig 2a) are clearly observed in the image. These fluorescence measurements confirm that (i) biotinylated ssDNA was indeed transferred from the gold nanowires to the PDMS substrate, and (ii) the transferred ssDNA nanolines were able to hybridize with ssDNA to form double stranded DNA (dsDNA) monolayers.
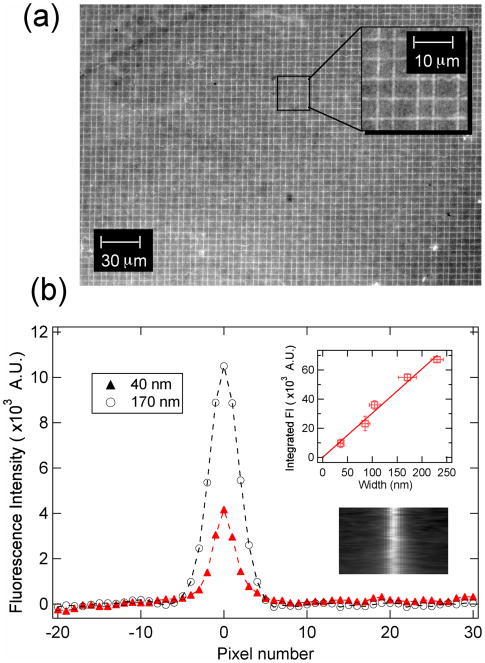
The gold nanowire master surface could be reinked and used again to create either additional PDMS surfaces or more complex nanoscale DNA patterns. Figure 3a shows the 20x fluorescence image from a PDMS substrate that was stamped twice in perpendicular directions, and then exposed to complementary ssDNA. A dsDNA pattern of two sets of nanolines clearly fills the entire fluorescence image; in fact, this high fidelity dsDNA nanoscale pattern was observed for virtually the entire sample surface. The fluorescence intensity observed from the second stamping (after re-inking) was slightly (4.0 ± 1) % less than the first stamping. In a separate experiment, the fluorescence from replicas obtained by stamping after re-inking a single master once and twice were observed to be (3.5 ± 1) % and (7.3 ± 1) % less than the original replica (data not shown). In principle, multiple stampings with different nanowire master surfaces can be used to create arbitrarily complex ssDNA and dsDNA nanoline structures.
Figure 3.
Fluorescence image and fluorescence measurements. (a) A 20x fluorescence image of a PDMS replica that was stamped in perpendicular directions and then exposed to fluorescently labeled complementary ssDNA, D1-F. (b) Two representative line profiles of the fluorescence intensity that were obtained from two different samples of dsDNA nanolines created from masters with average nanowire width of 40 nm and 170 nm. The inset plot is the integrated fluorescence intensity of line profiles obtained from five different samples of dsDNA nanolines created from masters with gold nanowire widths varying from 40 to 250 nm. The widths of the master gold nanowires were obtained by SEM measurements. (Note: 10 pixels in the fluorescence image ≈ 1.6 μm.)
The integrated fluorescence signal from a nanoline could be used to ascertain its relative width. Of course, the widths of the nanolines in the fluorescence images are limited by the optical resolution of the microscope. However, if the fluorescence profile across the width of a nanoline was integrated to give a value of the total fluorescence, this integrated fluorescence was observed to vary systematically with the width of the gold nanowire, even for nanowires as thin as 40 nm. Figure 3b shows two representative line profiles of dsDNA nanolines stamped from two different master gold nanowire widths: 170 nm and 40 nm. The integrated fluorescence values from five line profiles including the two mentioned above are shown in the inset of Figure 3b. This inset