Abstract
The transcriptional mechanisms by which temporary exposure to developmental signals instigates adipocyte differentiation are unknown. During early adipogenesis, we find transient enrichment of the glucocorticoid receptor (GR), CCAAT/enhancer-binding protein β (CEBPβ), p300, mediator subunit 1, and histone H3 acetylation near genes involved in cell proliferation, development, and differentiation, including the gene encoding the master regulator of adipocyte differentiation, peroxisome proliferator-activated receptor γ2 (PPARγ2). Occupancy and enhancer function are triggered by adipogenic signals, and diminish upon their removal. GR, which is important for adipogenesis but need not be active in the mature adipocyte, functions transiently with other enhancer proteins to propagate a new program of gene expression that includes induction of PPARγ2, thereby providing a memory of the earlier adipogenic signal. Thus, the conversion of preadipocyte to adipocyte involves the formation of an epigenomic transition state that is not observed in cells at the beginning or end of the differentiation process.
Keywords: PPARγ, adipogenesis, differentiation, enhancer, epigenomics
Transcription factors control cell fate decisions by programming the expression of large gene sets, often through mechanisms impacting the post-translational modification of histone proteins in chromatin (Allis et al. 2007). Genome-wide changes to histone modification patterns occur as cells differentiate, such that distinct epigenomes are present in different cell types (Bernstein et al. 2007). A cell's epigenome is thought to be involved in establishing its identity, yet whether histone modifications transmit the memory of a given cell state or implement the memory once it is transmitted by a distinct mechanism is currently unclear.
The epigenomic factors controlling adipocyte cell differentiation are not known. Adipocytes, the major fat-containing component of adipose tissue, are terminally differentiated cells derived from mesenchymal precursors (Ailhaud et al. 1992). Study of adipocyte precursors in vivo is difficult because adipose tissue in rodents is undetectable macroscopically until shortly before birth. Thus, most of what is known about the molecular basis of adipocyte differentiation derives from tissue culture models, one of the best characterized being the mouse 3T3-L1 model (Green and Kehinde 1975, 1976). Temporary exposure to a mix of insulin, glucocorticoid, and an inducer of cAMP signaling triggers adipogenesis, changing the expression of hundreds of genes, including a variety of transcription factors that regulate one another as well as structural genes (Farmer 2006). One of these encodes peroxisome proliferator-activated receptor γ (PPARγ), a sequence-specific transcription factor that is necessary (Barak et al. 1999; Kubota et al. 1999; Rosen et al. 1999) and sufficient (Tontonoz et al. 1994; Hu et al. 1995; Shao and Lazar 1997) for adipogenesis, and is hence termed the master regulator (Rosen et al. 2002). CCAAT/enhancer-binding protein (CEBP) transcriptional regulators also play a major role, and each can induce adipocyte differentiation of fibroblasts, although none can induce differentiation in the absence of PPARγ (Rosen et al. 2002).
Genome-wide location analysis for PPARγ and CEBPα in 3T3-L1-differentiated adipocytes revealed that both occupy sites near most of the genes that are up-regulated during adipogenesis, suggesting that they coordinate expression of the majority of genes determining the adipocyte phenotype (Lefterova et al. 2008). Multiple binding sites for PPARγ and CEBPα are found at the Pparγ and Cebpα loci, consistent with the fact that each autoregulates its own expression as well as that of the other (Wu et al. 1999). These observations suggest that PPARγ and CEBPα form a feed-forward circuit defined by a regulator that controls a second regulator, with the added feature that both regulators bind a common set of target genes (Alon 2007). A feed-forward loop with positive regulators can provide consistent activity that is relatively insensitive to spurious changes in input (Mangan and Alon 2003). That PPARγ and CEBPα are inextricably linked by interconnected autoregulatory loops may provide additional advantages for cell fate decisions. For example, a transient signal can cause the loop to lock irreversibly into the “on” state so that the circuit structure can provide memory of a developmental signal even after it is gone (Alon 2007).
The autoregulatory and feed-forward circuitry formed by PPARγ and CEBPα may explain how adipocyte cell identity is established and maintained upon induction of Pparγ and Cebpα expression. However, our understanding of the transcriptional mechanism initiating Pparγ and Cebpα expression during adipogenesis is incomplete. There is no disputing a role for CEBP proteins. CEBPβ and CEBPδ function in adipocyte differentiation before PPARγ and CEBPα (Cao et al. 1991), and ectopic expression of CEBPβ in nonadipogenic fibroblasts up-regulates Pparγ and Cebpα transcription (Farmer 2006). Also, mouse embryonic fibroblasts (MEFs) obtained from mice lacking both CEBPβ and CEBPδ do not express Pparγ and Cebpα, and are unable to undergo adipogenesis in culture (Tanaka et al. 1997). Yet, understanding of the precise role of CEBPβ and CEBPδ in regulating Pparγ and Cebpα is challenged by the finding that Pparγ and Cebpα are expressed in the poorly differentiated adipose tissue of neonatal mice lacking CEBPβ and CEBPδ (Tanaka et al. 1997). Thus, additional regulators may serve redundant and/or cooperative functions with CEBPβ and CEBPδ to ensure transcriptional activation of Pparγ and Cebpα. Identifying these should help to clarify the mechanisms regulating adipocyte differentiation.
We report here that glucocorticoid receptor (GR) and CEBPβ bind cooperatively near Pparγ and genes involved in cell proliferation during the beginning stages of 3T3-L1 adipogenesis. Colocalized and coincident with GR and CEBPβ binding are p300 and Med1 transcriptional coactivators and histone modification associated with transcriptional activation. The data indicate that GR–CEBPβ-bound regions serve as transcriptional enhancers, which is supported further by the finding that they can activate transcription from a heterologous promoter during the early stages of adipogenesis. Occupancy and enhancer function are triggered in response to temporary developmental signals provided by insulin, glucocorticoid, and an inducer of cAMP signaling, and diminish upon their removal. GR, which is required for adipogenesis but need not be active in the mature adipocyte, functions transiently with other enhancer proteins to propagate a new program of gene expression that includes induction of Pparγ2. PPARγ regulates its own transcription as well as that of Cebpα to drive expression of adipocyte-specific genes, thereby providing a memory of the earlier adipogenic signal. Thus, the conversion of preadipocyte to adipocyte is directed by an epigenomic transition state that is not observed in cells at the beginning or end of the differentiation process.
Results
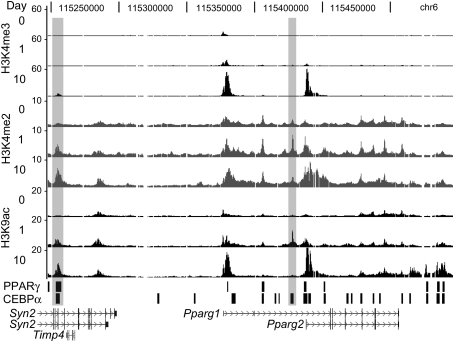
A transient peak of histone H3 acetylation appears early during adipogenesis at 10 kb upstream of Pparγ2
Genome-wide mapping studies reveal that transcriptionally active genes are enriched for specific post-translational histone modifications, and that some of these can be used to identify cis-regulatory sequences (Wang et al. 2008; Heintzman et al. 2009). Indeed, our previous studies mapping histone modifications in 3T3-L1-differentiated adipocytes identified intergenic regions enriched for histone marks that colocalize with PPARγ and CEBPα and confer enhancer activity (Steger et al. 2008; Tomaru et al. 2009). Therefore, to identify potential cis-acting sequences controlling adipoctye differentiation, we combined chromatin immunoprecipitation with microarray analysis (ChIP–chip) to map histone modifications at high resolution during different stages of adipogenesis. 3T3-L1 cells were induced to differentiate by exposure to insulin, dexamethasone, and 3-isobutyl-1-methylxanthine in 10% fetal bovine serum for 2 d. Six days to 8 d are required to form lipid-storing mature adipoctyes with this protocol, and histone modification profiles were generated at days 0, 1, and 10. Although the combinatorial patterns of histone modifications at active genes are complex (Wang et al. 2008), we chose to examine H3 Lys 4 trimethylation (H3K4me3) and dimethylation (H3K4me2) and H3K9 acetylation (H3K9ac). Genomic regions with high H3K4me3 levels are strongly correlated with transcription start sites (TSSs), whereas regions enriched with H3K4me2 and H3 acetylation, but containing relatively low H3K4me3, are often found to be enhancers (Bernstein et al. 2005; Heintzman et al. 2007; Roh et al. 2007).
Of the 74 genes represented on the custom microarray, 20 had greater levels of H3K4me3, H3K4me2, and H3K9ac in day 10 adipocytes compared with day 0 undifferentiated cells, consistent with induced transcription from these genes during adipogenesis (Steger et al. 2008). Interestingly, only Pparγ displayed increased levels for all three modifications at day 1 of differentiation. As shown in Figure 1, the TSS of Pparγ1 but not Pparγ2 contains a small peak of H3K4me3 on day 0, the Pparγ2 TSS is weakly marked on day 1, and both are strongly marked by day 10. Consistent with these data, Pparγ1 is expressed weakly in 3T3-L1-undifferentiated cells, Pparγ2 is induced ∼24 h after the initiation of differentiation, and both isoforms are significantly up-regulated in mature adipocytes (Supplemental Fig. S1). Profiles for H3K4me2 and H3K9ac show expected increases at the Pparγ TSSs for days 1 and 10. Outside of the TSSs, a notable region of enrichment is located 10 kb upstream of Pparγ2, where both H3K4me2 and H3K9ac levels increase from day 0 to day 1, then fall back to day 0 levels at day 10. A second region of interest is positioned 122 kb upstream of Pparγ1. Here, H3K4me2 and H3K9ac levels increase at day 1, but remain high in mature adipocytes. Thus, regions with relatively low H3K4me3 and high H3K4me2 and H3K9ac are detected near Pparγ during the time of adipogenesis when its expression is induced, suggesting that cis-regulatory sequences enhancing transcription reside there.
Figure 1.
Histone H3 modification profiles at Pparγ locus during 3T3-L1 cell differentiation. ChIP–chip mapping of H3K4me3, H3K4me2, and H3K9ac at days 0, 1, and 10 of 3T3-L1 differentiation. Fold enrichment values were plotted on a linear scale. Shaded rectangles indicate enrichment peaks for H3K4me2 and H3K9ac that are increased at day 1 relative to day 0 and not positioned at TSSs. Black bars indicate PPARγ- and CEBPα-binding regions in day 10 adipocytes.
Identification of GR and CEBPβ at a subset of genomic regions acetylated transiently during adipocyte differentiation
Sequence-specific transcription factors can direct histone modifications to particular genomic loci by recruiting chromatin-modifying activities when bound to cellular DNA (Hassan et al. 2001). Thus, we hypothesized that an activator binds a sequence motif located 10 kb upstream of Pparγ2 to produce the transient acetylation, and its binding to additional sites in the genome would produce similarly marked regions. To identify the protein(s), we performed genome-wide mapping of H3K9ac during 3T3-L1 differentiation, identified regions with more acetylation at day 1 compared with days 0 and 10, and examined these for enriched sequence motifs. Using a peak-calling method we developed termed STAR (Significance Tester for Accumulation of Reads), 1629 regions located outside of TSSs were determined to be significantly enriched for H3K9ac at day 1 but not days 0 and 10 (Supplemental Table S1). Gene ontology (GO) analysis of the closest genes determined that the most enriched biological processes were development (P = 3E-10) and cell proliferation and differentiation (P = 3E-10) (Supplemental Tables S2, S3). These reflect the early events taking place in 3T3-L1 cell differentiation. At day 0, a confluent population of quiescent cells is induced to re-enter the cell cycle and carry out at least two rounds of cell division before giving rise to post-mitotic terminally differentiated adipocytes (Farmer 2006). Thus, the GO analysis indicates that the transiently acetylated regions are likely functional.