graph clearly shows a linear relationship between the integrated fluorescence intensity and the master gold nanowire width over the entire range from 40 to 250 nm. This linear relationship demonstrates that the number of adsorbed ssDNA molecules in the ssDNA nanolines could indeed be controlled by the width of the gold nanowires in the master pattern, and strongly suggests that the width of the nanolines on the replica surface match the width of the gold nanowires on the master surface. Similar measurements of the total integrated fluorescence from optical images to ascertain the properties of sub-optical surface features have been used previously in single molecule fluorescence measurements.24
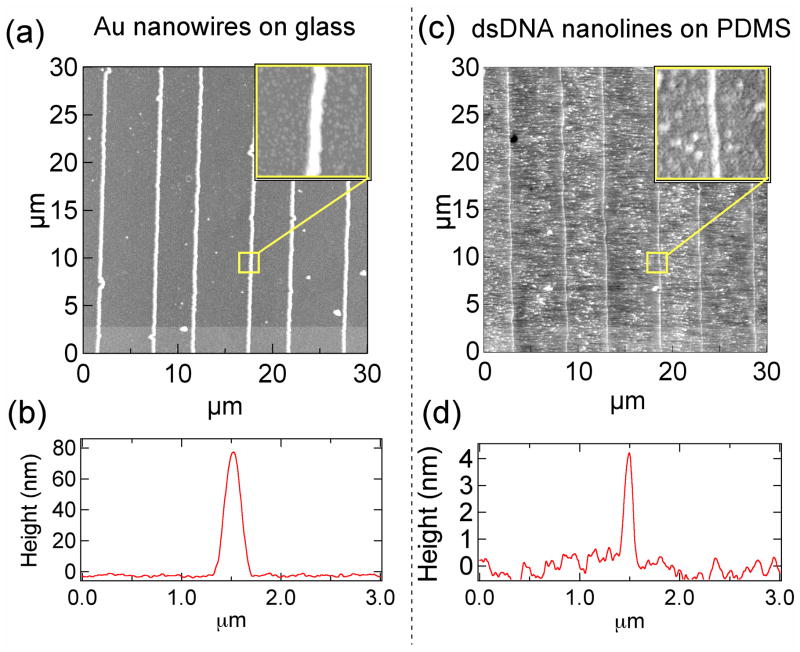
AFM measurements
Although the fluorescence data unequivocally demonstrates the successful fabrication of ssDNA nanoline replica patterns, we also wish to establish the physical dimensions of the transferred DNA relative to those of the metal nanowire “masters”. Atomic force microscopy (AFM) was used for this purpose (Fig 4). The limitation of AFM is that although it accurately measures the height of the gold nanowires and the transferred DNA, the apparent widths of these nanostructures are artificially broadened because of the convolution of an AFM tip of unknown shape25–27. The AFM image of the master gold nanowire arrays and the height profile of a single gold nanowire (Fig 4a and b) show the nanowires have a height of 80 nm and an average spacing of 5 μm, just as seen by SEM (Fig 2a). The AFM image of dsDNA nanolines on the PDMS replica surface obtained after the master-replica transfer process and hybridization with D1-F is shown in Fig 4c, and the representative height profile of a single dsDNA nanoline shows the transferred DNA nanolines have an average height of 4.50 +/− 0.2 nm on the surface (Fig 4d). The characteristic line spacing pattern seen for the gold nanowires (Fig 4a) is faithfully reproduced in the DNA nanolines (Fig 4c). The measured height of the transferred DNA of 4.5 nm height is less than the 9 nm expected from a full monolayer of hybridized 30mer dsDNA; this suggests that not all of the biotinylated ssDNA on the gold nanowire is transferred to the replica surface. Indeed, multiple stamped replicas could be created from a single inked gold nanowire master;13, 14 however, fluorescence measurements showed that the first stamped replica always had significantly more transferred ssDNA.
Figure 4.
AFM images and measurements. (a) AFM image of a master gold nanowire array at low and high (inset) magnification and (b) a representative height profile of a single gold nanowire. (c) AFM image of dsDNA nanolines on the PDMS replica surface obtained after the master-replica transfer process and hybridization with D1-F and (d) a representative height profile of a single dsDNA nanoline.
Optical diffraction measurements
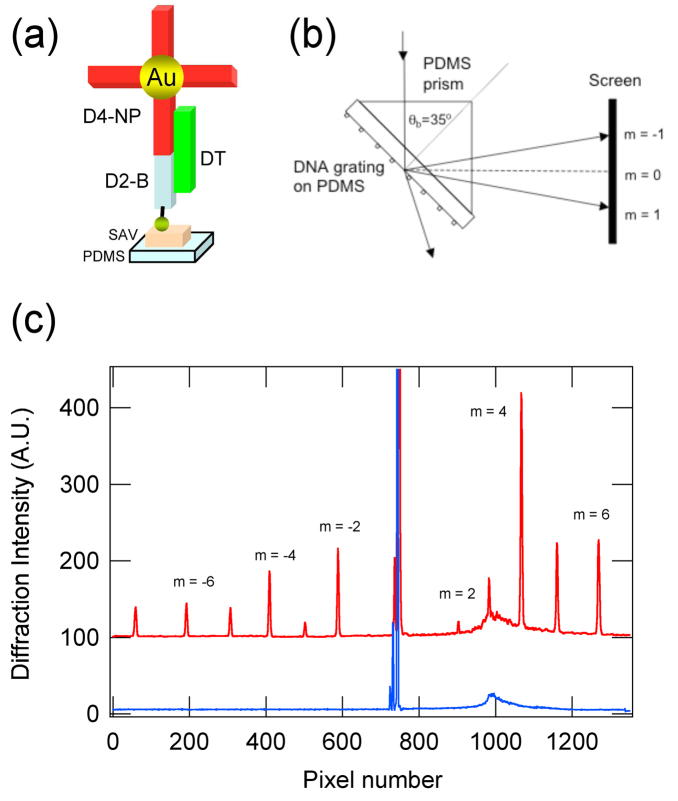
As a demonstration of how these ssDNA nanoline arrays can be applied to bioaffinity sensing, optical diffraction patterns were used to detect the hybridization adsorption of unlabeled ssDNA target molecules in solution. For these experiments, a 50mer target ssDNA in solution (sequence DT in Table I) was observed by a three sequence hybridization adsorption reaction in which the target ssDNA (DT) simultaneously hybridized to a 20mer ssDNA nanoline array (D2-B) and to a 30mer ssDNA sequence attached to a 13 nm gold nanoparticle (D4-NP) to form the surface structure shown schematically in Figure 5a. The surface grating used in the diffraction measurements consisted of an array of 230 nm ssDNA nanolines that were spaced by an average of 5 μm. This grating was placed in contact with a PDMS prism as shown in Figure 5b, and diffraction was observed from a 633 nm HeNe laser that was incident on the surface in total internal reflection (TIR) geometry at Brewster’s angle for the PDMS-air interface (35.0°). The presence of DNA-coated gold nanoparticles adsorbed onto this grating surface resulted in a strong optical diffraction signal.17 The results of the diffraction measurements are shown in Figure 5c; the red trace in the figure is the intensity of the laser light scattering observed in reflection from the PDMS prism after exposing the grating surface to a 1μM sample of the target ssDNA, followed by a 1 nM solution of ssDNA-coated gold nanoparticles. Three features stand out in the reflectance data from the grating: (i) a strong specular reflection peak at an output angle equal to the incident angle, (ii) a weak diffuse background due to scattering from imperfections in the PDMS that resulted in a broad peak at the PDMS-air critical angle (44.4°), and (iii) a series of diffraction peaks (where m ranges in value from +6 down to −7) at various angles due to the hybridization adsorption of the DNA-coated nanoparticles. The alternating intensities of the diffraction peaks follow the same pattern as those observed previously from gold nanowire arrays20 and are due to the alternating line spacing of 5 +/− Δ microns that occurs in the gold nanowire array on the master surface where Δ=0.25 microns as obtained from SEM measurements. Note that the intensities of the diffraction peaks in Figure 5c increase at output angles equal to or greater than the critical angle; this has also been observed previously in the diffraction patterns from gold nanowire arrays.20 The amount of TIR enhancement depends on the ratio of the refractive index of the prism and the adjacent medium (air, in this case); replacing PDMS with a higher refractive index polymer should result in greater TIR enhancement of the diffraction. As a control experiment, we repeated the diffraction measurements without the DT sequence present in solution, and no diffraction peaks were observed. The blue curve in Figure 5c corresponds to the reflection data from this control experiment; only a specular reflection peak and the diffuse background scattering are evident. Any nonspecific adsorption of nanoparticles onto the PDMS surface did not result in diffraction; this is an inherent advantage of all diffraction-based biosensing methods.
Figure 5.
Optical diffraction measurements of the hybridization adsorption of DNA-coated gold nanoparticles onto the ssDNA nanoline arrays. (a) Schematic diagram showing a three sequence hybridization adsorption reaction in which the target 50mer ssDNA, DT, simultaneously hybridized to a 20mer ssDNA nanoline array (D2-B) on PDMS replica and to a 30mer ssDNA sequence attached to a 13 nm gold nanoparticle (D4-NP). (b) The nanoparticle-DNA grating on PDMS was placed in contact with a PDMS prism, and diffraction was observed from a 633 nm HeNe laser that was incident on the surface in TIR geometry at Brewster’s angle for the PDMS-air interface (35.0°). (c) Diffraction intensity line profile (red) observed after exposing the PDMS grating surface to a 1μM sample of the target ssDNA, followed by a 1 nM solution of ssDNA-coated gold nanoparticles. The blue curve corresponded to the line profile of the control experiment that was repeated without the target sequence DT.