Next, the Asap program (Marstrand et al. 2008) was used to determine whether transcription factor-binding sites were overrepresented in the transiently acetylated regions. Furthermore, the search was narrowed based on the observation that a CEBP-binding site colocalizes with the transient acetylation near Pparγ2 (Fig. 1). Although CEBP proteins on their own are unlikely to cause the transient acetylation, because the vast majority of CEBP-binding sites at Pparγ are not marked in this manner, we reasoned that the acetylation could result from the combined function of CEBP proteins and a specific activator. Intersection of the CEBPα-binding regions (Lefterova et al. 2008) with the transiently acetylated regions uncovered 146 colocalizing regions. Within these, the top-scoring motifs from Asap were CEBP (P = 5E-05) and GR (P = 6E-05) (Supplemental Table S4).
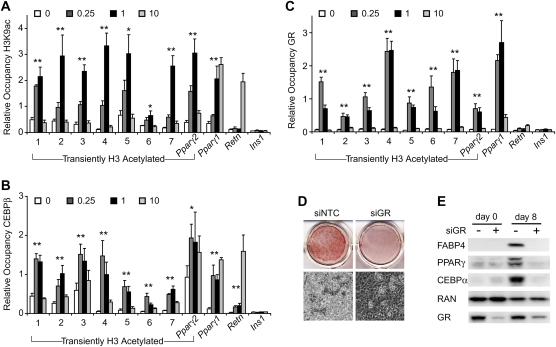
To determine whether GR and CEBP proteins could occupy the motifs in vivo, multiple regions were examined by ChIP. Regions with a range of false discovery rate (FDR) values were selected (Supplemental Table S5), and H3K9ac ChIP validated the genome-wide findings for all (Fig. 2A). Each has increased acetylation at day 1 relative to day 0, and all but the region upstream of Pparγ1 have more acetylation at day 1 compared with day 10. Acetylation remains high on day 10 upstream of Pparγ1, with a level similar to the Retn enhancer (Tomaru et al. 2009), possibly because PPARγ and CEBPα occupy both regions in adipocytes. CEBPβ drives 3T3-L1 differentiation before CEBPα, and CEBPβ ChIP revealed variable levels of occupancy for each region at day 0 that increase at day 1 for all and decrease by day 10 for most (Fig. 2B). CEBPδ, which plays a more limited role in adipogenesis, has a similar binding pattern, while CEBPα displays increased occupancy at day 10 relative to earlier times (Supplemental Fig. S2). Importantly, GR was found to bind to all regions during the early stages of adipogenesis, with little or no binding detected at days 0 and 10 (Fig. 2C). Note that GR is present in mature adipocytes (day 10), and thus its lack of binding is due to loss of the adipogenic signal conferred by dexamethasone. Indeed, GR binding at several of the transient sites could be restored by treatment of mature adipocytes with glucocorticoid (Supplemental Fig. S3).
Figure 2.
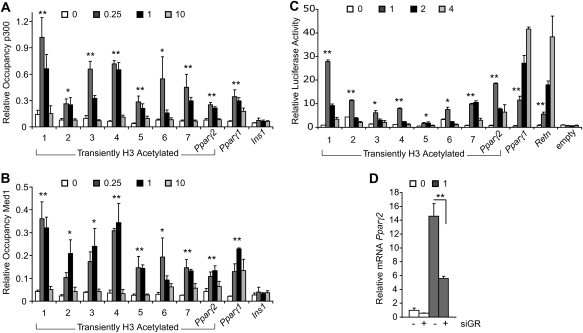
GR and CEBPβ target regions acetylated transiently during adipogenesis. ChIP at days 0, 0.25, 1, and 10 of 3T3-L1 differentiation for H3K9ac (A), CEBPβ (B), or GR (C). H3K9ac ChIP-seq regions are numbered. Also included are regions 10 and 122 kb upstream of Pparγ2 and Pparγ1, respectively, and regions from the Retn enhancer and Ins1. Region names remain constant for all successive figures. Data were plotted as the mean of at least three biological replicates ± SEM. Comparison between day 0 and either day 0.25 or day 1 was used for significance calculation. (*) P < 0.05; (**) P < 0.01. (D) 3T3-L1 cells treated with nontargeted control (NTC) or GR siRNAs were stained with Oil Red O (top panel) or were subjected to phase-contrast microscopy (bottom panel) 8 d after inducing adipocyte differentiation. (E). Western blot analysis of adipoctye-selective proteins 0 and 8 d after inducing differentiation of siRNA-treated cells.
The data suggest that GR colocalizes with CEBPβ at regions acetylated transiently during adipogenesis, with maximal binding coinciding temporally with maximal H3K9 acetylation. In addition, the data imply that GR regulates adipoctye differentiation. To directly assess its function in adipogenesis, we used siRNA to deplete GR in 3T3-L1 cells and examined their ability to differentiate. Cells with decreased GR levels were unable to efficiently store lipid and express adipocyte-selective proteins (Fig. 2D,E; Supplemental Fig. S4). Thus, although its occupancy at Pparγ2 and other regions is transient, GR is required for the adipogenic differentiation of 3T3-L1 cells.
GR and CEBPβ colocalize in differentiating 3T3-L1 cells
Multiple transcription factors can coordinate complex cell behavior by targeting the same genes (Boyer et al. 2005). The data thus far suggest that GR and CEBPβ may act in concert to facilitate adipocyte differentiation. To determine the extent to which they function together, we performed genome-wide mapping of GR and CEBPβ during an early stage of 3T3-L1 differentiation. Because GR and CEBPβ appear to occupy their sites similarly at 6 and 24 h of differentiation, ChIP-seq was performed at 6 h so that factor binding would be examined independently of cell division, which occurs later.