In this initial demonstration, the ssDNA target concentration was 1 μM. We can estimate the ultimate sensitivity of the nanoparticle-enhanced diffraction detection method by measuring of the lowest measurable surface coverage of target ssDNA. At very low concentrations, the relative surface coverage (θ = Γ/ΓMax) is linearly related to the solution concentration C by θ = KAdsC, where KAds is the Langmuir adsorption coefficient.23 We can estimate a limit of detection for target DNA concentration in solution (CLOD) from the lowest detectable surface coverage θ min: CLOD = θ min/KAds. From previous papers, we can obtain a θ min of 10−3 for SPRI,28 and 10−5 for nanoparticle-enhanced diffraction.17 For a Langmuir adsorption coefficient of 109 M−1, this corresponds to a CLOD of 1 pM and 10 fM respectively.
Conclusions
In this paper we have shown that nanoscale ssDNA patterns can be created on PDMS substrates by a master-replica transfer process from a LPNE nanowire pattern on glass substrates. Creating nanoscale patterns of metal nanowires on micron or even centimeter length scales by LPNE has previously led to a number of new devices and measurements.20, 22 Using this new transfer process, nanoscale patterns can now be constructed completely from ssDNA, and more complex nanoscale ssDNA surface patterns can be created by using multiple stamps on a single surface. These nanoparticle-enhanced diffraction measurements can be multiplexed in two ways: first, by creating an array of spots (the laser spot size is less than 1mm2), and secondly, by creating multiple diffraction patterns in one spot (for example, two orthogonal DNA nanoline arrays in one spot can be used to detect two different sequences via diffraction patterns in two orthogonal directions).
In addition, the total quantity of transferred ssDNA per unit length in these nanolines is directly proportional to the width of the gold nanowires in the master surface. As the thickness of the gold nanowires is reduced to 100 nm or below, they start to match the length scale of single DNA molecules (for example, a 1KB dsDNA molecule has a length of approximately 300 nm). As a first, very simple demonstration of the utility of this ssDNA nanoline fabrication process, we have created a ssDNA nanoline grating and then used optical diffraction measurements to detect unlabeled ssDNA target molecules in solution. The detection of ssDNA by measuring the diffraction from nanoparticle-decorated nanoline gratings is only a first example of how to use these nanoscale patterns. In the future, we intend to extend the transfer process to other biomolecules (e.g., antibodies, peptides) and we will use bioaffinity interactions with these surfaces to incorporate proteins, viruses and even cells into the nanoscale patterns.
Acknowledgments
This research was supported by the National Institute of Health (2RO1 GM059622), the National Science Foundation (RMC acknowledges funding from CHE-0551935 and RMP acknowledges funding from CHE-0956524), and the DARPA Micro/Nano Fluidics Fundamentals Focus MF3 Center at UCI.