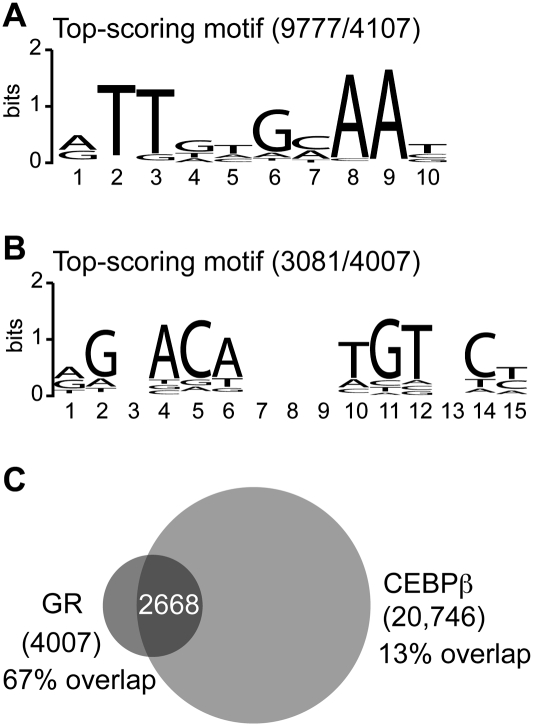
Peak calling with GLITR (Tuteja et al. 2009) identified 20,746 and 4007 high-confidence binding regions for CEBPβ and GR, respectively. A search within 4107 of the most highly enriched CEBPβ-bound regions for both de novo and known sequence motifs identified a de novo sequence that strongly resembles the CEBP recognition element as the top-scoring motif (Fig. 3A). Consistent with this, the highest-scoring motifs from the TRANSFAC database are matrices for CEBP proteins (Supplemental Table S6). A similar search within all GR-bound regions revealed the top-scoring motif as a de novo sequence for the GR-binding element (Fig. 3B). Binding sites for the androgen receptor, progesterone receptor, and GR are the most enriched motifs from TRANSFAC (Supplemental Table S7), which is not surprising given that these factors have similar DNA-binding specificities. Interestingly, CEBP matrices are next on the list of enriched TRANSFAC motifs, suggesting that many of the GR-bound regions may also be occupied by CEBP proteins.
Figure 3.
A majority of GR-bound regions overlap with CEBPβ-bound regions in differentiating 3T3-L1 cells. Top-scoring sequence motifs at 6 h of 3T3-L1 differentiation for CEBPβ-bound regions (A) or GR-bound regions (B). The number of binding regions containing the motif is indicated relative to the number examined. (C) Venn diagram representing the overlap of GR- and CEBPβ-bound regions.
To determine the degree of GR and CEBPβ co-occupancy on a genome-wide scale, we intersected the GR and CEBPβ cistromes to identify bound regions with any overlap. Remarkably, 67% of GR-bound regions were found to overlap with CEBPβ-bound regions (Fig. 3C), revealing that a majority of GR-bound regions are positioned near CEBPβ-bound sites and implying that the factors may influence each other's binding in vivo. To explore this further, we identified CEBPβ-bound regions in undifferentiated 3T3-L1 cells by ChIP-seq, because CEBPβ ChIP revealed robust occupancy at some sites prior to differentiation (Fig. 2B), and examined their locations relative to GR- and CEBPβ-bound regions at 6 h of differentiation. Peak calling identified 10,359 regions enriched with the same CEBP motifs as for 6 h (Supplemental Table S8), agreeing with the finding that 71% of the regions occupied at 0 h overlap those at 6 h. Interestingly, only 26% of the GR cistrome colocalizes with the CEBPβ cistrome at 0 h, indicating that most regions bound by both GR and CEBPβ are unoccupied in undifferentiated cells. In addition, nearly all (99%) of the 0 h CEBPβ-bound regions that colocalized with GR-bound regions are also occupied at 6 h, and 57% of these show at least a 1.5-fold increase in CEBPβ occupancy at 6 h based on enrichment or stack height from peak calling. Thus, where GR and CEBPβ colocalize at 6 h of differentiation, induced binding by GR is associated with induced CEBPβ binding at a majority of regions and increased CEBPβ binding at most of the remaining regions bound with CEBPβ in undifferentiated cells.
GR and CEBPβ facilitate each other's occupancy
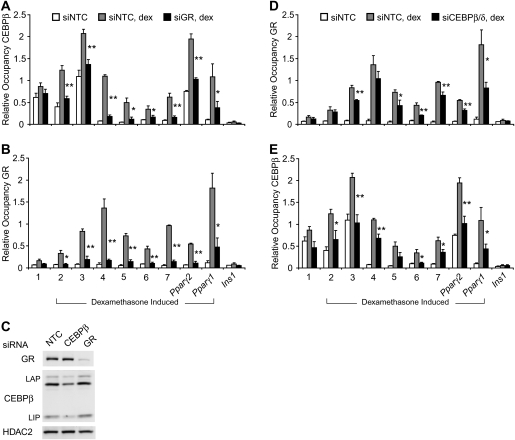
GR and CEBPβ may colocalize because the binding of one factor facilitates binding of the other. To investigate this, the ability of each factor to occupy native sites in the absence of the other was evaluated by ChIP in 3T3-L1 cells treated with siRNAs to reduce GR or CEBPβ protein levels. Because CEBPβ expression is increased during the early stages of adipogenesis, a response counteracting siRNA knockdown, we sought conditions that minimize this up-regulation yet maintain robust GR and CEBPβ binding. Without insulin and 3-isobutyl-1-methylxanthine, dexamethasone treatment for 2 h induced GR and CEBPβ binding at nearly all regions studied with little or no change to CEBPβ level (Supplemental Fig. S5). Interestingly, ChIP for CEBPβ revealed decreased occupancy in cells depleted for GR (Fig. 4A). As expected, GR binding in these cells was decreased (Fig. 4B), as was GR, but not CEBPβ protein (Fig. 4C). Thus, at the regions tested, GR binding is required for full dexamethasone-induced binding of CEBPβ. GR occupancy was significantly reduced at six of eight regions in cells with reduced CEBPβ and CEBPδ levels (Fig. 4D). CEBPβ binding was also significantly reduced in these cells (Fig. 4E), although the fold changes are moderate, likely because knockdown of CEBPβ decreased its expression by only 50% (Fig. 4C; Supplemental Fig. S6). Consistent with this, more dramatic effects were observed in GR knockdown cells in which GR was reduced to 10% of untreated cells. Taken together, the data suggest that GR and CEBPβ facilitate each other's binding at colocalized regions responsive to glucocorticoid.
Figure 4.
RNAi-mediated depletion of GR or CEBP proteins reduces binding of both GR and CEBPβ at colocalized Regions. ChIP for CEBPβ (A) or GR (B) in 3T3-L1 cells treated with GR siRNAs. (C) Western blot analysis detecting siRNA-mediated depletion of GR or CEBPβ. ChIP for GR (D) or CEBPβ (E) in cells treated with CEBPβ and CEBPδ siRNAs. Dexamethasone (1 μM) was added for 2 h. Data were plotted as the mean of three biological replicates ± SEM. Comparison between dexamethasone-treated nontargeted control (NTC) and either siGR or siCEBPβ was used for significance calculation. (*) P < 0.05; (**) P < 0.01.
Transiently acetylated regions bound by GR and CEBPβ display enhancer properties
The coactivators p300 and Mediator commonly associate with enhancers (Heintzman et al. 2009; Visel et al. 2009). Therefore, to further assess the enhancer qualities of the transiently acetylated regions targeted by GR and CEBPβ, we performed ChIP for p300 and MED1. All regions show increased occupancy of p300 and MED1 during the beginning stages of adipocyte differentiation when compared with day 0, and all but the region upstream of Pparγ1 have decreased occupancy at day 10 (Fig. 5A,B). To determine whether the regions can regulate transcription in cis, we cloned them upstream of the luciferase gene containing a minimal promoter. Clones were transfected into 3T3-L1 cells and assayed for reporter activity during the early stages of adipogenesis. With the exception of the region located 122 kb upstream of Pparγ1, all confer increased reporter activity at day 1 compared with days 0 and 4 (Fig. 5C). It is possible that decreased reporter activity after day 1 is due to plasmid dilution or loss over time, because the cells undergo at least two cell divisions by day 4. However, results from the Retn enhancer and the region upstream of Pparγ1 argue against this because reporter activity is highest at day 4. Unlike the other regions, these are targeted by PPARγ, which may explain their ability to activate expression later, given that PPARγ levels increase as differentiation proceeds. As a whole, the data demonstrate that the cloned regions are sufficient to drive transcription when occupied by GR, CEBPβ, p300, and MED1 during adipocyte differentation.
Figure 5.
Enhancer properties of transiently acetylated regions occupied by GR and CEBPβ. ChIP at days 0, 0.25, 1, and 10 of 3T3-L1 differentiation for p300 (A) or MED1 (B). Data are represented as the mean of three biological replicates ± SEM. Comparison between day 0 and either day 0.25 or day 1 was used for significance calculation. (C) Validation of predicted enhancers by the luciferase reporter assay. Reporter activity was measured in transfected cells at days 0, 1, 2, and 4 of 3T3-L1 differentiation. Average fold activation relative to empty vector at day 0 and standard deviation from triplicate samples were plotted. Comparison between days 0 and 1 was used for significance calculation. (D) Pparγ2 gene expression at days 0 and 1 in cells treated with nontargeted control (−) or GR siRNAs. Standard deviation from triplicate samples was plotted. (*) P < 0.05; (**) P < 0.01.
A prediction from these results is that GR participates in the early activation of Pparγ2. To test this, we examined Pparγ2 induction in cells with a reduced GR level. Although considerably less than the level in mature adipocytes, Pparγ2 expression increased dramatically from days 0 to 1, and this was markedly blunted by GR knockdown (Fig. 5D). Thus, GR is critical in initiating Pparγ2 expression during the beginning stages of adipocyte differentiation.
Discussion
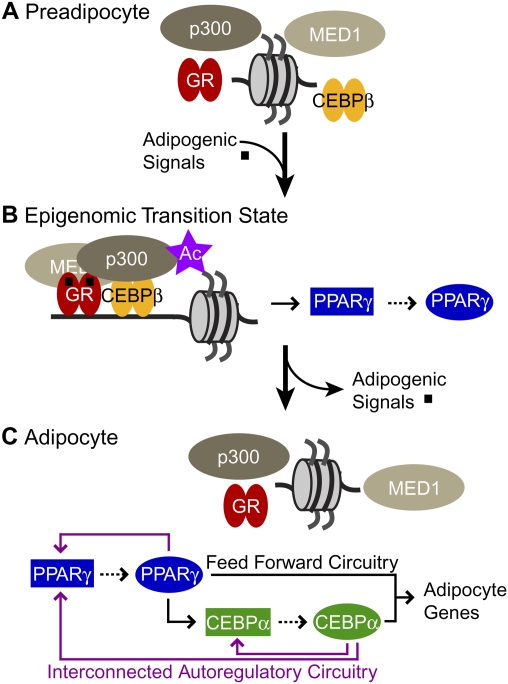
Our data show that adipogenic signals mobilize GR and CEBPβ present in preadipocytic cells (Fig. 6A) to transiently assemble with p300 and MED1 and induce H3 acetylation at many enhancers throughout the genome. Colocalization of these enhancer activities for only a short time comprises an epigenomic transition state that launches adipogenesis by activating transcription of Pparγ and potentially other genes involved in cell development, differentiation, and proliferation (Fig. 6B). PPARγ activates its own expression as well as that of CEBPα, and together they establish the adipocyte phenotype by driving transcription of adipocyte-specific genes (Fig. 6C). Thus, the memory of temporary exposure to adipogenic signals is propagated by an epigenomic transition state during the conversion of preadipocytes to adipoctyes.
Figure 6.