References
- 1.Schena M, Shalon D, Davis RW, Brown PO. Science. 1995;270(5235):467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 2.Wacker R, Schröder H, Niemeyer CM. Anal Biochem. 2004;330(2):281–287. doi: 10.1016/j.ab.2004.03.017. [DOI] [PubMed] [Google Scholar]
- 3.Yurke B, Turberfield AJ, Mills AP, Simmel FC, Neumann JL. Nature. 2000;406(6796):605–608. doi: 10.1038/35020524. [DOI] [PubMed] [Google Scholar]
- 4.Friedrich CS, Bernard Y. Appl Phys Lett. 2002;80(5):883–885. [Google Scholar]
- 5.Winfree E, Liu F, Wenzler LA, Seeman NC. Nature. 1998;394(6693):539–544. doi: 10.1038/28998. [DOI] [PubMed] [Google Scholar]
- 6.Fujibayashi K, Hariadi R, Park SH, Winfree E, Murata S. Nano Lett. 2007;8(7):1791–1797. doi: 10.1021/nl0722830. [DOI] [PubMed] [Google Scholar]
- 7.Zhang G-J, Tanii T, Miyake T, Funatsu T, Ohdomari I. Thin Solid Films. 2004:464–465. 452–455. [Google Scholar]
- 8.Kershner RJ, Bozano LD, Micheel CM, Hung AM, Fornof AR, Cha JN, Rettner CT, Bersani M, Frommer J, Rothemund PWK, Wallraff GM. Nat Nanotechnol. 2009;4(9):557–561. doi: 10.1038/nnano.2009.220. [DOI] [PubMed] [Google Scholar]
- 9.Piner RD, Zhu J, Xu F, Hong S, Mirkin CA. Science. 1999;283(5402):661–663. doi: 10.1126/science.283.5402.661. [DOI] [PubMed] [Google Scholar]
- 10.Lin H, Sun L, Crooks RM. J Am Chem Soc. 2005;127(32):11210–11211. doi: 10.1021/ja051914u. [DOI] [PubMed] [Google Scholar]
- 11.Kim J, Crooks RM. J Am Chem Soc. 2006;128(37):12076–12077. doi: 10.1021/ja0646139. [DOI] [PubMed] [Google Scholar]
- 12.Lin H, Kim J, Sun L, Crooks RM. J Am Chem Soc. 2006;128(10):3268–3272. doi: 10.1021/ja0578799. [DOI] [PubMed] [Google Scholar]
- 13.Kim J, Crooks RM. Anal Chem. 2007;79(23):8994–8999. doi: 10.1021/ac7015954. [DOI] [PubMed] [Google Scholar]
- 14.Kim J, Crooks RM. Anal Chem. 2007;79(19):7267–7274. doi: 10.1021/ac0715219. [DOI] [PubMed] [Google Scholar]
- 15.Yu AA, Savas TA, Taylor GS, Guiseppe-Elie A, Smith HI, Stellacci F. Nano Letters. 2005;5(6):1061–1064. doi: 10.1021/nl050495w. [DOI] [PubMed] [Google Scholar]
- 16.Yu AA, Savas T, Cabrini S, diFabrizio E, Smith HI, Stellacci F. J Am Chem Soc. 2005;127(48):16774–16775. doi: 10.1021/ja055762e. [DOI] [PubMed] [Google Scholar]
- 17.Sendroiu IE, Corn RM. Biointerphases. 2008;3(3):FD23–FD29. doi: 10.1116/1.2994689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Menke EJ, Thompson MA, Xiang C, Yang LC, Penner RM. Nat Mater. 2006;5(11):914–919. doi: 10.1038/nmat1759. [DOI] [PubMed] [Google Scholar]
- 19.Xiang C, Kung SC, Taggart DK, Yang F, Thompson MA, Güell AG, Yang Y, Penner RM. ACS Nano. 2008;2(9):1939–1949. doi: 10.1021/nn800394k. [DOI] [PubMed] [Google Scholar]
- 20.Halpern AR, Nishi N, Wen J, Yang F, Xiang C, Penner RM, Corn RM. Anal Chem. 2009;81(14):5585–5592. doi: 10.1021/ac900938t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiang C, Yang Y, Penner RM. Chem Commun. 2009;(8):859–873. doi: 10.1039/b815603d. [DOI] [PubMed] [Google Scholar]
- 22.Yang F, Taggart DK, Penner RM. Nano Lett. 2009;9(5):2177–2182. doi: 10.1021/nl9008474. [DOI] [PubMed] [Google Scholar]
- 23.Chen Y, Nguyen A, Niu L, Corn RM. Langmuir. 2009;25(9):5054–5060. doi: 10.1021/la804021t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muller JD. Biophys J. 2004;86:3981–3992. doi: 10.1529/biophysj.103.037887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Allen MJ, Hud NV, Balooch M, Tench RJ, Siekhaus WJ, Balhorn R. Ultramicroscopy. 1992;42–44(Part 2):1095–1100. doi: 10.1016/0304-3991(92)90408-c. [DOI] [PubMed] [Google Scholar]
- 26.Markiewicz P, Goh MC. J Vac Sci Technol B. 1995;13(3):1115–1118. [Google Scholar]
- 27.Thundat T, Warmack RJ, Allison DP, Bottomley LA, Lourenco AJ, Ferrell TL. J Vac Sci Technol A. 1992;10(4):630–635. [Google Scholar]
- 28.Li Y, Wark AW, Lee HJ, Corn RM. Analytical Chemistry. 2006;78(9):3158–3164. doi: 10.1021/ac0600151. [DOI] [PMC free article] [PubMed] [Google Scholar]