Propagation of adipogenic signals by an epigenomic transition state converting preadipocyte to adipocyte. Schematic summarizing the transcriptional mechanisms at early and end stages of 3T3-L1 cell differentiation. (A) GR, CEBPβ, p300, and MED1 are present in the preadipocyte, but are not colocalized at specific sites throughout the genome. (B) Their assembly and H3 acetylation (purple star) is induced at many enhancers, including Pparγ by treatment with insulin, cAMP inducers, and glucocorticoids (black squares). Binding of a transcription factor to a cis-regulatory sequence is indicated by a solid arrow. Genes encoding regulators are connected by dashed arrows. (C) Mature adipoctyes emerge 4–6 d after removal of the adipogenic signals. In their absence, factor occupancy, enhancer function, and H3 acetylation associated with GR are abolished, yet Pparγ transcription remains robust because it regulates its own expression as well as that for Cebpα. The transcriptional circuitry formed by PPARγ and CEBPα establishes and maintains adipocyte-selective gene expression. CEBPβ colocalizes with CEBPα in mature adipocytes (Lefterova et al. 2008), suggesting that it also contributes to this process.
The role we uncovered for GR in adipogenesis has important implications. The number of sequence-specific regulators impacting Pparγ expression has proliferated in recent years (Farmer 2006), yet identification and functional analysis of binding sites for many of these factors have not been determined. Our work shows that GR is required for adipogenesis, and glucocorticoids act by inducing sequence-specific binding of GR to DNA, despite complicated models suggesting otherwise (Wiper-Bergeron et al. 2007). Importantly, GR does not function in isolation, but cooperates with CEBPβ. An earlier study temporally uncoupling adipogenic signals found that dexamethasone treatment followed by IBMX, but not IBMX followed by dexamethasone, was sufficient for adipogenesis (Pantoja et al. 2008). Interestingly, the nondifferentiating IBMX-then-dexamethasone treatment transiently induced Cebpβ and Cebpδ, but not Pparγ and Cebpα, consistent with the idea that CEBP proteins must synergize with GR to activate target genes.
GR function is also likely related to metabolic physiology and pathophysiology. In humans, Cushing's syndrome due to endogenous or exogenous glucocorticoid excess leads to central adiposity (Peeke and Chrousos 1995; Pijl and Meinders 1996). A direct role for GR in mouse adipose physiology is unclear because GR-null mice die at birth (Cole et al. 1995). Mice carrying a targeted mutation that impairs GR dimerization in vitro are of normal weight (Reichardt et al. 1998); however, it has not been tested whether this GR mutant, which contains the wild-type DNA-binding domain, can occupy sites in native chromatin. Recent studies implicating PRDM16 and CEBPβ in brown fat differentiation from myoblastic precursors used a GR ligand for in vitro differentiation (Seale et al. 2008; Kajimura et al. 2009). Given the close similarity between brown and white fat, it is plausible that control of these cell fate decisions also involves a GR–CEBPβ transition state.
In general, understanding of cell development and differentiation requires understanding of the intermediate steps involved in the process. Importantly, as our study explicitly reveals, transcription factors, coactivators, and histone modifications can assemble simultaneously at specific sites throughout the genome in response to developmental signals, constituting a transition state that is required for differentiation. Although functioning for only a short time, identification of such transition states is critical, given that they can determine the outcome of developmentally induced cell fate decisions. The fact that the cellular transition state is critical but observed in neither the precursor nor the differentiated state is analogous to the transition state of a chemical reaction. Enzymes function by stabilizing the chemical transition state, and this forms the therapeutic basis for drugs that inhibit or activate enzymes (Berg et al. 2007). Likewise, the ability to facilitate or block the formation of cellular transition states based on an understanding of their molecular components could constitute novel modes of therapy for a wide range of diseases.
Materials and methods
Antibodies, ChIP, and Western analysis
The following antibodies were used in this study: histone H3 (Abcam, ab1791), H3K9ac (Upstate Biotechnologies, 06-942), H3K4me2 (Abcam, ab7766), H3K4me3 (Upstate Biotechnologies, 07-473), CEBPα (Santa Cruz Biotechnologies, sc-61), CEBPβ (Santa Cruz Biotechnologies, sc-150), CEBPδ (Santa Cruz Biotechnologies, sc-151 and sc-636), GR (Santa Cruz Biotechnologies, sc-1004; and Affinity BioReagents, PA1-511A), p300 (Santa Cruz Biotechnologies, sc-585), MED1 (TRAP220; Santa Cruz Biotechnologies, sc-8998), HDAC2 (Santa Cruz Biotechnologies, sc-7899). ChIP with 3T3-L1 cells was performed as described previously (Steger et al. 2008). Enrichments were plotted as a percentage of input. Primer sequences are provided in the Supplemental Material (Supplemental Table S9). Western blots were exposed and quantified using the ChemiDoc XRS and Quantity One 1-D analysis software (Bio-Rad).
ChIP–chip and ChIP-seq
ChIP–chip was performed as described previously (Steger et al. 2008). In short, input and ChIP DNA were amplified linearly using in vitro transcription, and were hybridized to a custom DNA microarray tiled with 60-base-pair (bp) oligonucleotides centered every 25 bp at select genes. ChIP–chip signals (IP/input) were normalized to total histone H3 occupancy, and were plotted as a moving average of 19 contiguous probes spanning 510 bp.
For ChIP-seq, ∼10 ng of ChIP DNA was prepared for sequencing according to the amplification protocol from Illumina. Sequence reads of 36 bp were obtained using the Solexa Analysis Pipeline, and were mapped to the mouse genome (mm8). Uniquely matching reads were retained with the following exceptions: Reads mapping to regions enriched in input DNA were removed, and all but one of the identically aligning reads were removed to minimize PCR bias. MACS (Zhang et al. 2008) and GLITR (Tuteja et al. 2009) were used to identify enriched regions within the CEBPβ and GR ChIP-seq data sets. Transcription factor cistromes were intersected using the University of California at Santa Cruz Genome Browser, and sites were considered overlapping if at least 1 bp was shared between the regions.
In order to identify regions enriched for H3K9ac, we developed an algorithm that determines whether local accumulations of reads are statistically significant while controlling for false positives. The algorithm, STAR, takes as input the genomic locations and read lengths of all mapped reads. Sequence reads from control (i.e., nonimmunoprecipitated input DNA) samples are expected to be uniformly distributed; however, these samples tend to show a consistent pattern of bias in Illumina Genome Analyzer data. These were characterized by comparing with simulated data sets of uniformly distributed reads, and regions of bias consistent across control samples were filtered out from all (noncontrol) samples. For a given (noncontrol) sample and a window W of L bases, we let N(W) be the number of reads overlapping the window. For each positive integer n we computed the number of windows of length L in uniformly randomized data for which N(W) > n. This gives an estimation of the number of false-positive windows with N(W) > n. Using these estimates, we computed for each n the proportion F(n) of false-positive windows in the set of all windows for which N(W) > n. Each window, W, was given a score: S(W) = F[N(W)]. A minimum n0 was chosen so that F(n0) < 0.05. Peaks were then called by merging into spans all windows in the sample with S(W) > n0. Peaks were reported as genomic spans with a score given by the average S(W) across all windows, W, contributing to the peak. This score should decrease as the strength of evidence for the region to have been correctly determined increases. STAR is implemented in Java and is freely available as open source at http://www.cbil.upenn.edu/STAR. Detailed strategies for data analysis can be found in the Supplemental Material. The ChIP-seq data have been deposited in Gene Expression Omnibus.
GO analysis
For each enriched ChIP-seq region, the nearest gene was determined using CisGenome (Ji et al. 2008). Only genes with an enriched region within 50 kb of the TSS were used for GO analysis in PANTHER (Thomas et al. 2003, 2006) to determine statistically significant biological processes. The entire mouse genome (NCBI: Mus musculus genes) was used as a reference list, and Bonferroni correction for multiple testing was applied.
Enriched motif analysis
The enrichment of JASPAR motifs within the H3K9ac ChIP-seq data was calculated by applying a Fisher's exact test to Asap, a software package for searching with position weight matrices (Marstrand et al. 2008). A negative sequence file of matched controls was generated by CisGenome. The enrichment of TRANSFAC motifs within the GR and CEBPβ ChIP-seq data sets was calculated based on a test for positional bias relative to the center of the ChIP-enriched regions analogous to what has been described previously for FoxA1 genome-wide binding (Lupien et al. 2008). Sequence motifs were discovered de novo using MDscan (Liu et al. 2002), and were assessed for significance using the same positional bias statistic.
Transfection and reporter gene assay
Genomic regions examined for cis-acting activity were PCR-amplified and subcloned into pGL4.24 (Promega). Amplification primers are listed in the Supplemental Material (Supplemental Table S9). 3T3-L1 cells were transfected by electroporation as described previously (Tomaru et al. 2009). Luciferase activity was normalized to β-galactosidase activity. RNAi-mediated silencing of GR and CEBP proteins was performed by transfecting 3T3-L1 cells with 2–4 nmol of siRNA oligos (Dharmacon SMART pool) and growing the cells to confluency for differentiation studies.
Acknowledgments
We thank Clifford A. Meyer, Josiah Altschuler, Hyunjin Shin, Tao Liu, and X. Shirley Liu (Dana-Farber Cancer Institute) for enriched motif analysis tools. We also thank Sam John (NIH) for GR ChIP advice, David Zhuo for technical help, and members of the Lazar laboratory for helpful discussion. This work was supported by NIH R01 DK49780 (to M.A.L.), by the George S. Cox Medical Research Institute, and by the Picower Foundation.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1907110.
Supplemental material is available at http://www.genesdev.org.
References
- Ailhaud G, Grimaldi P, Negrel R 1992. Cellular and molecular aspects of adipose tissue development. Annu Rev Nutr 12: 207–233 [DOI] [PubMed] [Google Scholar]
- Allis CD, Jenuwein T, Reinberg D 2007. Epigenetics Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Alon U 2007. Network motifs: Theory and experimental approaches. Nat Rev Genet 8: 450–461 [DOI] [PubMed] [Google Scholar]
- Barak Y, Nelson MC, Ong ES, Jones YZ, Ruiz-Lozano P, Chien KR, Koder A, Evans RM 1999. PPARγ is required for placental, cardiac, and adipose tissue development. Mol Cell 4: 585–595 [DOI] [PubMed] [Google Scholar]
- Berg JM, Tymoczko JL, Stryer L, Stryer L 2007. Biochemistry W.H. Freeman, New York [Google Scholar]
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ III, Gingeras TR, et al. 2005. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120: 169–181 [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Meissner A, Lander ES 2007. The mammalian epigenome. Cell 128: 669–681 [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. 2005. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122: 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z, Umek RM, McKnight SL 1991. Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes & Dev 5: 1538–1552 [DOI] [PubMed] [Google Scholar]
- Cole TJ, Blendy JA, Monaghan AP, Krieglstein K, Schmid W, Aguzzi A, Fantuzzi G, Hummler E, Unsicker K, Schutz G 1995. Targeted disruption of the glucocorticoid receptor gene blocks adrenergic chromaffin cell development and severely retards lung maturation. Genes & Dev 9: 1608–1621 [DOI] [PubMed] [Google Scholar]
- Farmer SR 2006. Transcriptional control of adipocyte formation. Cell Metab 4: 263–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green H, Kehinde O 1975. An established preadipose cell line and its differentiation in culture. II. Factors affecting the adipose conversion. Cell 5: 19–27 [DOI] [PubMed] [Google Scholar]
- Green H, Kehinde O 1976. Spontaneous heritable changes leading to increased adipose conversion in 3T3 cells. Cell 7: 105–113 [DOI] [PubMed] [Google Scholar]
- Hassan AH, Neely KE, Vignali M, Reese JC, Workman JL 2001. Promoter targeting of chromatin-modifying complexes. Front Biosci 6: D1054–D1064 [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. 2007. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39: 311–318 [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. 2009. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459: 108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu E, Tontonoz P, Spiegelman BM 1995. Transdifferentiation of myoblasts by the adipogenic transcription factors PPARγ and C/EBPα. Proc Natl Acad Sci 92: 9856–9860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Jiang H, Ma W, Johnson DS, Myers RM, Wong WH 2008. An integrated software system for analyzing ChIP–chip and ChIP-seq data. Nat Biotechnol 26: 1293–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajimura S, Seale P, Kubota K, Lunsford E, Frangioni JV, Gygi SP, Spiegelman BM 2009. Initiation of myoblast to brown fat switch by a PRDM16-C/EBP-β transcriptional complex. Nature 460: 1154–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota N, Terauchi Y, Miki H, Tamemoto H, Yamauchi T, Komeda K, Satoh S, Nakano R, Ishii C, Sugiyama T, et al. 1999. PPARγ mediates high-fat diet-induced adipocyte hypertrophy and insulin resistance. Mol Cell 4: 597–609 [DOI] [PubMed] [Google Scholar]
- Lefterova MI, Zhang Y, Steger DJ, Schupp M, Schug J, Cristancho A, Feng D, Zhuo D, Stoeckert CJ Jr, Liu XS, et al. 2008. PPARγ and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes & Dev 22: 2941–2952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XS, Brutlag DL, Liu JS 2002. An algorithm for finding protein–DNA binding sites with applications to chromatin-immunoprecipitation microarray experiments. Nat Biotechnol 20: 835–839 [DOI] [PubMed] [Google Scholar]
- Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, Carroll JS, Liu XS, Brown M 2008. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell 132: 958–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan S, Alon U 2003. Structure and function of the feed-forward loop network motif. Proc Natl Acad Sci 100: 11980–11985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marstrand TT, Frellsen J, Moltke I, Thiim M, Valen E, Retelska D, Krogh A 2008. Asap: A framework for over-representation statistics for transcription factor binding sites. PLoS One 3: e1623 doi: 10.1371/journal.pone.0001623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantoja C, Huff JT, Yamamoto KR 2008. Glucocorticoid signaling defines a novel commitment state during adipogenesis in vitro. Mol Biol Cell 19: 4032–4041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeke PM, Chrousos GP 1995. Hypercortisolism and obesity. Ann N Y Acad Sci 771: 665–676 [DOI] [PubMed] [Google Scholar]
- Pijl H, Meinders AE 1996. Bodyweight change as an adverse effect of drug treatment. Mechanisms and management. Drug Saf 14: 329–342 [DOI] [PubMed] [Google Scholar]
- Reichardt HM, Kaestner KH, Tuckermann J, Kretz O, Wessely O, Bock R, Gass P, Schmid W, Herrlich P, Angel P, et al. 1998. DNA binding of the glucocorticoid receptor is not essential for survival. Cell 93: 531–541 [DOI] [PubMed] [Google Scholar]
- Roh TY, Wei G, Farrell CM, Zhao K 2007. Genome-wide prediction of conserved and nonconserved enhancers by histone acetylation patterns. Genome Res 17: 74–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen ED, Sarraf P, Troy AE, Bradwin G, Moore K, Milstone DS, Spiegelman BM, Mortensen RM 1999. PPARγ is required for the differentiation of adipose tissue in vivo and in vitro. Mol Cell 4: 611–617 [DOI] [PubMed] [Google Scholar]
- Rosen ED, Hsu CH, Wang X, Sakai S, Freeman MW, Gonzalez FJ, Spiegelman BM 2002. C/EBPα induces adipogenesis through PPARγ: A unified pathway. Genes & Dev 16: 22–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Bjork B, Yang W, Kajimura S, Chin S, Kuang S, Scime A, Devarakonda S, Conroe HM, Erdjument-Bromage H, et al. 2008. PRDM16 controls a brown fat/skeletal muscle switch. Nature 454: 961–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao D, Lazar MA 1997. Peroxisome proliferator activated receptor γ, CCAAT/enhancer-binding protein α, and cell cycle status regulate the commitment to adipocyte differentiation. J Biol Chem 272: 21473–21478 [DOI] [PubMed] [Google Scholar]
- Steger DJ, Lefterova MI, Ying L, Stonestrom AJ, Schupp M, Zhuo D, Vakoc AL, Kim JE, Chen J, Lazar MA, et al. 2008. DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Mol Cell Biol 28: 2825–2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Yoshida N, Kishimoto T, Akira S 1997. Defective adipocyte differentiation in mice lacking the C/EBPβ and/or C/EBPδ gene. EMBO J 16: 7432–7443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, Narechania A 2003. PANTHER: A library of protein families and subfamilies indexed by function. Genome Res 13: 2129–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PD, Kejariwal A, Guo N, Mi H, Campbell MJ, Muruganujan A, Lazareva-Ulitsky B 2006. Applications for protein sequence-function evolution data: mRNA/protein expression analysis and coding SNP scoring tools. Nucleic Acids Res 34: W645–W650 doi: 10.1093/nar/gkl229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaru T, Steger DJ, Lefterova MI, Schupp M, Lazar MA 2009. Adipocyte-specific expression of murine resistin is mediated by synergism between peroxisome proliferator-activated receptor γ and CCAAT/enhancer-binding proteins. J Biol Chem 284: 6116–6125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tontonoz P, Hu E, Spiegelman BM 1994. Stimulation of adipogenesis in fibroblasts by PPARγ2, a lipid-activated transcription factor. Cell 79: 1147–1156 [DOI] [PubMed] [Google Scholar]
- Tuteja G, White P, Schug J, Kaestner KH 2009. Extracting transcription factor targets from ChIP-seq data. Nucleic Acids Res 37: e113 doi: 10.1093/nar/gkp536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. 2009. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457: 854–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, et al. 2008. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet 40: 897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiper-Bergeron N, Salem HA, Tomlinson JJ, Wu D, Hache RJ 2007. Glucocorticoid-stimulated preadipocyte differentiation is mediated through acetylation of C/EBPβ by GCN5. Proc Natl Acad Sci 104: 2703–2708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Rosen ED, Brun R, Hauser S, Adelmant G, Troy AE, McKeon C, Darlington GJ, Spiegelman BM 1999. Cross-regulation of C/EBPα and PPARγ controls the transcriptional pathway of adipogenesis and insulin sensitivity. Mol Cell 3: 151–158 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based analysis of ChIP-seq (MACS). Genome Biol 9: R137 doi: 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]